Carbon Fluxes in Potato (Solanum tuberosum) Remain Stable in Cell Cultures Exposed to Nutritional Phosphate Deficiency
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Plant Material
2.2. Isotopic Labeling Experiments
2.3. Statistics
3. Results
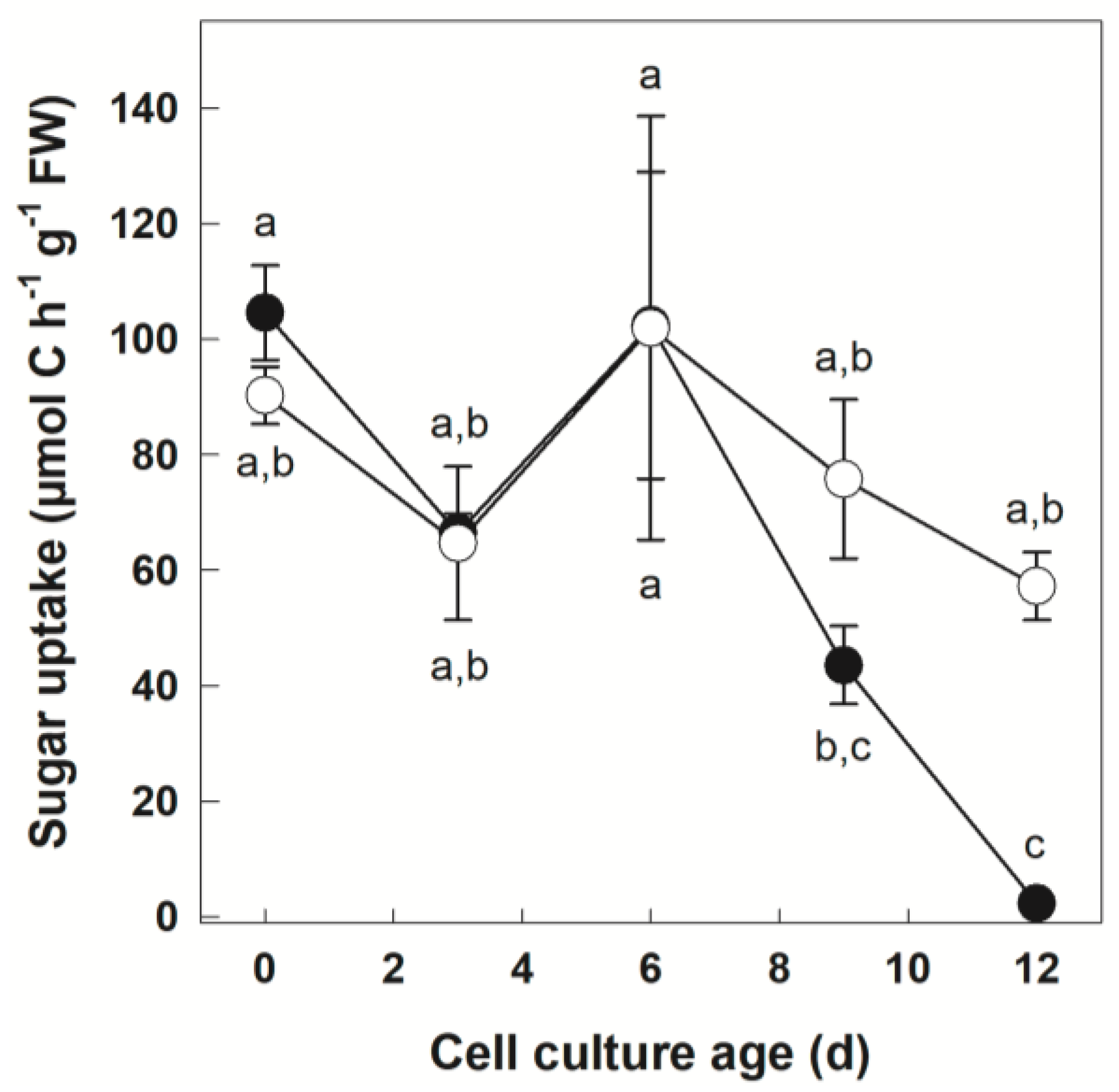
3.1. Tracer Uptake by +Pi and −Pi Cells
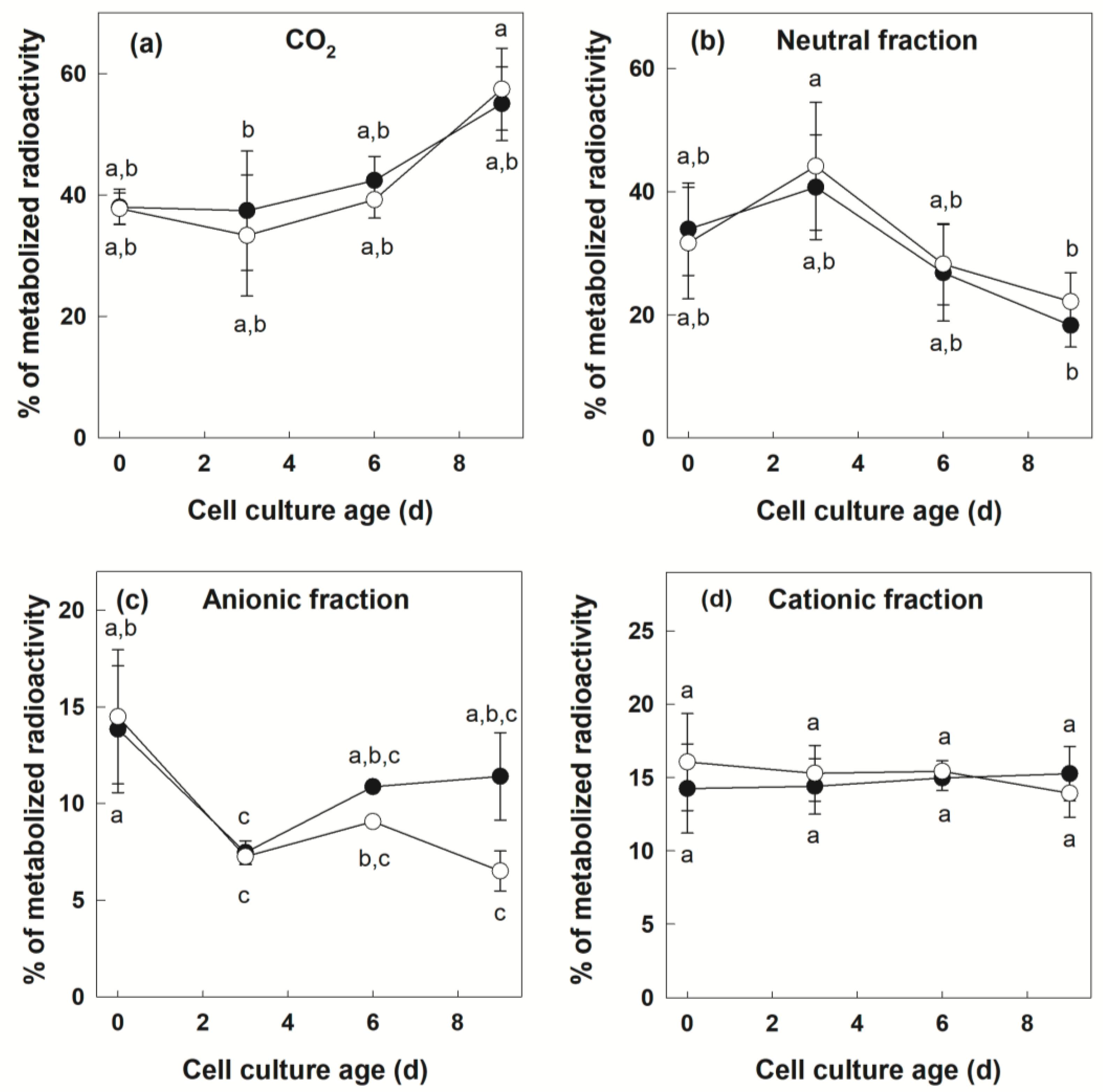
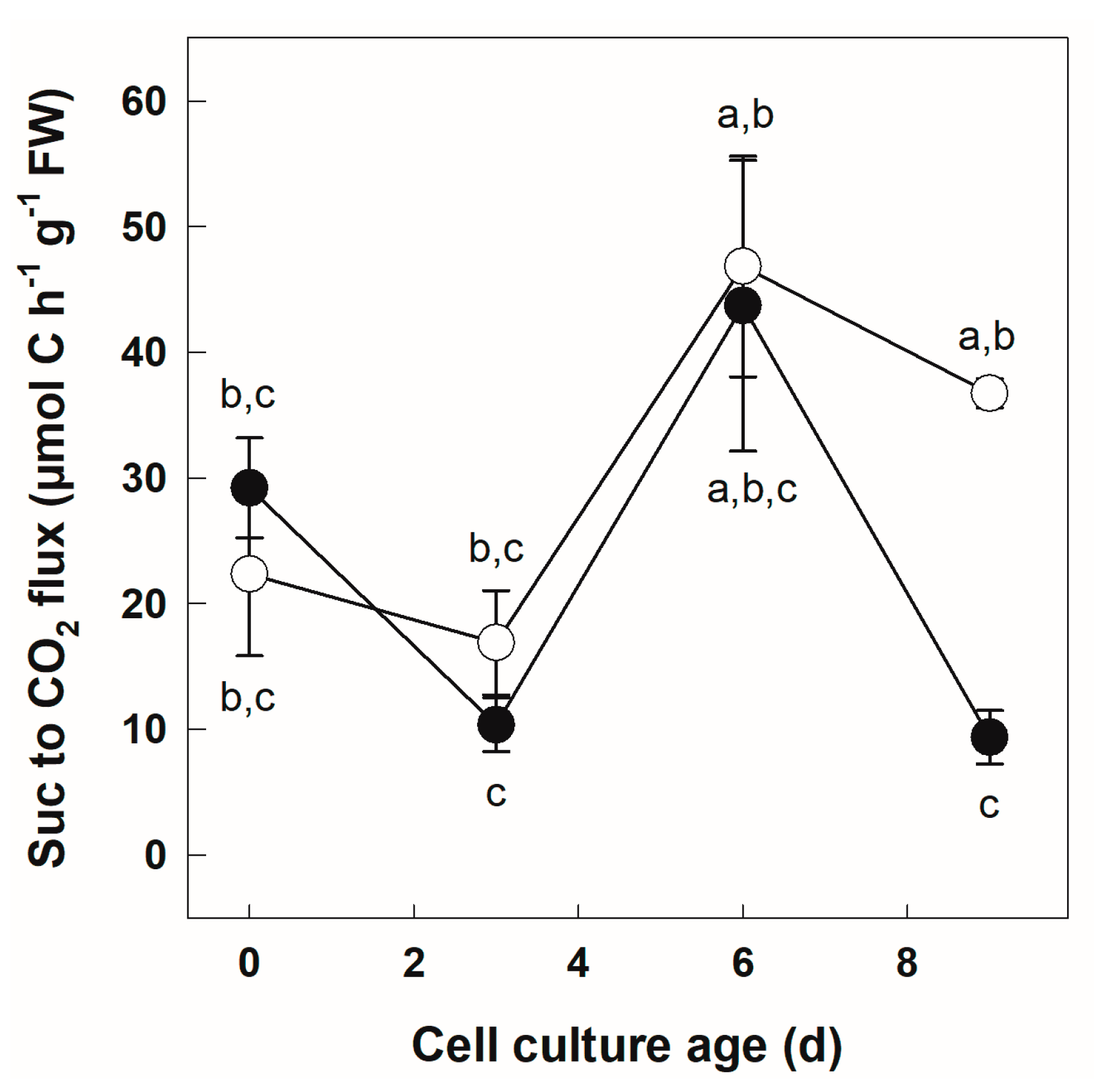
3.2. Metabolization of [U-14C]Suc by +Pi and −Pi Cells
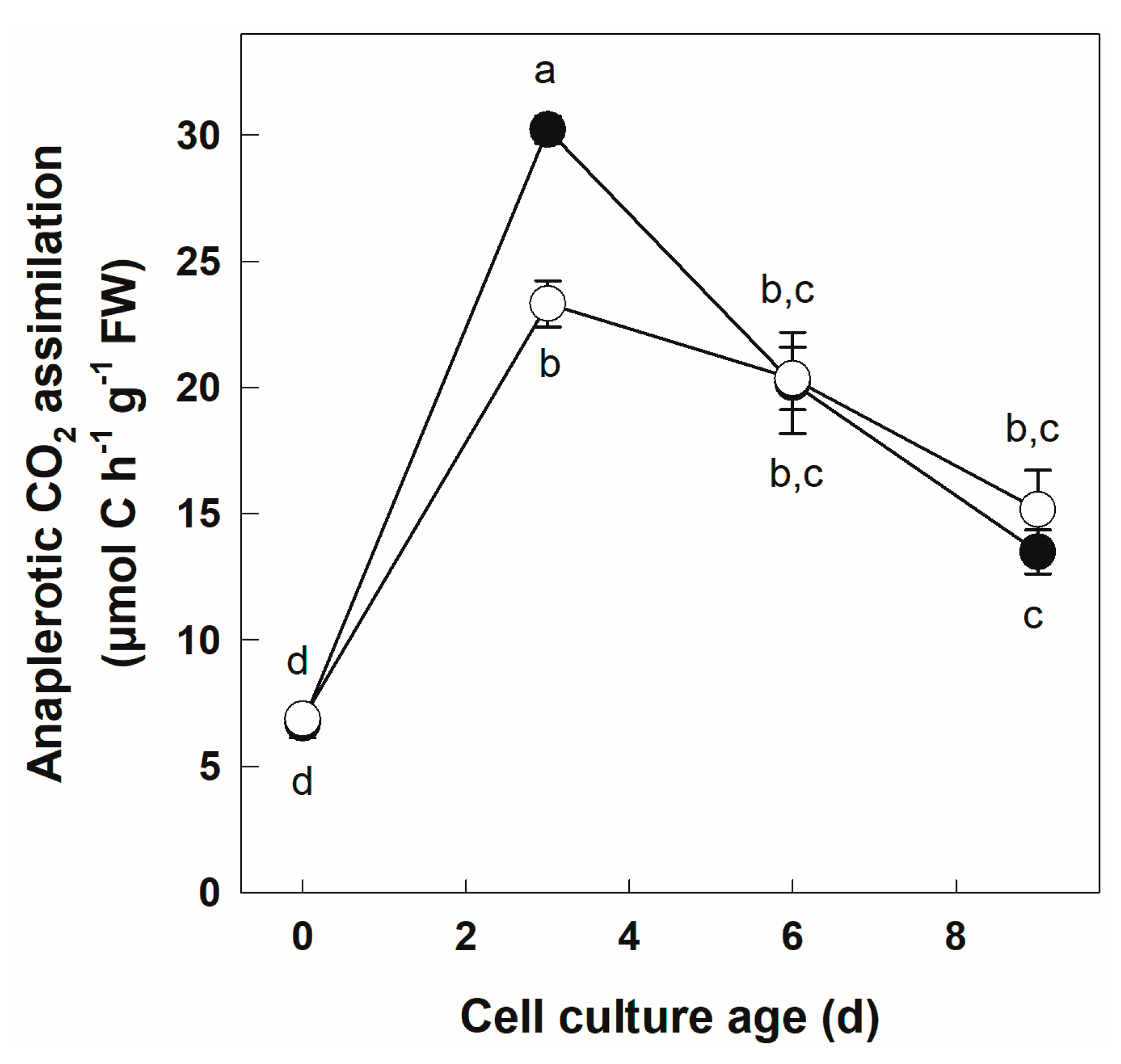
3.3. Labeling of +Pi and −Pi Cells with NaH14CO3
3.4. Analysis of the Interaction between Cell Culture Age and C Fluxes
4. Discussion
4.1. Sugar Uptake Is Maintained in Potato Cell Cultures Subjected to Pi Deficiency
4.2. Sugar Catabolic Fluxes Are Similar in +Pi and −Pi Cells
4.3. Metabolism of NaH14CO3 in +Pi and −Pi Cells Allows a Better Understanding of the Role of PEPC in the Stability of Anaplerotic C Flux under Pi Deficiency
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Malhotra, H.; Vandana; Sharma, S.; Pandey, R. Phosphorus Nutrition: Plant Growth in Response to Deficiency and Excess. In Plant Nutrients and Abiotic Stress Tolerance; Hasanuzzaman, M., Fujita, M., Oku, H., Nahar, K., Hawrylak-Nowak, B., Eds.; Springer: Singapore, 2018; pp. 171–190. [Google Scholar]
- Bechtaoui, N.; Rabiu, M.K.; Raklami, A.; Oufdou, K.; Hafidi, M.; Jemo, M. Phosphate-Dependent Regulation of Growth and Stresses Management in Plants. Front. Plant Sci. 2021, 12, 679916. [Google Scholar] [CrossRef]
- Raghothama, K.G. Phosphate acquisition. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 665–693. [Google Scholar] [CrossRef]
- Nussaume, L.; Kanno, S.; Javot, H.; Marin, E.; Pochon, N.; Ayadi, A.; Nakanishi, T.M.; Thibaud, M.-C. Phosphate Import in Plants: Focus on the PHT1 Transporters. Front. Plant Sci. 2011, 2, 83. [Google Scholar] [CrossRef] [PubMed]
