Inserting CTL Epitopes of the Viral Nucleoprotein to Improve Immunogenicity and Protective Efficacy of Recombinant Protein against Influenza A Virus
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
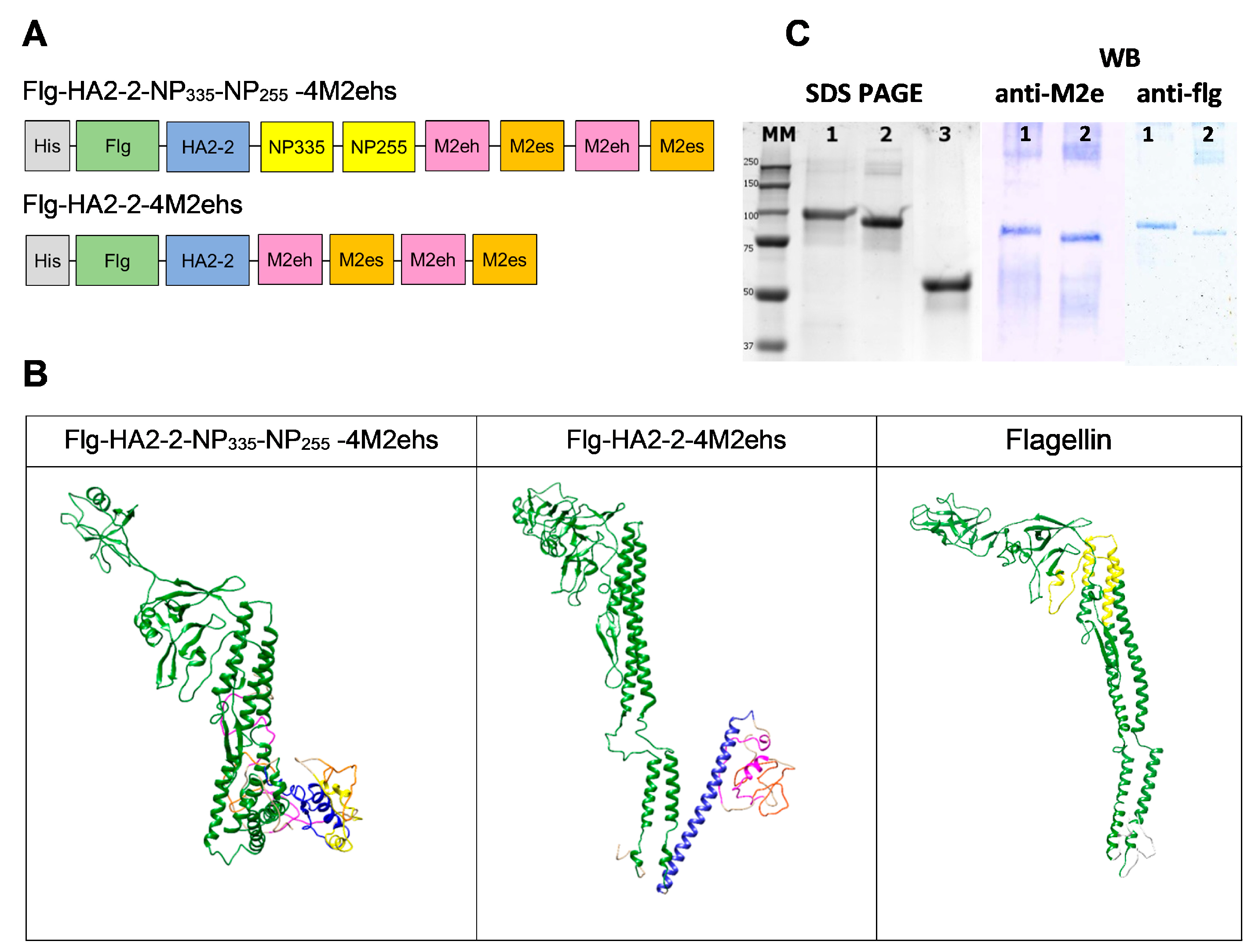
2.1. Recombinant Proteins
- -
- M2eh (SLLTEVETPIRNEWGSRSNDSSD)—the consensus sequence of the extracellular domain of the M2 protein of the human influenza A viruses;
- -
- M2es (SLLTEVETPTRSEWESRSSDSSD)—the M2e sequence of influenza A/California/07/09 (H1N1pdm09) virus;
- -
- HA2-2 (RIQDLEKYVEDTKIDLWSYNAELLVALENQHTIDLTDSEMNKLFE KTRRQLRENA)—a consensus sequence of the conserved region of HA2 (aa 76–130) of influenza A/H3N2 and A/H7N9 viruses;
- -
- NP255 (aa 255–275) DLIFLARSALILRGSVAHKS—a highly conserved region with more than 70% identity; a consensus sequence for influenza A viruses. It contains many human CTL epitopes, confirmed both experimentally (HLA-A 02, 03, 11) and theoretically (HLA-A 24, HLA-B 07, 08, 27), as well as T-cell epitopes of mice [27];
- -
- NP335 (aa 335–350) SAAFEDLRVLSFIRGY—a highly conserved region, homologous among influenza A viruses of subtypes H1, H2, H3, and H9. It contains human CTL epitopes, confirmed both experimentally (HLA-A 02, 03, HLA-B 18, 27, 37, 44) and theoretically (HLA-A 01, 26, HLA-B 15, 40, 58), as well as T-cell epitopes of mice [28].
