Linking Vertebrate Gene Duplications to the New Head Hypothesis
Abstract
Simple Summary
Abstract
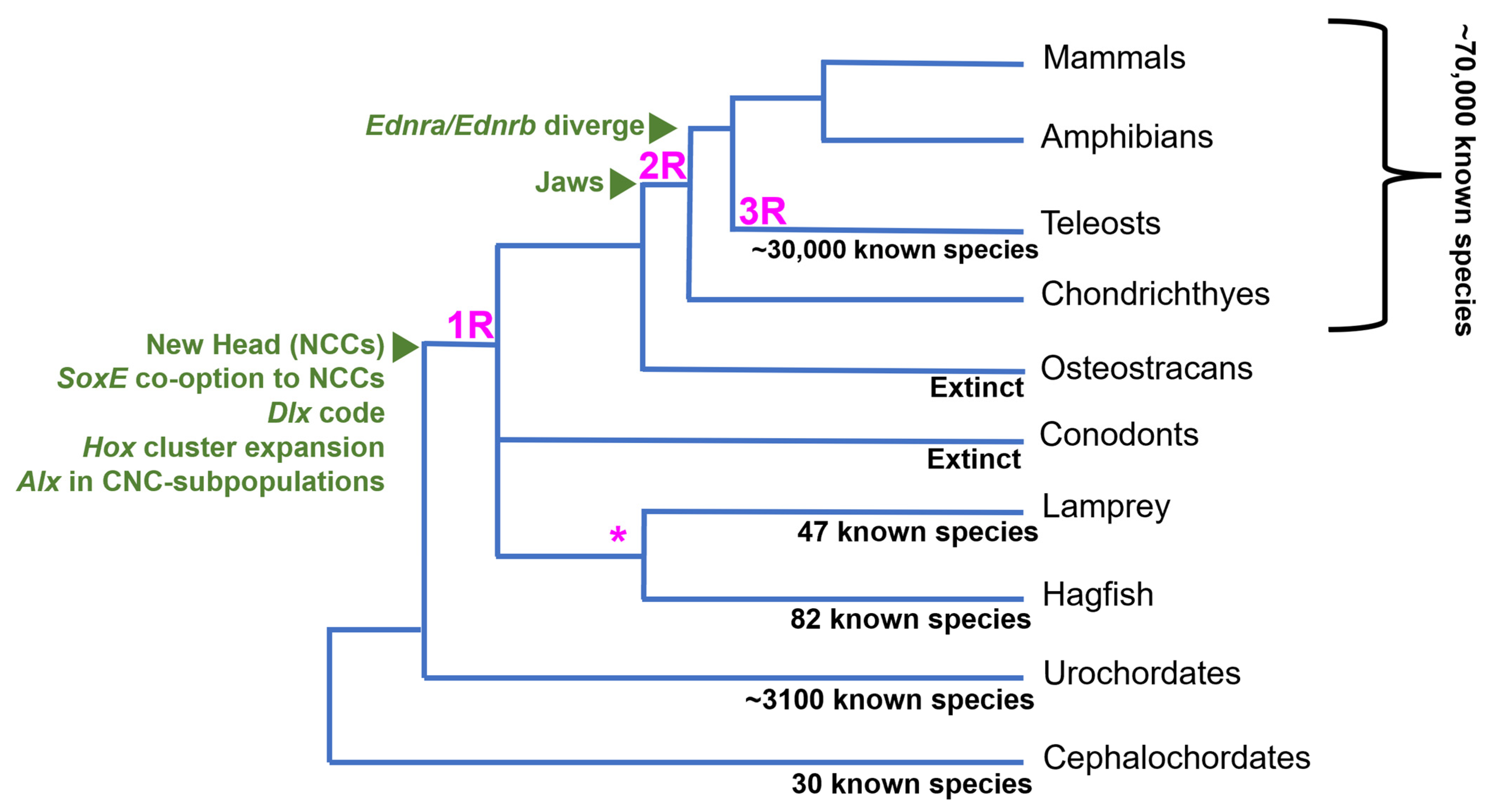
1. Introduction
2. The NC GRN
2.1. Neural Crest Establishment and Migration
2.2. Neural Crest Derivatives in the New Head
3. Duplicated Genes within the NC GRN

3.1. SoxE
3.2. Dlx
3.3. Hox Clusters
3.4. EdnR
3.5. Alx
4. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bánki, O.; Roskov, Y.; Döring, M.; Ower, G.; Hernández Robles, D.R.; Plata Corredor, C.A.; Stjernegaard Jeppesen, T.; Örn, A.; Vandepitte, L.; Hobern, D.; et al. Catalogue of Life Checklist. Cat. Life 2023. [Google Scholar] [CrossRef]
- Medeiros, D.M. The evolution of the neural crest: New perspectives from lamprey and invertebrate neural crest-like cells. WIREs Dev. Biol. 2013, 2, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Van Otterloo, E.; Cornell, R.A.; Medeiros, D.M.; Garnett, A.T. Gene regulatory evolution and the origin of macroevolutionary novelties: Insights from the neural crest. Genesis 2013, 51, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Jandzik, D.; Garnett, A.T.; Square, T.A.; Cattell, M.V.; Yu, J.-K.; Medeiros, D.M. Evolution of the new vertebrate head by co-option of an ancient chordate skeletal tissue. Nature 2015, 518, 534–537. [Google Scholar] [CrossRef]
- Martik, M.L.; Bronner, M.E. Riding the crest to get a head: Neural crest evolution in vertebrates. Nat. Rev. Neurosci. 2021, 22, 616–626. [Google Scholar] [CrossRef] [PubMed]
- Cheung, M.; Tai, A.; Lu, P.J.; Cheah, K.S. Acquisition of multipotent and migratory neural crest cells in vertebrate evolution. Curr. Opin. Genet. Dev. 2019, 57, 84–90. [Google Scholar] [CrossRef]
- York, J.R.; McCauley, D.W. The origin and evolution of vertebrate neural crest cells. Open Biol. 2020, 10, 190285. [Google Scholar] [CrossRef]
- Gans, C.; Northcutt, R.G. Neural Crest and the Origin of Vertebrates: A New Head. Science 1983, 220, 268–273. [Google Scholar] [CrossRef]
- Sauka-Spengler, T.; Meulemans, D.; Jones, M.; Bronner-Fraser, M. Ancient Evolutionary Origin of the Neural Crest Gene Regulatory Network. Dev. Cell 2007, 13, 405–420. [Google Scholar] [CrossRef]
- Martik, M.L.; Gandhi, S.; Uy, B.R.; Gillis, J.A.; Green, S.A.; Simoes-Costa, M.; Bronner, M.E. Evolution of the new head by gradual acquisition of neural crest regulatory circuits. Nature 2019, 574, 675–678. [Google Scholar] [CrossRef]
- Betancur, P.; Bronner-Fraser, M.; Sauka-Spengler, T. Assembling Neural Crest Regulatory Circuits into a Gene Regulatory Network. Annu. Rev. Cell Dev. Biol. 2010, 26, 581–603. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S. Evolution by Gene Duplication; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Putnam, N.H.; Butts, T.; Ferrier, D.E.K.; Furlong, R.F.; Hellsten, U.; Kawashima, T.; Robinson-Rechavi, M.; Shoguchi, E.; Terry, A.; Yu, J.-K.; et al. The amphioxus genome and the evolution of the chordate karyotype. Nature 2008, 453, 1064–1071. [Google Scholar] [CrossRef] [PubMed]