- López-Arredondo, D.L.; Leyva-González, M.A.; González-Morales, S.I.; López-Bucio, J.; Herrera-Estrella, L. Phosphate nutrition: Improving low phosphate tolerance in crops. Annu. Rev. Plant Biol. 2014, 65, 95–123. [Google Scholar] [CrossRef] [PubMed]
- Cordell, D.; Drangert, J.-O.; White, S. The story of phosphorus: Global food security and food for thought. Glob. Environ. Chang. Hum. Policy Dimens. 2009, 19, 292–305. [Google Scholar] [CrossRef]
- Reijnders, L. Phosphorus resources, their depletion and conservation, a review. Resour. Conserv. Recycl. 2014, 93, 32–49. [Google Scholar] [CrossRef]
- Wu, P.; Ma, L.; Hou, X.; Wang, M.; Wu, Y.; Liu, F.; Deng, X.W. Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol. 2003, 132, 1260–1271. [Google Scholar] [CrossRef]
- Chiou, T.J.; Aung, K.; Lin, S.I.; Wu, C.C.; Chiang, S.F.; Su, C.L. Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 2006, 18, 412–421. [Google Scholar] [CrossRef]
- Veljanovski, V.; Vanderbeld, B.; Knowles, V.L.; Snedden, W.A.; Plaxton, W.C. Biochemical and molecular characterization of AtPAP26, a vacuolar purple acid phosphatase up-regulated in phosphate-deprived arabidopsis suspension cells and seedlings. Plant Physiol. 2006, 142, 1282–1293. [Google Scholar] [CrossRef]
- Li, K.P.; Xu, C.Z.; Li, Z.X.; Zhang, K.W.; Yang, A.F.; Zhang, J.R. Comparative proteome analyses of phosphorus responses in maize (Zea mays L.) roots of wild-type and a low-P-tolerant mutant reveal root characteristics associated with phosphorus efficiency. Plant J. 2008, 55, 927–939. [Google Scholar] [CrossRef]
- Alexova, R.; Millar, A.H. Proteomics of phosphate use and deprivation in plants. Proteomics 2013, 13, 609–623. [Google Scholar] [CrossRef]
- Gregory, A.L.; Hurley, B.A.; Tran, H.T.; Valentine, A.J.; She, Y.-M.; Knowles, V.L.; Plaxton, W.C. In vivo regulatory phosphorylation of the phosphoenolpyruvate carboxylase AtPPC1 in phosphate-starved Arabidopsis thaliana. Biochem. J. 2009, 420, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Li, K.P.; Xu, C.Z.; Fan, W.M.; Zhang, H.L.; Hou, J.J.; Yang, A.F.; Zhang, K.W. Phosphoproteome and proteome analyses reveal low-phosphate mediated plasticity of root developmental and metabolic regulation in maize (Zea mays L.). Plant Physiol. Biochem. 