2.2. Sodium Dodecyl Sulfate–Polyacrylamide Gel (SDS-PAGE) Electrophoresis and Western Blot
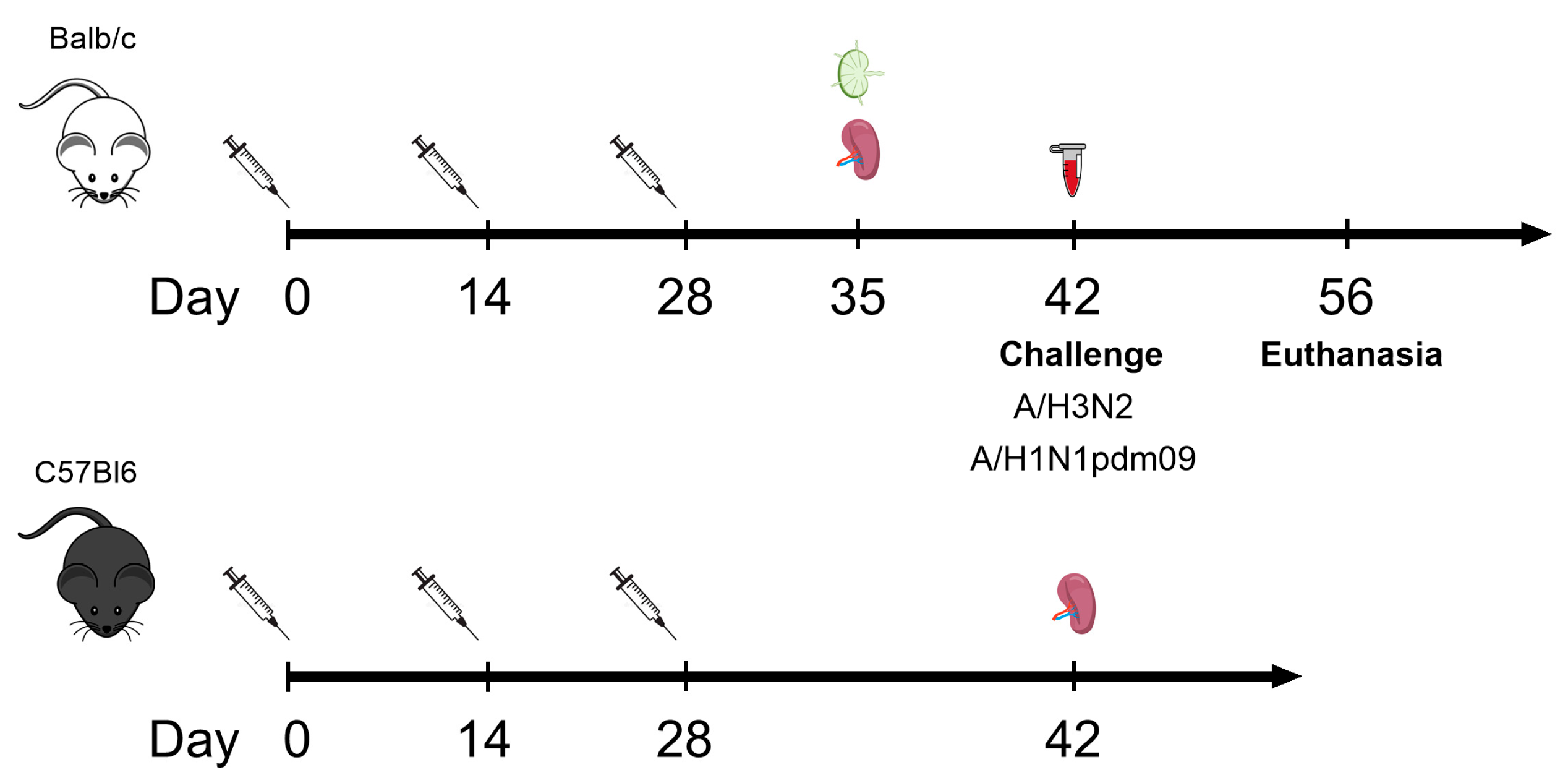
2.3. Mice
2.4. Immunization
2.5. Isolation of Cells from the Spleen and Lymph Nodes
2.6. Serum Antibody Detection by ELISA
2.7. Flow Cytometry and Intracellular Cytokine Staining (ICS)
2.8. Challenge with Influenza Viruses
2.9. Statistical Analysis
3. Results
3.1. Characterisation of Recombinant Proteins
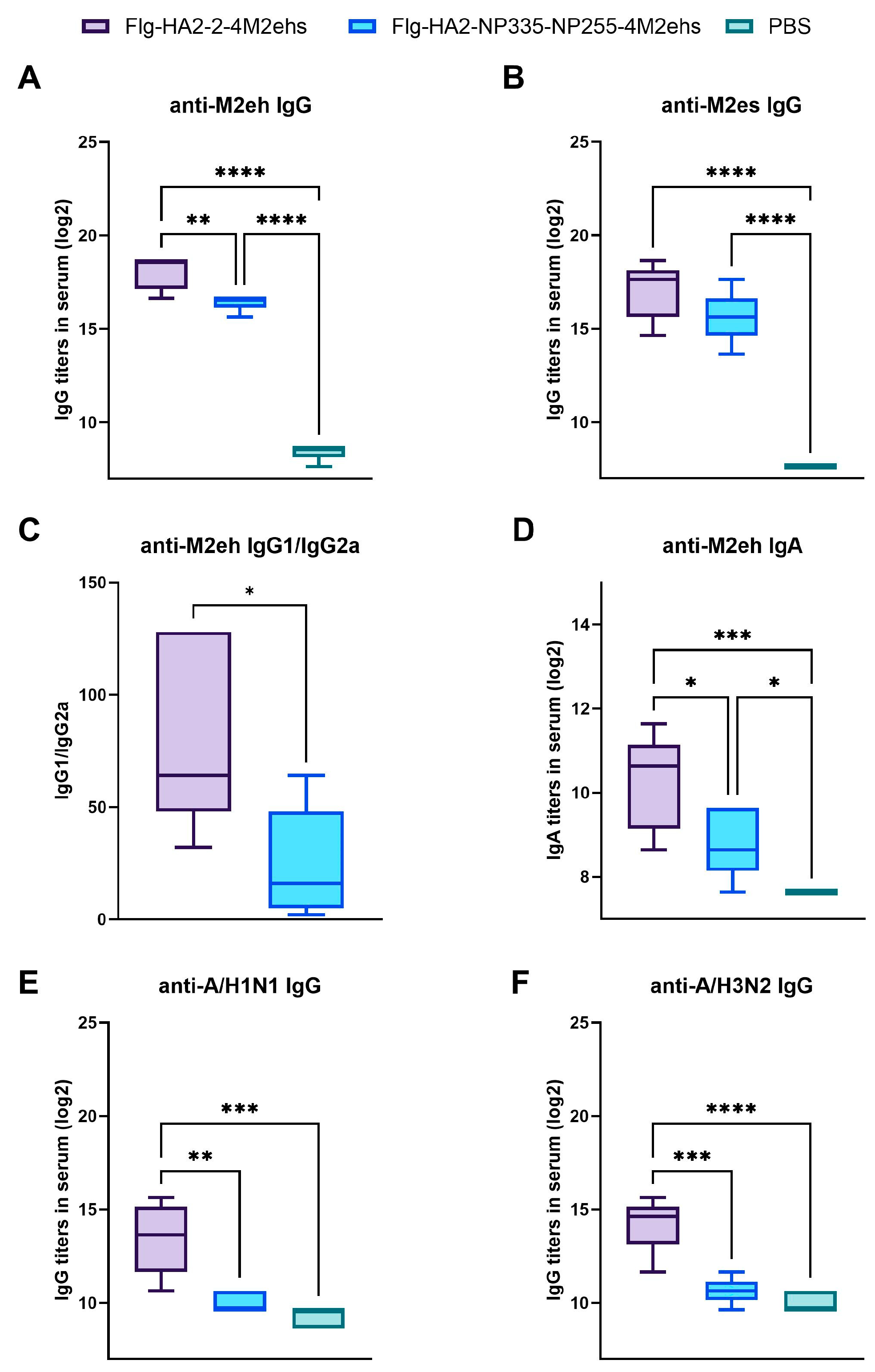
3.2. Antigen-Specific Antibody Response in Sera of BALB/c Mice after Immunization
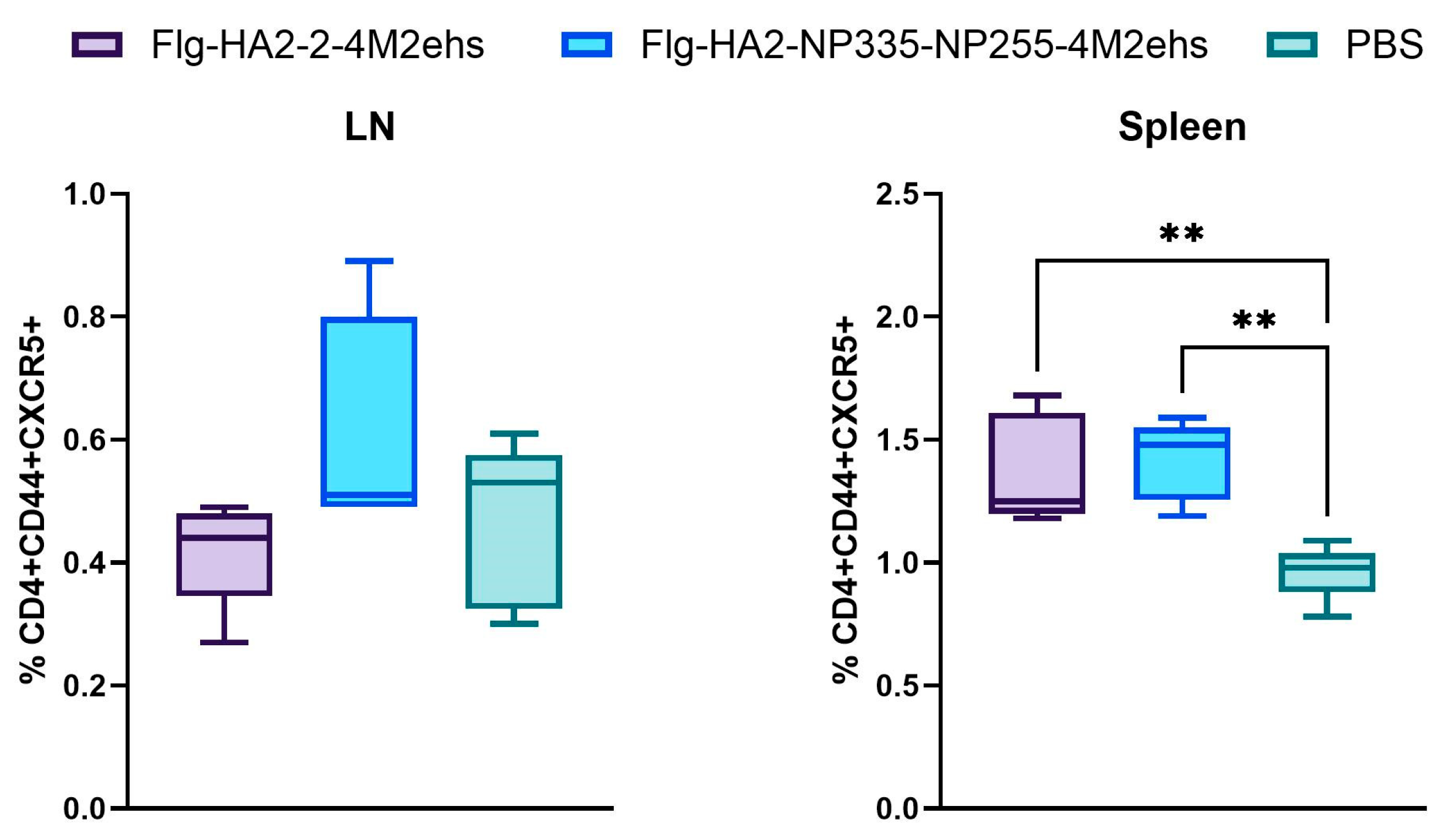
3.3. Population of Tfh Cells in Spleens and Lymph Nodes in BALB/c Mice
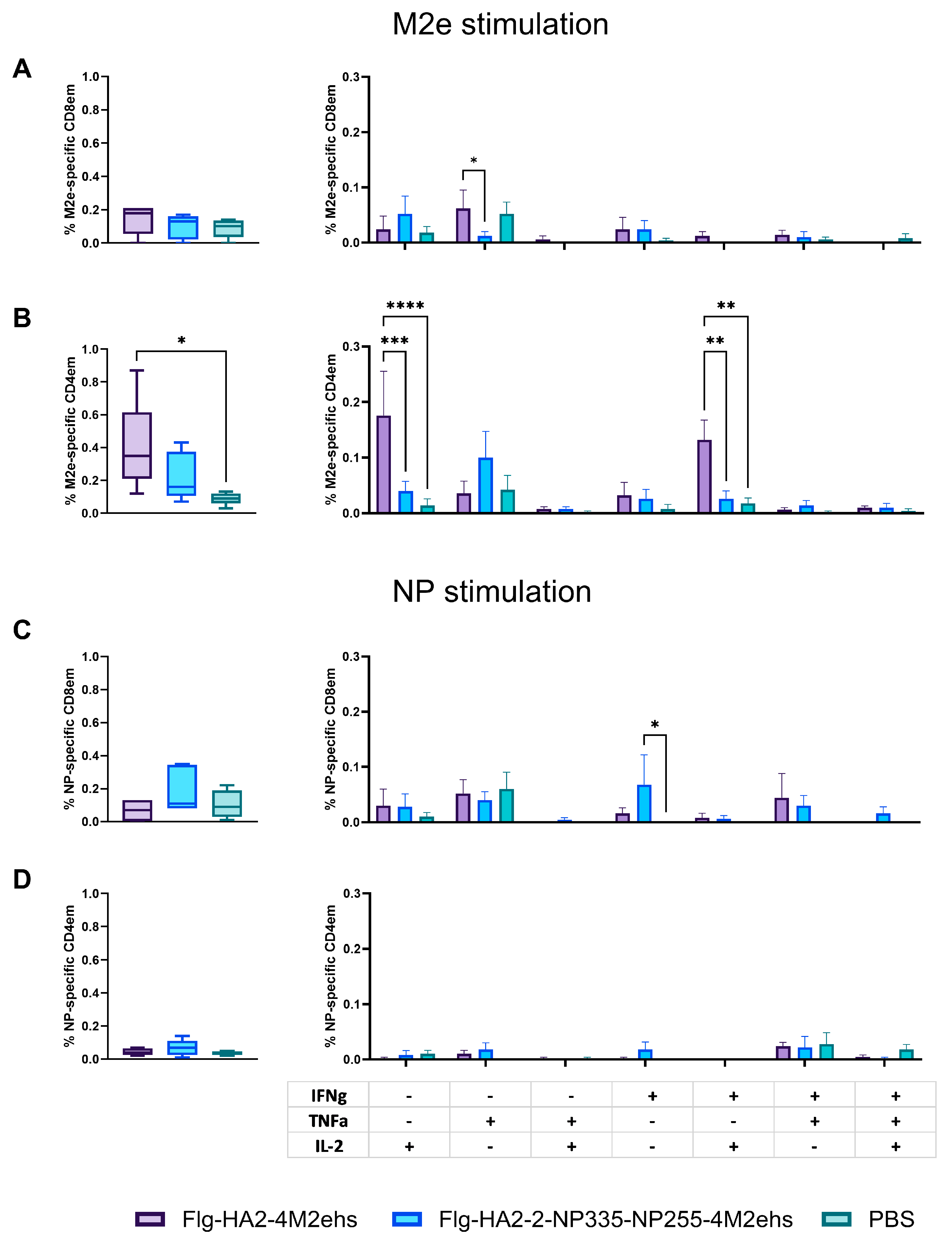
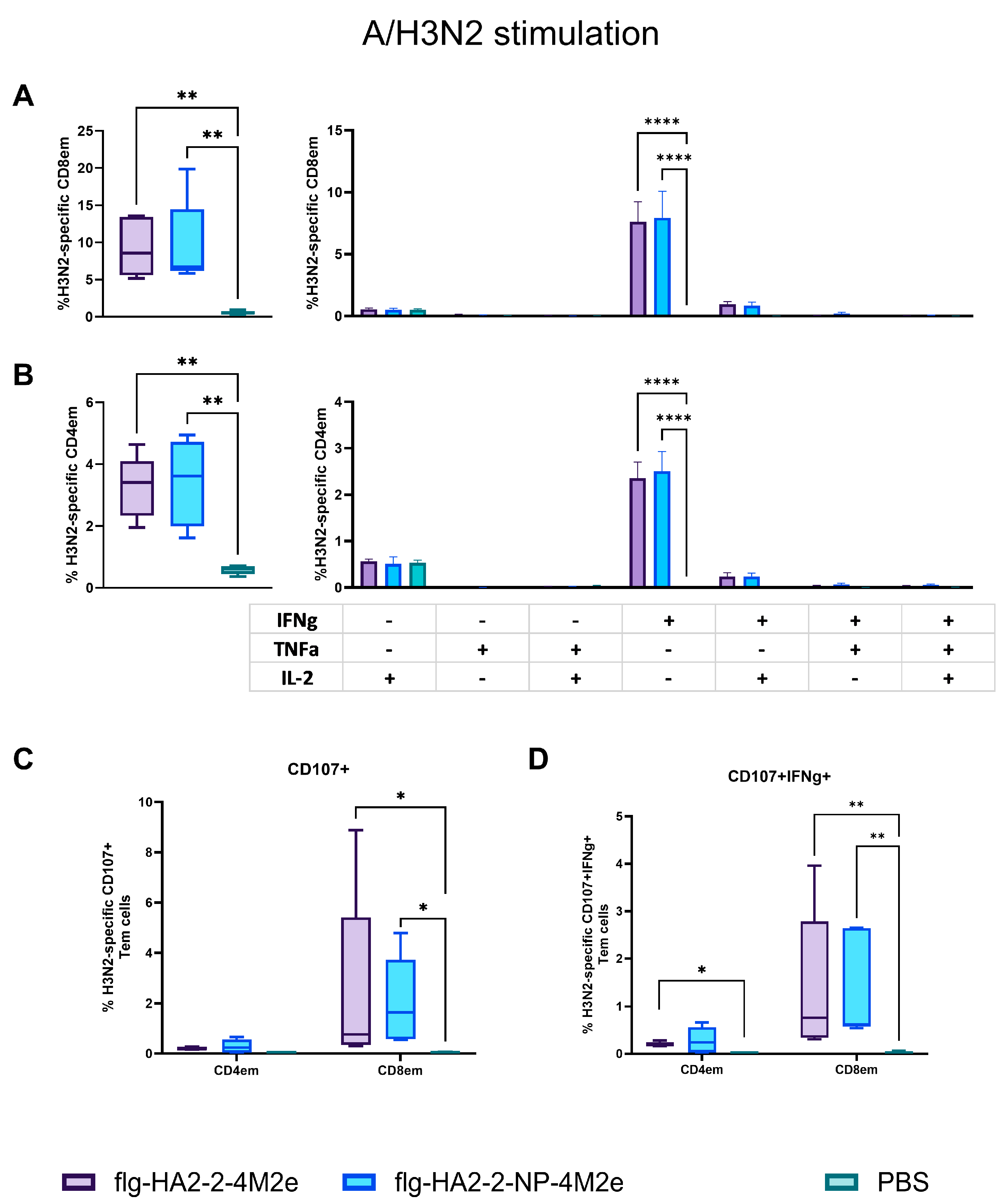
3.4. Antigen-Specific T-Cell Response in Spleens in C57Bl6 Mice
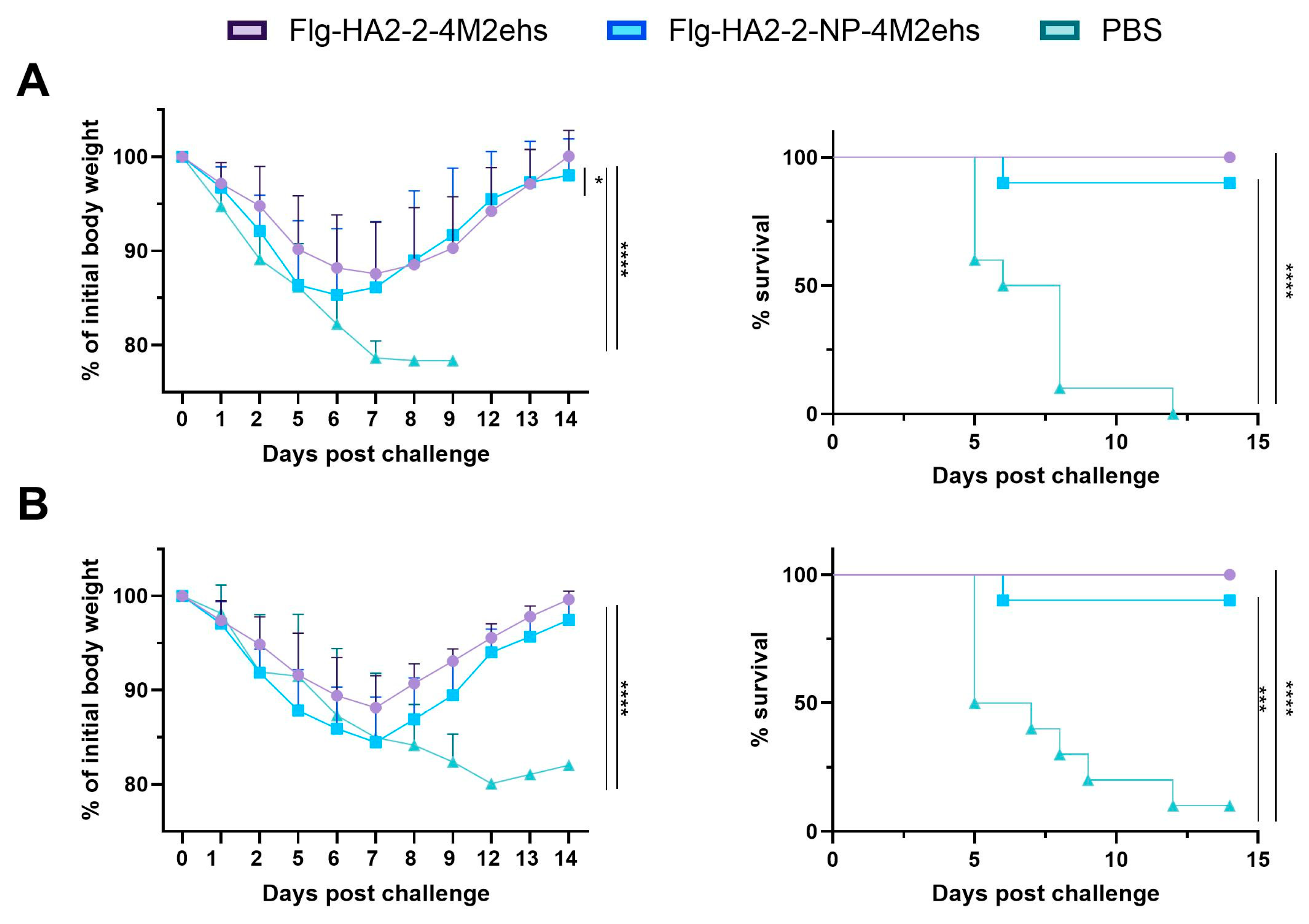
3.5. Protection against Lethal Influenza Challenge
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Universal Influenza Vaccine Technology Landscape. Available online: https://ivr.cidrap.umn.edu/universal-influenza-vaccine-technology-landscape (accessed on 29 July 2024).
- Sah, P.; Alfaro-Murillo, J.A.; Fitzpatrick, M.C.; Neuzil, K.M.; Meyers, L.A.; Singer, B.H.; Galvani, A.P. Future epidemiological and economic impacts of universal influenza vaccines. Proc. Natl. Acad. Sci. USA 2019, 116, 20786–20792. [Google Scholar] [CrossRef]
- Taylor, D.N.; Treanor, J.J.; Strout, C.; Johnson, C.; Fitzgerald, T.; Kavita, U.; Ozer, K.; Tussey, L.; Shaw, A. Induction of a potent immune response in the elderly using the TLR-5 agonist, flagellin, with a recombinant hemagglutinin influenza–flagellin fusion vaccine (VAX125, STF2. HA1 SI). Vaccine 2011, 29, 4897–4902. [Google Scholar] [CrossRef]
- Turley, C.B.; Rupp, R.E.; Johnson, C.; Taylor, D.N.; Wolfson, J.; Tussey, L.; Kavita, U.; Stanberry, L.; Shaw, A. Safety and immunogenicity of a recombinant M2e–flagellin influenza vaccine (STF2. 4xM2e) in healthy adults. Vaccine 2011, 29, 5145–5152. [Google Scholar] [CrossRef]
- Wang, W.-C.; Sayedahmed, E.E.; Sambhara, S.; Mittal, S.K. Progress towards the development of a universal influenza vaccine. Viruses 2022, 14, 1684. [Google Scholar] [CrossRef]
- Grant, E.J.; Josephs, T.M.; Loh, L.; Clemens, E.B.; Sant, S.; Bharadwaj, M.; Chen, W.; Rossjohn, J.; Gras, S.; Kedzierska, K. Broad CD8+ T cell cross-recognition of distinct influenza A strains in humans. Nat. Commun. 2018, 9, 5427. [Google Scholar] [CrossRef]
- Sridhar, S.; Begom, S.; Bermingham, A.; Hoschler, K.; Adamson, W.; Carman, W.; Bean, T.; Barclay, W.; Deeks, J.J.; Lalvani, A. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 2013, 19, 1305–1312. [Google Scholar] [CrossRef]
- Altenburg, A.F.; Rimmelzwaan, G.F.; de Vries, R.D. Virus-specific T cells as correlate of (cross-)protective immunity against influenza. Vaccine 2015, 33, 500–506. [Google Scholar] [CrossRef]