- Davesne, D.; Friedman, M.; Schmitt, A.D.; Fernandez, V.; Carnevale, G.; Ahlberg, P.E.; Sanchez, S.; Benson, R.B.J. Fossilized cell structures identify an ancient origin for the teleost whole-genome duplication. Proc. Natl. Acad. Sci. USA 2021, 118, e2101780118. [Google Scholar] [CrossRef] [PubMed]
- Donoghue, P.C.J.; Keating, J.N. Early vertebrate evolution. Palaeontology 2014, 57, 879–893. [Google Scholar] [CrossRef]
- Square, T.; Jandzik, D.; Romášek, M.; Cerny, R.; Medeiros, D.M. The origin and diversification of the developmental mechanisms that pattern the vertebrate head skeleton. Dev. Biol. 2017, 427, 219–229. [Google Scholar] [CrossRef]
- Randle, E.; Sansom, R.S. Bite marks and predation of fossil jawless fish during the rise of jawed vertebrates. Proc. R. Soc. B Biol. Sci. 2019, 286, 20191596. [Google Scholar] [CrossRef]
- Glenn Northcutt, R. The new head hypothesis revisited. J. Exp. Zool. B Mol. Dev. Evol. 2005, 304, 274–297. [Google Scholar] [CrossRef]
- Purnell, M.A. Feeding in extinct jawless heterostracan fishes and testing scenarios of early vertebrate evolution. Proc. R. Soc. Lond. B Biol. Sci. 2002, 269, 83–88. [Google Scholar] [CrossRef]
- Marlétaz, F.; Firbas, P.N.; Maeso, I.; Tena, J.J.; Bogdanovic, O.; Perry, M.; Wyatt, C.D.R.; de la Calle-Mustienes, E.; Bertrand, S.; Burguera, D.; et al. Amphioxus functional genomics and the origins of vertebrate gene regulation. Nature 2018, 564, 64–70. [Google Scholar] [CrossRef]
- Holland, L.Z.; Ocampo Daza, D. A new look at an old question: When did the second whole genome duplication occur in vertebrate evolution? Genome Biol. 2018, 19, 209. [Google Scholar] [CrossRef]
- Tai, A.; Cheung, M.; Huang, Y.-H.; Jauch, R.; Bronner, M.E.; Cheah, K.S.E. SOXE neofunctionalization and elaboration of the neural crest during chordate evolution. Sci. Rep. 2016, 6, 34964. [Google Scholar] [CrossRef] [PubMed]
- Square, T.A.; Jandzik, D.; Massey, J.L.; Romášek, M.; Stein, H.P.; Hansen, A.W.; Purkayastha, A.; Cattell, M.V.; Medeiros, D.M. Evolution of the endothelin pathway drove neural crest cell diversification. Nature 2020, 585, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Simakov, O.; Marlétaz, F.; Yue, J.-X.; O’Connell, B.; Jenkins, J.; Brandt, A.; Calef, R.; Tung, C.-H.; Huang, T.-K.; Schmutz, J.; et al. Deeply conserved synteny resolves early events in vertebrate evolution. Nat. Ecol. Evol. 2020, 4, 820–830. [Google Scholar] [CrossRef] [PubMed]