2014, 83, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.; Hurley, B.A.; Plaxton, W.C. Feeding hungry plants: The role of purple acid phosphatases in phosphate nutrition. Plant Sci. 2010, 179, 14–27. [Google Scholar] [CrossRef]
- Abel, S. Phosphate scouting by root tips. Curr. Opin. Plant Biol. 2017, 39, 168–177. [Google Scholar] [CrossRef]
- del Pozo, J.C.; Allona, I.; Rubio, V.; Leyva, A.; de la Pena, A.; Aragoncillo, C.; Paz-Ares, J. A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J. 1999, 19, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Bozzo, G.G.; Raghothama, K.G.; Plaxton, W.C. Purification and characterization of two secreted purple acid phosphatase isozymes from phosphate-starved tomato (Lycopersicon esculentum) cell cultures. Eur. J. Biochem. 2002, 269, 6278–6286. [Google Scholar] [CrossRef]
- Plaxton, W.C.; Tran, H.T. Metabolic adaptations of phosphate-starved plants. Plant Physiol. 2011, 156, 1006–1015. [Google Scholar] [CrossRef]
- Duff, S.M.G.; Moorhead, G.B.; Lefebvre, D.D.; Plaxton, W.C. Phosphate starvation inducible ‘bypasses’ of adenylate and phosphate dependent glycolytic enzymes in Brassica nigra suspension cells. Plant Physiol. 1989, 90, 1275–1278. [Google Scholar] [CrossRef]
- Johnson, J.F.; Allan, D.L.; Vance, C.P. Phosphorus Stress-Induced Proteoid Roots Show Altered Metabolism in Lupinus albus. Plant Physiol. 1994, 104, 657–665. [Google Scholar] [CrossRef]
- Uhde-Stone, C.; Gilbert, G.; Johnson, J.M.F.; Litjens, R.; Zinn, K.E.; Temple, S.J.; Vance, C.P.; Allan, D.L. Acclimation of white lupin to phosphorus deficiency involves enhanced expression of genes related to organic acid metabolism. Plant Soil 2003, 248, 99–116. [Google Scholar] [CrossRef]
- Le Roux, M.R.; Ward, C.L.; Botha, F.C.; Valentine, A.J. Routes of pyruvate synthesis in phosphorus-deficient lupin roots and nodules. New Phytol. 2006, 169, 399–408. [Google Scholar] [CrossRef]