- van de Ven, K.; de Heij, F.; van Dijken, H.; Ferreira, J.A.; de Jonge, J. Systemic and respiratory T-cells induced by seasonal H1N1 influenza protect against pandemic H2N2 in ferrets. Commun. Biol. 2020, 3, 564. [Google Scholar] [CrossRef]
- Li, Z.R.T.; Zarnitsyna, V.I.; Lowen, A.C.; Weissman, D.; Koelle, K.; Kohlmeier, J.E.; Antiaet, R. Why are CD8 T cell epitopes of human influenza A virus conserved? J. Virol. 2019, 93, e01534-18. [Google Scholar] [CrossRef]
- Forrest, B.D.; Pride, M.W.; Dunning, A.J.; Capeding, M.R.Z.; Chotpitayasunondh, T.; Tam, J.S.; Rappaport, R.; Eldridge, J.H.; Grube, W.C. Correlation of cellular immune responses with protection against culture-confirmed influenza virus in young children. Clin. Vaccine Immunol. 2008, 15, 1042–1053. [Google Scholar] [CrossRef]
- Yuan, Y.; Li, B.; Zhou, L.; Luo, J.; Liu, X.; Wang, S.; Lu, Q.; Tan, W.; Chen, Z. Protein transduction domain-mediated influenza NP subunit vaccine generates a potent immune response and protection against influenza virus in mice. Emerg. Microbes Infect 2020, 9, 1933–1942. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Luo, J.; Chen, Z. Development of universal influenza vaccines based on influenza virus M and NP genes. Infection 2014, 42, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Eickhoff, C.S.; Terry, F.E.; Peng, L.; Meza, K.A.; Sakala, I.G.; Van Aartsen, D.; Moise, L.; Martin, W.D.; Schriewer, J.; Buller, R.M.; et al. Highly conserved influenza T cell epitopes induce broadly protective immunity. Vaccine 2019, 37, 5371–5381. [Google Scholar] [CrossRef] [PubMed]
- van de Sandt, C.E.; Clemens, E.B.; Grant, E.J.; Rowntree, L.C.; Sant, S.; Halim, H.; Crowe, J.; Cheng, A.C.; Kotsimbos, T.C.; Richards, M.; et al. Challenging immunodominance of influenza-specific CD8+ T cell responses restricted by the risk-associated HLA-A* 68: 01 allomorph. Nat. Commun. 2019, 10, 5579. [Google Scholar] [CrossRef] [PubMed]
- Heiny, A.T.; Miotto, O.; Srinivasan, K.N.; Khan, A.M.; Zhang, G.L.; Brusic, V.; Tan, T.W.; August, J.T. Evolutionarily conserved protein sequences of influenza a viruses, avian and human, as vaccine targets. PLoS ONE 2007, 2, e1190. [Google Scholar] [CrossRef] [PubMed]
- Balz, K.; Trassl, L.; Härtel, V.; Nelson, P.P.; Skevaki, C. Virus-induced T cell-mediated heterologous immunity and vaccine development. Front. Immunol. 2020, 11, 513. [Google Scholar] [CrossRef]
- Romeli, S.; Hassan, S.S.; Yap, W.B. Multi-Epitope Peptide-Based and Vaccinia-Based Universal Influenza Vaccine Candidates Subjected to Clinical Trials. Malays. J. Med. Sci. 2020, 27, 10–20. [Google Scholar] [CrossRef]
- Bates, J.T.; Honko, A.N.; Graff, A.H.; Kock, N.D.; Mizel, S.B. Mucosal adjuvant activity of flagellin in aged mice. Mech. Ageing Dev. 2008, 129, 271–281. [Google Scholar] [CrossRef]