- Schock, E.N.; York, J.R.; LaBonne, C. The developmental and evolutionary origins of cellular pluripotency in the vertebrate neural crest. Semin. Cell Dev. Biol. 2023, 138, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Thawani, A.; Groves, A.K. Building the Border: Development of the Chordate Neural Plate Border Region and Its Derivatives. Front. Physiol. 2020, 11, 608880. [Google Scholar] [CrossRef]
- Simões-Costa, M.; Bronner, M.E. Establishing neural crest identity: A gene regulatory recipe. Development 2015, 142, 242–257. [Google Scholar] [CrossRef]
- Rothstein, M.; Simoes-Costa, M. On the evolutionary origins and regionalization of the neural crest. Semin. Cell Dev. Biol. 2023, 138, 28–35. [Google Scholar] [CrossRef]
- Yu, J.-K.; Meulemans, D.; McKeown, S.J.; Bronner-Fraser, M. Insights from the amphioxus genome on the origin of vertebrate neural crest. Genome Res. 2008, 18, 1127–1132. [Google Scholar] [CrossRef]
- Leathers, T.A.; Rogers, C.D. Time to go: Neural crest cell epithelial-to-mesenchymal transition. Dev. Camb. Engl. 2022, 149, dev200712. [Google Scholar] [CrossRef]
- Taneyhill, L.A.; Schiffmacher, A.T. Should I stay or should I go? Cadherin function and regulation in the neural crest. Genesis 2017, 55, e23028. [Google Scholar] [CrossRef]
- Rothstein, M.; Bhattacharya, D.; Simoes-Costa, M. The molecular basis of neural crest axial identity. Dev. Biol. 2018, 444, S170–S180. [Google Scholar] [CrossRef]
- Srinivasan, A.; Toh, Y.-C. Human Pluripotent Stem Cell-Derived Neural Crest Cells for Tissue Regeneration and Disease Modeling. Front. Mol. Neurosci. 2019, 12, 39. [Google Scholar] [CrossRef] [PubMed]
- McCauley, D.W.; Bronner-Fraser, M. Neural crest contributions to the lamprey head. Development 2003, 130, 2317–2327. [Google Scholar] [CrossRef] [PubMed]
- Zalc, A.; Sinha, R.; Gulati, G.S.; Wesche, D.J.; Daszczuk, P.; Swigut, T.; Weissman, I.L.; Wysocka, J. Reactivation of the pluripotency program precedes formation of the cranial neural crest. Science 2021, 371, eabb4776. [Google Scholar] [CrossRef] [PubMed]
- York, J.R.; Yuan, T.; McCauley, D.W. Evolutionary and Developmental Associations of Neural Crest and Placodes in the Vertebrate Head: Insights from Jawless Vertebrates. Front. Physiol. 2020, 11, 986. [Google Scholar] [CrossRef]
- Green, S.A.; Uy, B.R.; Bronner, M.E. Ancient evolutionary origin of vertebrate enteric neurons from trunk-derived neural crest. Nature 2017, 544, 88–91. [Google Scholar] [CrossRef]
- Nikitina, N.V.; Bronner-Fraser, M. Gene regulatory networks that control the specification of neural-crest cells in the lamprey. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2009, 1789, 274–278. [Google Scholar] [CrossRef]
- Martin, W.M.; Bumm, L.A.; McCauley, D.W. Development of the viscerocranial skeleton during embryogenesis of the sea lamprey, Petromyzon Marinus. Dev. Dyn. 2009, 238, 3126–3138. [Google Scholar] [CrossRef]