- He, J.Z.; Dorion, S.; Lacroix, M.; Rivoal, J. Sustained substrate cycles between hexose phosphates and free sugars in phosphate-deficient potato (Solanum tuberosum) cell cultures. Planta 2019, 249, 1319–1336. [Google Scholar] [CrossRef]
- Masakapalli, S.K.; Bryant, F.M.; Kruger, N.J.; Ratcliffe, R.G. The metabolic flux phenotype of heterotrophic Arabidopsis cells reveals a flexible balance between the cytosolic and plastidic contributions to carbohydrate oxidation in response to phosphate limitation. Plant J. 2014, 78, 964–977. [Google Scholar] [CrossRef]
- O’Leary, B.; Park, J.; Plaxton, W.C. The remarkable diversity of plant PEPC (phosphoenolpyruvate carboxylase): Recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs. Biochem. J. 2011, 436, 15–34. [Google Scholar] [CrossRef] [PubMed]
- Stevens, G.G.; Pérez-Fernández, M.A.; Morcillo, R.J.L.; Kleinert, A.; Hills, P.; Brand, D.J.; Steenkamp, E.T.; Valentine, A.J. Roots and Nodules Response Differently to P Starvation in the Mediterranean-Type Legume Virgilia divaricata. Front. Plant Sci. 2019, 10, 73. [Google Scholar] [CrossRef]
- Lan, P.; Li, W.; Schmidt, W. Complementary Proteome and Transcriptome Profiling in Phosphate-deficient Arabidopsis Roots Reveals Multiple Levels of Gene Regulation. Mol. Cell. Proteom. 2012, 11, 1156–1166. [Google Scholar] [CrossRef]
- Birch, P.R.J.; Bryan, G.; Fenton, B.; Gilroy, E.M.; Hein, I.; Jones, J.T.; Prashar, A.; Taylor, M.A.; Torrance, L.; Toth, I.K. Crops that feed the world 8: Potato: Are the trends of increased global production sustainable? Food Secur. 2012, 4, 477–508. [Google Scholar] [CrossRef]
- Koch, M.; Naumann, M.; Pawelzik, E.; Gransee, A.; Thiel, H. The importance of nutrient management for potato production Part I: Plant nutrition and yield. Potato Res. 2020, 63, 97–119. [Google Scholar] [CrossRef]