- Huleatt, J.W.; Nakaar, V.; Desai, P.; Huang, Y.; Hewitt, D.; Jacobs, A.; Tang, J.; McDonald, W.; Song, L.; Evans, R.K.; et al. Potent immunogenicity and efficacy of a universal influenza vaccine candidate comprising a recombinant fusion protein linking influenza M2e to the TLR5 ligand flagellin. Vaccine 2008, 26, 201–214. [Google Scholar] [CrossRef]
- Song, L.; Zhang, Y.; Yun, N.E.; Poussard, A.L.; Smith, J.N.; Smith, J.K.; Borisevich, V.; Linde, J.J.; Zacks, M.A.; Li, H.; et al. Superior efficacy of a recombinant flagellin: H5N1 HA globular head vaccine is determined by the placement of the globular head within flagellin. Vaccine 2009, 27, 5875–5884. [Google Scholar] [CrossRef]
- Liu, G.; Tarbet, B.; Song, L.; Reiserova, L.; Weaver, B.; Chen, Y.; Li, H.; Hou, F.; Liu, X.; Parent, J.; et al. Immunogenicity and efficacy of flagellin-fused vaccine candidates targeting 2009 pandemic H1N1 influenza in mice. PLoS ONE 2011, 6, e20928. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Song, L.; Reiserova, L.; Trivedi, U.; Li, H.; Liu, X.; Noah, D.; Hou, F.; Weaver, B.; Tussey, L. Flagellin-HA vaccines protect ferrets and mice against H5N1 highly pathogenic avian influenza virus (HPAIV) infections. Vaccine 2012, 30, 6833–6838. [Google Scholar] [CrossRef]
- Stepanova, L.A.; Mardanova, E.S.; Shuklina, M.A.; Blokhina, E.A.; Kotlyarov, R.Y.; Potapchuk, M.V.; Kovaleva, A.A.; Vidyaeva, I.G.; Korotkov, A.V.; Eletskaya, E.I.; et al. Flagellin-fused protein targeting M2e and HA2 induces potent humoral and T-cell response and protects mice against various influenza viruses a subtypes. J. Biomed. Sci. 2018, 25, 33. [Google Scholar] [CrossRef]
- Shuklina, M.A.; Stepanova, L.A.; Kovaleva, A.A.; Korotkov, A.V.; Shaldzhyan, A.A.; Zaitseva, M.V.; Eletskaya, E.I.; Tsybalova, L.M. Intranasal immunization with a recombinant protein based on the M2e peptide and second subunit of influenza A viral hemagglutinin fragment induces a cross-protective humoral and T cell response in mice. Med. Immunol. 2020, 22, 357–370. [Google Scholar] [CrossRef]
- Tsybalova, L.M.; Stepanova, L.A.; Shuklina, M.A.; Mardanova, E.S.; Kotlyarov, R.Y.; Potapchuk, M.V.; Petrov, S.A.; Blokhina, E.A.; Ravin, N.V. Combination of M2e peptide with stalk HA epitopes of influenza A virus enhances protective properties of recombinant vaccine. PLoS ONE 2018, 13, e0201429. [Google Scholar] [CrossRef]
- Atsmon, J.; Kate-Ilovitz, E.; Shaikevich, D.; Singer, Y.; Volokhov, I.; Haim, K.Y.; Ben-Yedidia, T. Safety and Immunogenicity of 653 Multimeric-001—A Novel Universal Influenza Vaccine. J. Clin. Immunol. 2012, 32, 595–603. [Google Scholar] [CrossRef]
- Stoloff, G.A.; Caparros-Wanderley, W. Synthetic Multi-Epitope Peptides Identified in Silico Induce Protective Immunity against Multiple Influenza Serotypes. Eur. J. Immunol. 2007, 37, 2441–2449. [Google Scholar] [CrossRef]