- Jandzik, D.; Hawkins, M.B.; Cattell, M.V.; Cerny, R.; Square, T.A.; Medeiros, D.M. Roles for FGF in lamprey pharyngeal pouch formation and skeletogenesis highlight ancestral functions in the vertebrate head. Development 2014, 141, 629–638. [Google Scholar] [CrossRef]
- Cattell, M.; Lai, S.; Cerny, R.; Medeiros, D.M. A New Mechanistic Scenario for the Origin and Evolution of Vertebrate Cartilage. PLoS ONE 2011, 6, e22474. [Google Scholar] [CrossRef]
- Matsuoka, T.; Ahlberg, P.E.; Kessaris, N.; Iannarelli, P.; Dennehy, U.; Richardson, W.D.; McMahon, A.P.; Koentges, G. Neural crest origins of the neck and shoulder. Nature 2005, 436, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Parker, H.J.; Pushel, I.; Krumlauf, R. Coupling the roles of Hox genes to regulatory networks patterning cranial neural crest. Dev. Biol. 2018, 444, S67–S78. [Google Scholar] [CrossRef]
- Parker, H.J.; De Kumar, B.; Green, S.A.; Prummel, K.D.; Hess, C.; Kaufman, C.K.; Mosimann, C.; Wiedemann, L.M.; Bronner, M.E.; Krumlauf, R. A Hox-TALE regulatory circuit for neural crest patterning is conserved across vertebrates. Nat. Commun. 2019, 10, 1189. [Google Scholar] [CrossRef]
- Lakiza, O.; Miller, S.; Bunce, A.; Lee, E.M.-J.; McCauley, D.W. SoxE gene duplication and development of the lamprey branchial skeleton: Insights into development and evolution of the neural crest. Dev. Biol. 2011, 359, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Sumiyama, K.; Irvine, S.Q.; Ruddle, F.H. The role of gene duplication in the evolution and function of the vertebrate Dlx/distal-less bigene clusters. In Genome Evolution: Gene and Genome Duplications and the Origin of Novel Gene Functions; Meyer, A., Van de Peer, Y., Eds.; Springer: Dordrecht, The Netherlands, 2003; pp. 151–159. [Google Scholar] [CrossRef]
- Yu, J.-K.; Holland, N.D.; Holland, L.Z. Tissue-specific expression of FoxD reporter constructs in amphioxus embryos. Dev. Biol. 2004, 274, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Tocchini-Valentini, G.D.; Rochel, N.; Escriva, H.; Germain, P.; Peluso-Iltis, C.; Paris, M.; Sanglier-Cianferani, S.; Dorsselaer, A.V.; Moras, D.; Laudet, V. Structural and Functional Insights into the Ligand-binding Domain of a Nonduplicated Retinoid X Nuclear Receptor from the Invertebrate Chordate Amphioxus. J. Biol. Chem. 2009, 284, 1938–1948. [Google Scholar] [CrossRef] [PubMed]
- Meulemans, D.; Bronner-Fraser, M. Insights from Amphioxus into the Evolution of Vertebrate Cartilage. PLoS ONE 2007, 2, e787. [Google Scholar] [CrossRef] [PubMed]
- Satou, Y.; Imai, K.S. Ascidian Zic Genes. In Zic Family: Evolution, Development and Disease; Aruga, J., Ed.; Advances in Experimental Medicine and Biology; Springer: Singapore, 2018; pp. 87–106. [Google Scholar] [CrossRef]
- Kim, K.; Orvis, J.; Stolfi, A. Pax3/7 regulates neural tube closure and patterning in a non-vertebrate chordate. Front. Cell Dev. Biol. 2022, 10, 999511. [Google Scholar] [CrossRef]