- Rosen, C.J.; Bierman, P.M. Potato yield and tuber set as affected by phosphorus fertilization. Am. J. Potato Res. 2008, 85, 110–120. [Google Scholar] [CrossRef]
- Dorion, S.; Rivoal, J. Quantification of uridine 5’-diphosphate (UDP)-glucose by high-performance liquid chromatography and its application to nonradioactive assay for nucleoside diphosphate kinase using UDP-glucose pyrophosphorylase as a coupling enzyme. Anal. Biochem. 2003, 323, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Dorion, S.; Clendenning, A.; Jeukens, J.; Salas, J.J.; Parveen, N.; Haner, A.A.; Law, R.D.; Martinez-Force, E.; Rivoal, J. A large decrease of cytosolic triosephosphate isomerase in transgenic potato roots affects the distribution of carbon in primary metabolism. Planta 2012, 236, 1177–1190. [Google Scholar] [CrossRef] [PubMed]
- Rivoal, J.; Hanson, A.D. Evidence for a large and sustained glycolytic flux to lactate in anoxic roots of some members of the halophytic genus Limonium. Plant Physiol. 1993, 101, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Dorion, S.; Clendenning, A.; Rivoal, J. Engineering the expression level of cytosolic nucleoside diphosphate kinase in transgenic Solanum tuberosum roots alters growth, respiration and carbon metabolism. Plant J. 2017, 89, 914–926. [Google Scholar] [CrossRef] [PubMed]
- Claeyssen, E.; Dorion, S.; Clendenning, A.; He, J.Z.; Wally, O.; Chen, J.; Auslender, E.L.; Moisan, M.-C.; Jolicoeur, M.; Rivoal, J. The futile cycling of hexose phosphates could account for the fact that hexokinase exerts a high control on glucose phosphorylation but not on glycolytic rate in transgenic potato (Solanum tuberosum) Roots. PLoS ONE 2013, 8, e53898. [Google Scholar] [CrossRef]
- Rolletschek, H.; Borisjuk, L.; Radchuk, R.; Miranda, M.; Heim, U.; Wobus, U.; Weber, H. Seed-specific expression of a bacterial phosphoenolpyruvate carboxylase in Vicia narbonensis increases protein content and improves carbon economy. Plant Biotechnol. J. 2004, 2, 211–219. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 17 July 2023).