- De Filette, M.; Min Jou, W.; Birkett, A.; Lyons, K.; Schultz, B.; Tonkyro, A.; Resch, S.; Fiers, W. Universal Influenza A Vaccine: Optimization of M2-Based Constructs. Virology 2005, 337, 149–161. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Wille, M.; Holmes, E.C. The Ecology and Evolution of Influenza Viruses. Cold Spring Harb. Perspect. Med. 2020, 10, a038489. [Google Scholar]
- Babar, M.M.; Zaidi, N.-U.-S.S. Protein sequence conservation and stable molecular evolution reveals influenza virus nucleoprotein as a universal druggable target. Infect. Genet. Evol. 2015, 34, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Kang, J.O.; Chang, J. Nucleoprotein vaccine induces cross-protective cytotoxic T lymphocytes against both lineages of influenza B virus. Clin. Exp. Vaccine Res. 2019, 8, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Jegaskanda, S.; Job, E.R.; Kramski, M.; Laurie, K.; Isitman, G.; de Rose, R.; Winnall, W.R.; Stratov, I.; Brooks, A.G.; Reading, P.C.; et al. Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity antibodies in the absence of neutralizing antibodies. J. Immunol. 2013, 190, 1837–1848. [Google Scholar] [CrossRef]
- Liao, H.-Y.; Wang, S.-C.; Ko, Y.-A.; Lin, K.-I.; Ma, C.; Cheng, T.-J.R.; Wong, C.-H. Chimeric hemagglutinin vaccine elicits broadly protective CD4 and CD8 T cell responses against multiple influenza strains and subtypes. Proc. Natl. Acad. Sci. USA 2020, 117, 17757–17763. [Google Scholar] [CrossRef]
- Stepanova, L.A.; Shuklina, M.A.; Vasiliev, K.A.; Kovaleva, A.A.; Vidyaeva, I.G.; Zabrodskaya, Y.A.; Korotkov, A.V.; Tsybalova, L.M. Flagellin-Fused Protein Targeting M2e and HA2 Induces Innate and T-Cell Responses in Mice of Different Genetic Lines. Vaccines 2022, 10, 2098. [Google Scholar] [CrossRef]
- Zykova, A.A.; Blokhina, E.A.; Stepanova, L.A.; Shuklina, M.A.; Ozhereleva, O.O.; Tsybalova, L.M.; Kuprianov, V.V.; Ravin, N.V. Nanoparticles Carrying Conserved Regions of Influenza A Hemagglutinin, Nucleoprotein, and M2 Protein Elicit a Strong Humoral and T Cell Immune Response and Protect Animals from Infection. Molecules 2023, 28, 6441. [Google Scholar] [CrossRef]
- El Bakkouri, K.; Descamps, F.; de Filette, M.; Smet, A.; Festjens, E.; Birkett, A.; van Rooijen, N.; Verbeek, S.; Fiers, W.; Saelens, X. Universal vaccine based on ectodomain of matrix protein 2 of influenza a: Fc receptors and alveolar macrophages mediate protection. J. Immunol. 2011, 186, 1022–1031. [Google Scholar] [CrossRef]
- Kolpe, A.; Schepens, B.; Fiers, W.; Saelens, X. M2-based influenza vaccines: Recent advances and clinical potential. Expert Rev. Vaccines 2017, 16, 123–136. [Google Scholar] [CrossRef]
- Lee, Y.-N.; Kim, M.-C.; Lee, Y.-T.; Kim, Y.-J.; Kang, S.-M. Mechanisms of Cross-protection by Influenza Virus M2-based Vaccines. Immune Netw. 2015, 15, 213–221. [Google Scholar] [CrossRef]