- Caracciolo, A.; Di Gregorio, A.; Aniello, F.; Di Lauro, R.; Branno, M. Identification and developmental expression of three Distal-less homeobox containing genes in the ascidian Ciona intestinalis. Mech. Dev. 2000, 99, 173–176. [Google Scholar] [CrossRef]
- Aniello, F.; Locascio, A.; Villani, M.G.; Di Gregorio, A.; Fucci, L.; Branno, M. Identification and developmental expression of Ci-msxb: A novel homologue of Drosophila msh gene in Ciona intestinalis. Mech. Dev. 1999, 88, 123–126. [Google Scholar] [CrossRef]
- Jeffery, W.R.; Chiba, T.; Krajka, F.R.; Deyts, C.; Satoh, N.; Joly, J.-S. Trunk lateral cells are neural crest-like cells in the ascidian Ciona intestinalis: Insights into the ancestry and evolution of the neural crest. Dev. Biol. 2008, 324, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Kerner, P.; Hung, J.; Béhague, J.; Le Gouar, M.; Balavoine, G.; Vervoort, M. Insights into the evolution of the snail superfamily from metazoan wide molecular phylogenies and expression data in annelids. BMC Evol. Biol. 2009, 9, 94. [Google Scholar] [CrossRef] [PubMed]
- Yamada, L.; Kobayashi, K.; Degnan, B.; Satoh, N.; Satou, Y. A genomewide survey of developmentally relevant genes in Ciona intestinalis. Dev. Genes Evol. 2003, 213, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Nagatomo, K.; Ishibashi, T.; Satou, Y.; Satoh, N.; Fujiwara, S. Retinoic acid affects gene expression and morphogenesis without upregulating the retinoic acid receptor in the ascidian Ciona intestinalis. Mech. Dev. 2003, 120, 363–372. [Google Scholar] [CrossRef]
- McGonnell, I.M.; Graham, A.; Richardson, J.; Fish, J.L.; Depew, M.J.; Dee, C.T.; Holland, P.W.; Takahashi, T. Evolution of the Alx homeobox gene family: Parallel retention and independent loss of the vertebrate Alx3 gene. Evol. Dev. 2011, 13, 343–351. [Google Scholar] [CrossRef]
- Imai, K.S.; Satoh, N.; Satou, Y. A Twist-like bHLH gene is a downstream factor of an endogenous FGF and determines mesenchymal fate in the ascidian embryos. Development 2003, 130, 4461–4472. [Google Scholar] [CrossRef]
- Meulemans, D.; Bronner-Fraser, M. Amphioxus and lamprey AP-2 genes: Implications for neural crest evolution and migration patterns. Development 2002, 129, 4953–4962. [Google Scholar] [CrossRef]
- York, J.R.; Zehnder, K.; Yuan, T.; Lakiza, O.; McCauley, D.W. Evolution of Snail-mediated regulation of neural crest and placodes from an ancient role in bilaterian neurogenesis. Dev. Biol. 2019, 453, 180–190. [Google Scholar] [CrossRef]
- Manzon, L.A.; Youson, J.H.; Holzer, G.; Staiano, L.; Laudet, V.; Manzon, R.G. Thyroid hormone and retinoid X receptor function and expression during sea lamprey (Petromyzon marinus) metamorphosis. Gen. Comp. Endocrinol. 2014, 204, 211–222. [Google Scholar] [CrossRef]