- Péret, B.; Clément, M.; Nussaume, L.; Desnos, T. Root developmental adaptation to phosphate starvation: Better safe than sorry. Trends Plant Sci. 2011, 16, 442–450. [Google Scholar] [CrossRef]
- Veneklaas, E.J.; Lambers, H.; Bragg, J.; Finnegan, P.M.; Lovelock, C.E.; Plaxton, W.C.; Price, C.A.; Scheible, W.R.; Shane, M.W.; White, P.J.; et al. Opportunities for improving phosphorus-use efficiency in crop plants. New Phytol. 2012, 195, 306–320. [Google Scholar] [CrossRef]
- O’Rourke, J.A.; Yang, S.S.; Miller, S.S.; Bucciarelli, B.; Liu, J.Q.; Rydeen, A.; Bozsoki, Z.; Uhde-Stone, C.; Tu, Z.J.; Allan, D.; et al. An RNA-Seq Transcriptome Analysis of Orthophosphate-Deficient White Lupin Reveals Novel Insights into Phosphorus Acclimation in Plants. Plant Physiol. 2013, 161, 705–724. [Google Scholar] [CrossRef]
- Baker, A.; Ceasar, S.A.; Palmer, A.J.; Paterson, J.B.; Qi, W.; Muench, S.P.; Baldwin, S.A. Replace, reuse, recycle: Improving the sustainable use of phosphorus by plants. J. Exp. Bot. 2015, 66, 3523–3540. [Google Scholar] [CrossRef]
- Paul, M.J.; Stitt, M. Effects of nitrogen and phosphorus deficiencies an levels of carbohydrates, respiratory enzymes and metabolites in seedlings of tobacco and their response to exogenous sucrose. Plant Cell Environ. 1993, 16, 1047–1057. [Google Scholar] [CrossRef]
- Morcuende, R.; Bari, R.; Gibon, Y.; Zheng, W.; Pant, B.D.; Bläsing, O.; Usadel, B.; Czechowski, T.; Udvardi, M.K.; Stitt, M.; et al. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 2007, 30, 85–112. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Roessner, U.; Eickmeier, I.; Genc, Y.; Callahan, D.L.; Shirley, N.; Langridge, P.; Bacic, A. Metabolite profiling reveals distinct changes in carbon and nitrogen metabolism in phosphate-deficient barley plants (Hordeum vulgare L.). Plant Cell Physiol. 2008, 49, 691–703. [Google Scholar] [CrossRef] [PubMed]
- Hoffland, E.; Vandenboogaard, R.; Nelemans, J.; Findenegg, G. Biosynthesis and root exudation of citric and malic-acids in phosphate-starved rape plants. New Phytol. 1992, 122, 675–680. [Google Scholar] [CrossRef]
- Neumann, G.; Massonneau, A.; Martinoia, E.; Römheld, V. Physiological adaptations to phosphorus deficiency during proteoid root development in white lupin. Planta 1999, 208, 373–382. [Google Scholar] [CrossRef]
- Neumann, G.; Römheld, V. Root excretion of carboxylic acids and protons in phosphorus-deficient plants. Plant Soil 1999, 211, 121–130. [Google Scholar] [CrossRef]
- Ryan, P.R.; Delhaize, E.; Jones, D.L. Function and mechanism of organic anion exudation from plant roots. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 527–560. [Google Scholar] [CrossRef]
- Johnson, J.F.; Allan, D.L.; Vance, C.P.; Weiblen, G. Root Carbon Dioxide Fixation by Phosphorus-Deficient Lupinus albus (Contribution to Organic Acid Exudation by Proteoid Roots). Plant Physiol. 1996, 112, 19–30. [Google Scholar] [CrossRef]
- Le Roux, M.; Phiri, E.; Khan, W.; Şakiroğlu, M.; Valentine, A.; Khan, S. Expression of novel cytosolic malate dehydrogenases (cMDH) in Lupinus angustifolius nodules during phosphorus starvation. J. Plant Physiol. 2014, 171, 1609–1618. [Google Scholar] [CrossRef]
- Liang, C.; Wang, J.; Zhao, J.; Tian, J.; Liao, H. Control of phosphate homeostasis through gene regulation in crops. Curr. Opin. Plant Biol. 2014, 21, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Plaxton, W.C.; Podestà, F.E. The functional organization and control of plant respiration. Crit. Rev. Plant Sci. 2006, 25, 159–198. [Google Scholar] [CrossRef]
- Ding, Y.; Tang, Y.; Kwok, C.K.; Zhang, Y.; Bevilacqua, P.C.; Assmann, S.M. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 2013, 505, 696–700. [Google Scholar] [CrossRef] [PubMed]