- Wang, L.; Hess, A.; Chang, T.Z.; Wang, Y.-C.; Champion, J.A.; Compans, R.W.; Wang, B.-Z. Nanoclusters self-assembled from conformation-stabilized influenza M2e as broadly cross-protective influenza vaccines. Nanomedicine 2014, 10, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Mohan, T.; Chang, T.Z.; Gonzalez, G.X.; Wang, Y.; Kwon, Y.M.; Kang, S.M.; Compans, R.W.; Champion, J.A.; Wang, B.Z. Double-layered protein nanoparticles induce broad protection against divergent influenza A viruses. Nat. Commun. 2018, 9, 359. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Chang, T.Z.; Wang, Y.; Li, S.; Wang, S.; Matsuyama, S.; Yu, G.; Compans, R.W.; Li, J.D.; Prausnitz, M.R.; et al. Heterosubtypic influenza protection elicited by double-layered polypeptide nanoparticles in mice. Proc. Natl. Acad. Sci. USA 2018, 115, E7758–E7767. [Google Scholar] [CrossRef] [PubMed]
- Epstein, S.L.; Price, G.E. Cross-protective immunity to influenza A viruses. Expert Rev. Vaccines 2010, 9, 1325–1341. [Google Scholar]
- Grant, E.; Wu, C.; Chan, K.F.; Eckle, S.; Bharadwaj, M.; Zou, Q.M.; Kedzierska, K.; Chen, W. Nucleoprotein of influenza A virus is a major target of immunodominant CD8+ T-cell responses. Immunol. Cell Biol. 2013, 91, 184–194. [Google Scholar] [CrossRef]
- Pizzolla, A.; Nguyen, T.H.O.; Smith, J.M.; Brooks, A.G.; Kedzierska, K.; Heath, W.R.; Reading, P.; Wakim, L.M. Resident memory CD8+T cells in the upper respiratory tract prevent pulmonary influenza virus infection. Sci. Immunol. 2017, 2, eaam6970. [Google Scholar] [CrossRef]
- Kwak, C.; Nguyen, Q.T.; Kim, J.; Kim, T.-H.; Poo, H. Influenza Chimeric Protein (3M2e-3HA2-NP) Adjuvanted with PGA/Alum Confers Cross-Protection against Heterologous Influenza A Viruses. J. Microbiol. Biotechnol. 2021, 31, 304–316. [Google Scholar]
- Ma, Y.; Wang, Y.; Dong, C.; Gonzalez, G.X.; Song, Y.; Zhu, W.; Kim, J.; Wei, L.; Wang, B.-Z. Influenza NP core and HA or M2e shell double-layered protein nanoparticles induce broad protection against divergent influenza A viruses. Nanomedicine 2022, 40, 102479. [Google Scholar] [CrossRef]
- Ekramy, E.; Sayedahmed, N.O.; Elshafie, A.P.; dos Santos, C.J.; Sambhara, S.; Mittal, S.K. Development of NP-Based Universal Vaccine for Influenza A Viruses. Vaccines 2024, 12, 157. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, C.; Ma, Y.; Zhu, W.; Gill, H.S.; Denning, T.L.; Kang, S.-M.; Wang, B.-Z. Monophosphoryl lipid A-adjuvanted nucleoprotein neuraminidase nanoparticles improve immune protection against divergent influenza viruses. Nanomedicine 2023, 47, 102614. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Huang, B.; Wang, X.; Tan, W.; Ruan, L. Improving Cross-Protection against Influenza Virus Using Recombinant Vaccinia Vaccine Expressing NP and M2 Ectodomain Tandem Repeats. Virol. Sin. 2019, 34, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Koutsakos, M.; Nguyen, T.H.O.; Kedzierska, K. With a little help from T follicular helper friends: Humoral immunity to influenza vaccination. J. Immunol. 2019, 202, 360–367. [Google Scholar] [CrossRef] [PubMed]