- Singh, P.P.; Isambert, H. OHNOLOGS v2: A comprehensive resource for the genes retained from whole genome duplication in vertebrates. Nucleic Acids Res. 2019, 48, gkz909. [Google Scholar] [CrossRef]
- Holland, P.W.; Garcia-Fernàndez, J.; Williams, N.A.; Sidow, A. Gene duplications and the origins of vertebrate development. Development 1994, 1994, 125–133. [Google Scholar] [CrossRef]
- Smith, J.J.; Timoshevskaya, N.; Ye, C.; Holt, C.; Keinath, M.C.; Parker, H.J.; Cook, M.E.; Hess, J.E.; Narum, S.R.; Lamanna, F.; et al. The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nat. Genet. 2018, 50, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.M.; Sucharov, J.; Pulvino, A.T.; Brooks, E.P.; Gillen, A.E.; Nichols, J.T. The alx3 gene shapes the zebrafish neurocranium by regulating frontonasal neural crest cell differentiation timing. Development 2021, 148, dev197483. [Google Scholar] [CrossRef]
- Beverdam, A.; Brouwer, A.; Reijnen, M.; Korving, J.; Meijlink, F. Severe nasal clefting and abnormal embryonic apoptosis in Alx3/Alx4 double mutant mice. Development 2001, 128, 3975–3986. [Google Scholar] [CrossRef]
- Iyyanar, P.P.R.; Wu, Z.; Lan, Y.; Hu, Y.-C.; Jiang, R. Alx1 Deficient Mice Recapitulate Craniofacial Phenotype and Reveal Developmental Basis of ALX1-Related Frontonasal Dysplasia. Front. Cell Dev. Biol. 2022, 10, 777887. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.M.; Yuan, T.; Ballim, R.D.; Nguyen, K.; Kelsh, R.N.; Medeiros, D.M.; McCauley, D.W. Functional constraints on SoxE proteins in neural crest development: The importance of differential expression for evolution of protein activity. Dev. Biol. 2016, 418, 166–178. [Google Scholar] [CrossRef]
- Cossais, F.; Sock, E.; Hornig, J.; Schreiner, S.; Kellerer, S.; Bösl, M.R.; Russell, S.; Wegner, M. Replacement of mouse Sox10 by the Drosophila ortholog Sox100B provides evidence for co-option of SoxE proteins into vertebrate-specific gene-regulatory networks through altered expression. Dev. Biol. 2010, 341, 267–281. [Google Scholar] [CrossRef][Green Version]
- Kellerer, S.; Schreiner, S.; Stolt, C.C.; Scholz, S.; Bösl, M.R.; Wegner, M. Replacement of the Sox10 transcription factor by Sox8 reveals incomplete functional equivalence. Development 2006, 133, 2875–2886. [Google Scholar] [CrossRef]
- Stock, D.W.; Ellies, D.L.; Zhao, Z.; Ekker, M.; Ruddle, F.H.; Weiss, K.M. The evolution of the vertebrate Dlx gene family. Proc. Natl. Acad. Sci. USA 1996, 93, 10858–10863. [Google Scholar] [CrossRef]
- Takechi, M.; Adachi, N.; Hirai, T.; Kuratani, S.; Kuraku, S. The Dlx genes as clues to vertebrate genomics and craniofacial evolution. Semin. Cell Dev. Biol. 2013, 24, 110–118. [Google Scholar] [CrossRef]