- Rivoal, J.; Hanson, A.D. Metabolic control of anaerobic glycolysis—Overexpression of lactate dehydrogenase in transgenic tomato roots supports the Davies-Roberts hypothesis and points to a critical role for lactate secretion. Plant Physiol. 1994, 106, 1179–1185. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Geigenberger, P.; Stitt, M. Flux an important, but neglected, component of functional genomics. Curr. Opin. Plant Biol. 2005, 8, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Gaudinier, A.; Tang, M.; Kliebenstein, D.J. Transcriptional networks governing plant metabolism. Curr. Plant Biol. 2015, 3–4, 56–64. [Google Scholar] [CrossRef]
- Cheung, C.Y.M.; Williams, T.C.R.; Poolman, M.G.; Fell, D.A.; Ratcliffe, R.G.; Sweetlove, L.J. A method for accounting for maintenance costs in flux balance analysis improves the prediction of plant cell metabolic phenotypes under stress conditions. Plant J. 2013, 75, 1050–1061. [Google Scholar] [CrossRef]
- Dong, N.Q.; Sun, Y.W.; Guo, T.; Shi, C.L.; Zhang, Y.M.; Kan, Y.; Xiang, Y.H.; Zhang, H.; Yang, Y.B.; Li, Y.C.; et al. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat. Commun. 2020, 11, 2629. [Google Scholar] [CrossRef]
- Qian, X.; Zhang, Y.; Lun, D.S.; Dismukes, G.C. Rerouting of Metabolism into Desired Cellular Products by Nutrient Stress: Fluxes Reveal the Selected Pathways in Cyanobacterial Photosynthesis. ACS Synth. Biol. 2018, 7, 1465–1476. [Google Scholar] [CrossRef]
- Heydarizadeh, P.; Veldl, B.; Huang, B.; Lukomska, E.; Wielgosz-Collin, G.; Couzinet-Mossion, A.; Bougaran, G.; Marchand, J.; Schoefs, B. Carbon Orientation in the Diatom Phaeodactylum tricornutum: The Effects of Carbon Limitation and Photon Flux Density. Front. Plant Sci. 2019, 10, 471. [Google Scholar] [CrossRef]
- Wang, Q.D.; Hu, J.K.; Hu, H.F.; Li, Y.; Xiang, M.L.; Wang, D.Z. Integrated eco-physiological, biochemical, and molecular biological analyses of selenium fortification mechanism in alfalfa. Planta 2022, 256, 114. [Google Scholar] [CrossRef] [PubMed]
- Sulieman, S.; Abdelrahman, M.; Tran, L.S.P. Carbon metabolic adjustment in soybean nodules in response to phosphate limitation: A metabolite perspective. Environ. Exp. Bot. 2022, 196, 104810. [Google Scholar] [CrossRef]
- Ruan, Y.-L. Sucrose Metabolism: Gateway to Diverse Carbon Use and Sugar Signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef] [PubMed]
- Komor, E.; Thom, M.; Maretzki, A. The mechanism of sugar uptake by sugarcane suspension cells. Planta 1981, 153, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Botha, F.C.; O’Kennedy, M.M. Carbohydrate utilisation by cell suspension cultures of Phaseolus vulgaris. Physiol. Plant. 1998, 102, 429–436. [Google Scholar] [CrossRef]
- Krook, J.; Vreugdenhil, D.; van der Plas, L.H.W. Uptake and phosphorylation of glucose and fructose in Daucus carota cell suspensions are differently regulated. Plant Physiol. Biochem. 2000, 38, 603–612. [Google Scholar] [CrossRef]
- Conde, C.; Agasse, A.; Glissant, D.; Tavares, R.; Gerós, H.; Delrot, S. Pathways of Glucose Regulation of Monosaccharide Transport in Grape Cells. Plant Physiol. 2006, 141, 1563–1577. [Google Scholar] [CrossRef]
- Felker, F.C.; Goodwin, J.C. Sugar uptake by maize endosperm suspension cultures. Plant Physiol. 1988, 88, 1235–1239. [Google Scholar] [CrossRef]
- Etxeberria, E.; Baroja-Fernandez, E.; Munoz, F.J.; Pozueta-Romero, J. Sucrose-inducible endocytosis as a mechanism for nutrient uptake in heterotrophic plant cells. Plant Cell Physiol. 2005, 46, 474–481. [Google Scholar] [CrossRef]
- Menges, M.; Hennig, L.; Gruissem, W.; Murray, J.A.H. Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol. Biol. 2003, 53, 423–442. [Google Scholar] [CrossRef]
- Ganie, A.H.; Ahmad, A.; Pandey, R.; Aref, I.M.; Yousuf, P.Y.; Ahmad, S.; Iqbal, M. Metabolite profiling of low-P tolerant and low-P sensitive maize genotypes under phosphorus starvation and restoration conditions. PLoS ONE 2015, 10, e0129520. [Google Scholar] [CrossRef]