- DiPiazza, A.T.; Fan, S.; Rattan, A.; DeDiego, M.L.; Chaves, F.; Neumann, G.; Kawaoka, Y.; Sant, A.S. A novel vaccine strategy to overcome poor immunogenicity of avian influenza vaccines through mobilization of memory CD4 T cells established by seasonal influenza. J. Immunol. 2019, 203, 1502–1508. [Google Scholar] [CrossRef] [PubMed]
- Richards, K.A.; Shannon, I.; Treanor, J.J.; Yang, H.; Nayak, J.L.; Sant, A.J. Evidence that blunted CD4 T-cell responses underlie deficient protective antibody responses to influenza vaccines in repeatedly vaccinated human subjects. J. Infect. Dis. 2020, 222, 273–277. [Google Scholar] [CrossRef]
- Ramiscal, R.R.; Vinuesa, C.G. T-cell subsets in the germinal center. Immunol. Rev. 2013, 252, 146–155. [Google Scholar] [CrossRef]
- Crotty, S.; Follicular Helper, T. Cell Biology: A decade of discovery and diseases. Immunity 2019, 50, 1132–1148. [Google Scholar] [CrossRef]
- Aljurayyan, A.; Puksuriwong, S.; Ahmed, M.; Sharma, R.; Krishnan, M.; Sood, S.; Davies, K.; Rajashekar, D.; Leong, S.; McNamara, P.S.; et al. Activation and Induction of Antigen-Specific T Follicular Helper Cells Play a Critical Role in Live-Attenuated Influenza Vaccine-Induced Human Mucosal Anti-influenza Antibody Response. J. Virol. 2018, 92, e00114-18. [Google Scholar] [CrossRef]
- Sánchez-Vargas, L.A.; Kounlavouth, S.; Smith, M.L.; Anderson, K.B.; Srikiatkhachorn, A.; Ellison, D.W.; Currier, J.R.; Endy, T.P.; Mathew, A.; Rothman, A.L. Longitudinal analysis of memory B and T cell responses to dengue virus in a 5-year prospective cohort study in Thailand. Front. Immunol. 2019, 10, 1359. [Google Scholar] [CrossRef]
- Krammer, F. The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 2019, 19, 383–397. [Google Scholar] [CrossRef]
- Jelley-Gibbs, D.M.; Brown, D.M.; Dibble, J.P.; Haynes, L.; Eaton, S.M.; Swain, S.L. Unexpected prolonged presentation of influenza antigens promotes CD4 T cell memory generation. J. Exp. Med. 2005, 202, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Bautista, B.L.; Devarajan, P.; McKinstry, K.K.; Strutt, T.M.; Vong, A.M.; Jones, M.C.; Kuang, Y.; Mott, D.; Swain, S.L. Short-lived antigen recognition but not viral infection at a defined checkpoint programs effector CD4 T cells to become protective memory. J. Immunol. 2016, 197, 3936–3949. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Kuang, Y.; Liang, J.; Jones, M.; Swain, S.L. Influenza vaccine-induced CD4 effectors require antigen recognition at an effector checkpoint to generate CD4 lung memory and antibody production. J. Immunol. 2020, 205, 2077–2090. [Google Scholar] [CrossRef] [PubMed]