- Kuraku, S.; Takio, Y.; Sugahara, F.; Takechi, M.; Kuratani, S. Evolution of oropharyngeal patterning mechanisms involving Dlx and endothelins in vertebrates. Dev. Biol. 2010, 341, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Cerny, R.; Cattell, M.; Sauka-Spengler, T.; Bronner-Fraser, M.; Yu, F.; Medeiros, D.M. Evidence for the prepattern/cooption model of vertebrate jaw evolution. Proc. Natl. Acad. Sci. USA 2010, 107, 17262–17267. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Cohn, M.J. Genome duplication and the origin of the vertebrate skeleton. Curr. Opin. Genet. Dev. 2008, 18, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Wada, H.; Makabe, K. Genome duplications of early vertebrates as a possible chronicle of the evolutionary history of the neural crest. Int. J. Biol. Sci. 2006, 2, 133–141. [Google Scholar] [CrossRef][Green Version]
- Carroll, S.B. Homeotic genes and the evolution of arthropods and chordates. Nature 1995, 376, 479–485. [Google Scholar] [CrossRef]
- Abbasi, A.A. Diversification of four human HOX gene clusters by step-wise evolution rather than ancient whole-genome duplications. Dev. Genes Evol. 2015, 225, 353–357. [Google Scholar] [CrossRef]
- Pervaiz, N.; Shakeel, N.; Qasim, A.; Zehra, R.; Anwar, S.; Rana, N.; Xue, Y.; Zhang, Z.; Bao, Y.; Abbasi, A.A. Evolutionary history of the human multigene families reveals widespread gene duplications throughout the history of animals. BMC Evol. Biol. 2019, 19, 128. [Google Scholar] [CrossRef]
- Abbasi, A.A. Unraveling ancient segmental duplication events in human genome by phylogenetic analysis of multigene families residing on HOX-cluster paralogons. Mol. Phylogenet. Evol. 2010, 57, 836–848. [Google Scholar] [CrossRef]
- Kanzler, B.; Kuschert, S.J.; Liu, Y.-H.; Mallo, M. Hoxa-2 restricts the chondrogenic domain and inhibits bone formation during development of the branchial area. Development 1998, 125, 2587–2597. [Google Scholar] [CrossRef]
- Pasqualetti, M.; Ori, M.; Nardi, I.; Rijli, F.M. Ectopic Hoxa2 induction after neural crest migration results in homeosis of jaw elements in Xenopus. Development 2000, 127, 5367–5378. [Google Scholar] [CrossRef]
- Parker, H.J.; Bronner, M.E.; Krumlauf, R. An atlas of anterior hox gene expression in the embryonic sea lamprey head: Hox-code evolution in vertebrates. Dev. Biol. 2019, 453, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Hunter, M.P.; Prince, V.E. Zebrafish Hox Paralogue Group 2 Genes Function Redundantly as Selector Genes to Pattern the Second Pharyngeal Arch. Dev. Biol. 2002, 247, 367–389. [Google Scholar] [CrossRef] [PubMed]
- Minoux, M.; Antonarakis, G.S.; Kmita, M.; Duboule, D.; Rijli, F.M. Rostral and caudal pharyngeal arches share a common neural crest ground pattern. Development 2009, 136, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Couly, G.; Grapin-Botton, A.; Coltey, P.; Ruhin, B.; Douarin, N.M.L. Determination of the identity of the derivatives of the cephalic neural crest: Incompatibility between Hox gene expression and lower jaw development. Development 1998, 125, 3445–3459. [Google Scholar] [CrossRef]