- Valentinuzzi, F.; Pii, Y.; Vigani, G.; Lehmann, M.; Cesco, S.; Mimmo, T. Phosphorus and iron deficiencies induce a metabolic reprogramming and affect the exudation traits of the woody plant Fragaria × ananassa. J. Exp. Bot. 2015, 66, 6483–6495. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.S.; Israel, D.W. Diurnal starch accumulation and utilization in phosphorus-deficient soybean plants. Plant Physiol. 1992, 98, 316–323. [Google Scholar] [CrossRef]
- Qiu, J.; Israel, D.W. Carbohydrate accumulation and utilization in soybean plants in response to altered phosphorus-nutrition. Physiol. Plant. 1994, 90, 722–728. [Google Scholar] [CrossRef]
- Ciereszko, I.; Barbachowska, A. Sucrose metabolism in leaves and roots of bean (Phaseolus vulgaris L.) during phosphate deficiency. J. Plant Physiol. 2000, 156, 640–644. [Google Scholar] [CrossRef]
- Rontein, D.; Dieuaide-Noubhani, M.; Dufourc, E.J.; Raymond, P.; Rolin, D. The metabolic architecture of plant cells. Stability of central metabolism and flexibility of anabolic pathways during the growth cycle of tomato cells. J. Biol. Chem. 2002, 277, 43948–43960. [Google Scholar] [CrossRef] [PubMed]
- Mori, S. A High Performance Liquid Chromatography-Radio-Organic Acid Analyzer for determining the specific activities of radioactive organic acids. Plant Cell Physiol. 1982, 23, 703–708. [Google Scholar] [CrossRef]
- Cramer, M.D.; Lewis, O.A.M.; Lips, S.H. Inorganic carbon fixation and metabolism in maize roots as affected by nitrate and ammonium nutrition. Physiol. Plant. 1993, 89, 632–639. [Google Scholar] [CrossRef]
- Wegner, A.; Meiser, J.; Weindl, D.; Hiller, K. How metabolites modulate metabolic flux. Curr. Opin. Biotechnol. 2015, 34, 16–22. [Google Scholar] [CrossRef]
- Clark, T.J.; Guo, L.; Morgan, J.; Schwender, J. Modeling plant metabolism: From network reconstruction to mechanistic models. Annu. Rev. Plant Biol. 2020, 71, 303–326. [Google Scholar] [CrossRef]
- De Nisi, P.; Zocchi, G. Phosphoenolpyruvate carboxylase in cucumber (Cucumis sativus L.) roots under iron deficiency: Activity and kinetic characterization. J. Exp. Bot. 2000, 51, 1903–1909. [Google Scholar] [CrossRef] [PubMed]
| Parameter | Cell Culture Age | Phosphate Regime | Phosphate Regime x Cell Culture Age | |||
|---|---|---|---|---|---|---|
| F | p | F | p | F | p | |
| C uptake (μmol C h−1·g−1 FW) | 6.22 | 0.002 ** | 2.15 | 0.157 | 1.43 | 0.260 |
| % CO2 in metabolized [U-14C]Suc | 4.92 | 0.013 * | 1.33 | 0.2667 | 0.849 | 0.4874 |
| % Neutral fraction in metabolized [U-14C]Suc | 4.14 | 0.023 * | 0.74 | 0.401 | 0.214 | 0.8850 |
| % Anionic fraction in metabolized [U-14C]Suc | 5.46 | 0.008 ** | 2.60 | 0.126 | 0.41 | 0.743 |
| % Cationic fraction in metabolized [U-14C]Suc | 0.14 | 0.932 | 0.030 | 0.865 | 0.188 | 0.903 |
| Suc to CO2 flux (μmol C h−1 g−1 FW) | 11.16 | 3.37 × 10−4 *** | 3.22 | 0.0916 | 2.94 | 0.064 |
| Anaplerotic CO2 assimilation (μmol C h−1 g−1 FW) | 83.9 | 5.25 × 10−10 *** | 3.03 | 0.100 | 5.25 | 0.010 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, J.Z.; Dorion, S.; Carmona-Rojas, L.M.; Rivoal, J. Carbon Fluxes in Potato (Solanum tuberosum) Remain Stable in Cell Cultures Exposed to Nutritional Phosphate Deficiency. Biology 2023, 12, 1190. https://doi.org/10.3390/biology12091190
He JZ, Dorion S, Carmona-Rojas LM, Rivoal J. Carbon Fluxes in Potato (Solanum tuberosum) Remain Stable in Cell Cultures Exposed to Nutritional Phosphate Deficiency. Biology. 2023; 12(9):1190. https://doi.org/10.3390/biology12091190
Chicago/Turabian StyleHe, Jiang Zhou, Sonia Dorion, Laura Michell Carmona-Rojas, and Jean Rivoal. 2023. "Carbon Fluxes in Potato (Solanum tuberosum) Remain Stable in Cell Cultures Exposed to Nutritional Phosphate Deficiency" Biology 12, no. 9: 1190. https://doi.org/10.3390/biology12091190
APA StyleHe, J. Z., Dorion, S., Carmona-Rojas, L. M., & Rivoal, J. (2023). Carbon Fluxes in Potato (Solanum tuberosum) Remain Stable in Cell Cultures Exposed to Nutritional Phosphate Deficiency. Biology, 12(9), 1190. https://doi.org/10.3390/biology12091190