- Eldi, P.; Chaudhri, G.; Nutt, S.L.; Newsome, T.P.; Karupiah, G. Viral replicative capacity, antigen availability via hematogenous spread, and high TFH:TFR ratios drive induction of potent neutralizing antibody responses. J. Virol. 2019, 93, e01795-18. [Google Scholar] [CrossRef] [PubMed]
- Baumjohann, D.; Preite, S.; Reboldi, A.; Ronchi, F.; Ansel, K.M.; Lanzavecchia, A.; Sallusto, F. Persistent antigen and germinal center B cells sustain T follicular helper cell responses and phenotype. Immunity 2013, 38, 596–605. [Google Scholar] [CrossRef]
- Tam, H.H.; Melo, M.B.; Kang, M.; Pelet, J.M.; Ruda, V.M.; Foley, M.H.; Hu, J.K.; Kumari, S.; Crampton, J.; Baldeon, A.D.; et al. Sustained antigen availability during germinal center initiation enhances antibody responses to vaccination. Proc. Natl. Acad. Sci. USA 2016, 113, E6639–E6648. [Google Scholar] [CrossRef]
- Savic, M.; Dembinski, J.L.; Laake, I.; Hungnes, O.; Cox, R.; Oftung, F.; Trogstad, L.; Mjaaland, S. Distinct T and NK cell populations may serve as immune correlates of protection against symptomatic pandemic influenza A(H1N1) virus infection during pregnancy. PLoS ONE 2017, 12, e0188055. [Google Scholar] [CrossRef]
- L’Huillier, A.G.; Ferreira, V.H.; Hirzel, C.; Nellimarla, S.; Ku, T.; Natori, Y.; Humar, A.; Kumar, D. T-cell responses following Natural Influenza Infection or Vaccination in Solid Organ Transplant Recipients. Sci. Rep. 2020, 10, 10104. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shuklina, M.; Stepanova, L.; Ozhereleva, O.; Kovaleva, A.; Vidyaeva, I.; Korotkov, A.; Tsybalova, L. Inserting CTL Epitopes of the Viral Nucleoprotein to Improve Immunogenicity and Protective Efficacy of Recombinant Protein against Influenza A Virus. Biology 2024, 13, 801. https://doi.org/10.3390/biology13100801
Shuklina M, Stepanova L, Ozhereleva O, Kovaleva A, Vidyaeva I, Korotkov A, Tsybalova L. Inserting CTL Epitopes of the Viral Nucleoprotein to Improve Immunogenicity and Protective Efficacy of Recombinant Protein against Influenza A Virus. Biology. 2024; 13(10):801. https://doi.org/10.3390/biology13100801
Chicago/Turabian StyleShuklina, Marina, Liudmila Stepanova, Olga Ozhereleva, Anna Kovaleva, Inna Vidyaeva, Alexandr Korotkov, and Liudmila Tsybalova. 2024. "Inserting CTL Epitopes of the Viral Nucleoprotein to Improve Immunogenicity and Protective Efficacy of Recombinant Protein against Influenza A Virus" Biology 13, no. 10: 801. https://doi.org/10.3390/biology13100801
APA StyleShuklina, M., Stepanova, L., Ozhereleva, O., Kovaleva, A., Vidyaeva, I., Korotkov, A., & Tsybalova, L. (2024). Inserting CTL Epitopes of the Viral Nucleoprotein to Improve Immunogenicity and Protective Efficacy of Recombinant Protein against Influenza A Virus. Biology, 13(10), 801. https://doi.org/10.3390/biology13100801