- Kitazawa, T.; Minoux, M.; Ducret, S.; Rijli, F.M. Different Ectopic Hoxa2 Expression Levels in Mouse Cranial Neural Crest Cells Result in Distinct Craniofacial Anomalies and Homeotic Phenotypes. J. Dev. Biol. 2022, 10, 9. [Google Scholar] [CrossRef]
- Singh, N.P.; Krumlauf, R. Diversification and Functional Evolution of HOX Proteins. Front. Cell Dev. Biol. 2022, 10, 798812. [Google Scholar] [CrossRef]
- Braasch, I.; Volff, J.-N.; Schartl, M. The Endothelin System: Evolution of Vertebrate-Specific Ligand–Receptor Interactions by Three Rounds of Genome Duplication. Mol. Biol. Evol. 2009, 26, 783–799. [Google Scholar] [CrossRef]
- Square, T.; Jandzik, D.; Cattell, M.; Hansen, A.; Medeiros, D.M. Embryonic expression of endothelins and their receptors in lamprey and frog reveals stem vertebrate origins of complex Endothelin signaling. Sci. Rep. 2016, 6, 34282. [Google Scholar] [CrossRef]
- Clouthier, D.E.; Garcia, E.; Schilling, T.F. Regulation of facial morphogenesis by endothelin signaling: Insights from mice and fish. Am. J. Med. Genet. A 2010, 152, 2962–2973. [Google Scholar] [CrossRef]
- Hirschberger, C.; Sleight, V.A.; Criswell, K.E.; Clark, S.J.; Gillis, J.A. Conserved and unique transcriptional features of pharyngeal arches in the skate (Leucoraja erinacea) and evolution of the jaw. Mol. Biol. Evol. 2021, 38, 4187–4204. [Google Scholar] [CrossRef]
- Medeiros, D.M.; Crump, J.G. New perspectives on pharyngeal dorsoventral patterning in development and evolution of the vertebrate jaw. Dev. Biol. 2012, 371, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.T.; Yelon, D.; Stainier, D.Y.R.; Kimmel, C.B. Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development 2003, 130, 1353–1365. [Google Scholar] [CrossRef] [PubMed]
- Khor, J.M.; Ettensohn, C.A. Transcription Factors of the Alx Family: Evolutionarily Conserved Regulators of Deuterostome Skeletogenesis. Front. Genet. 2020, 11, 569314. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ray, L.; Medeiros, D. Linking Vertebrate Gene Duplications to the New Head Hypothesis. Biology 2023, 12, 1213. https://doi.org/10.3390/biology12091213
Ray L, Medeiros D. Linking Vertebrate Gene Duplications to the New Head Hypothesis. Biology. 2023; 12(9):1213. https://doi.org/10.3390/biology12091213
Chicago/Turabian StyleRay, Lindsey, and Daniel Medeiros. 2023. "Linking Vertebrate Gene Duplications to the New Head Hypothesis" Biology 12, no. 9: 1213. https://doi.org/10.3390/biology12091213
APA StyleRay, L., & Medeiros, D. (2023). Linking Vertebrate Gene Duplications to the New Head Hypothesis. Biology, 12(9), 1213. https://doi.org/10.3390/biology12091213





