The Unique Roles of Ion Channels in Pluripotent Stem Cells in Response to Biological Stimuli
Simple Summary
Abstract
1. Introduction
2. Ion Channels
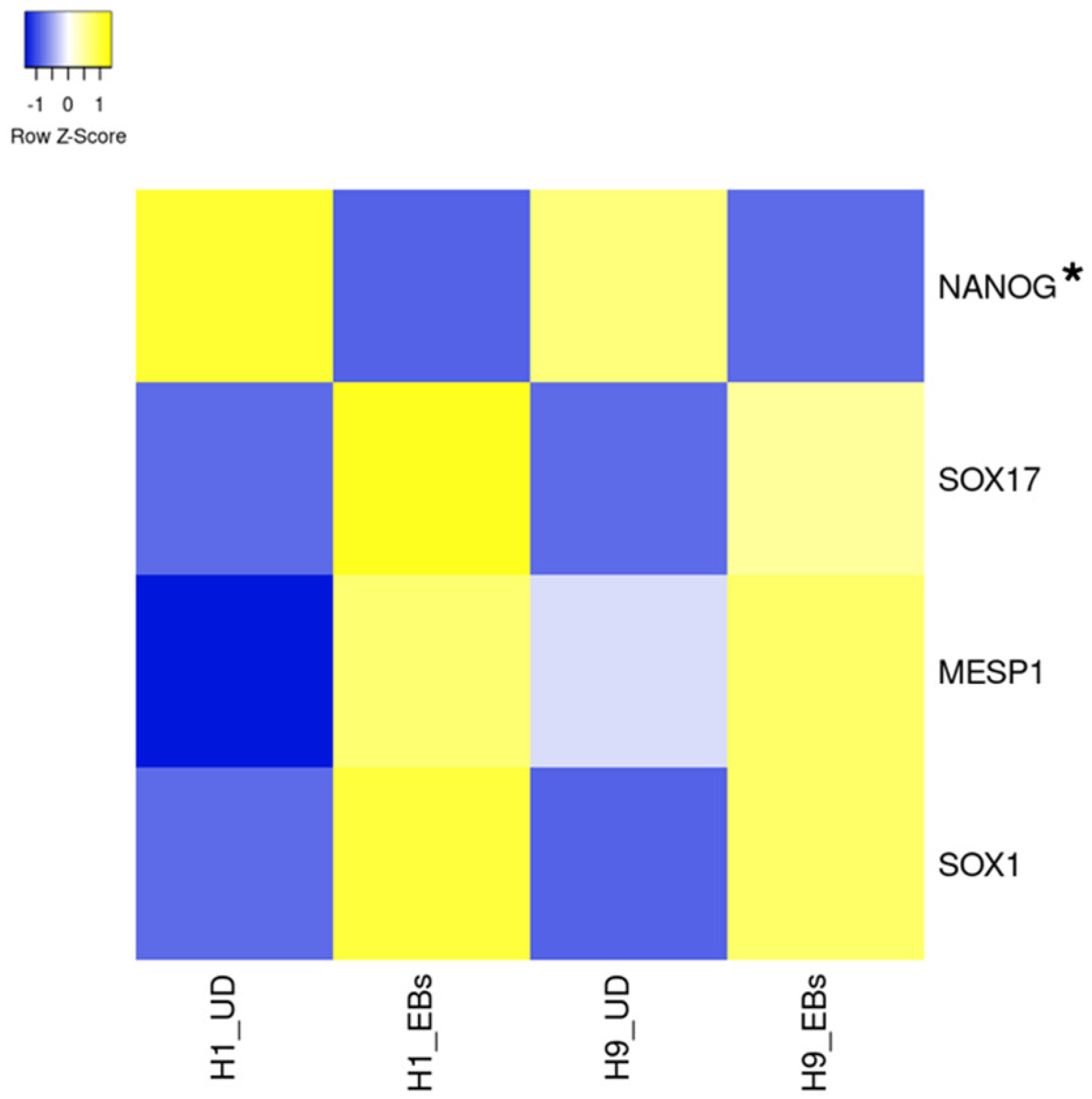
3. Ion Channels Expressed in Pluripotent Stem Cells
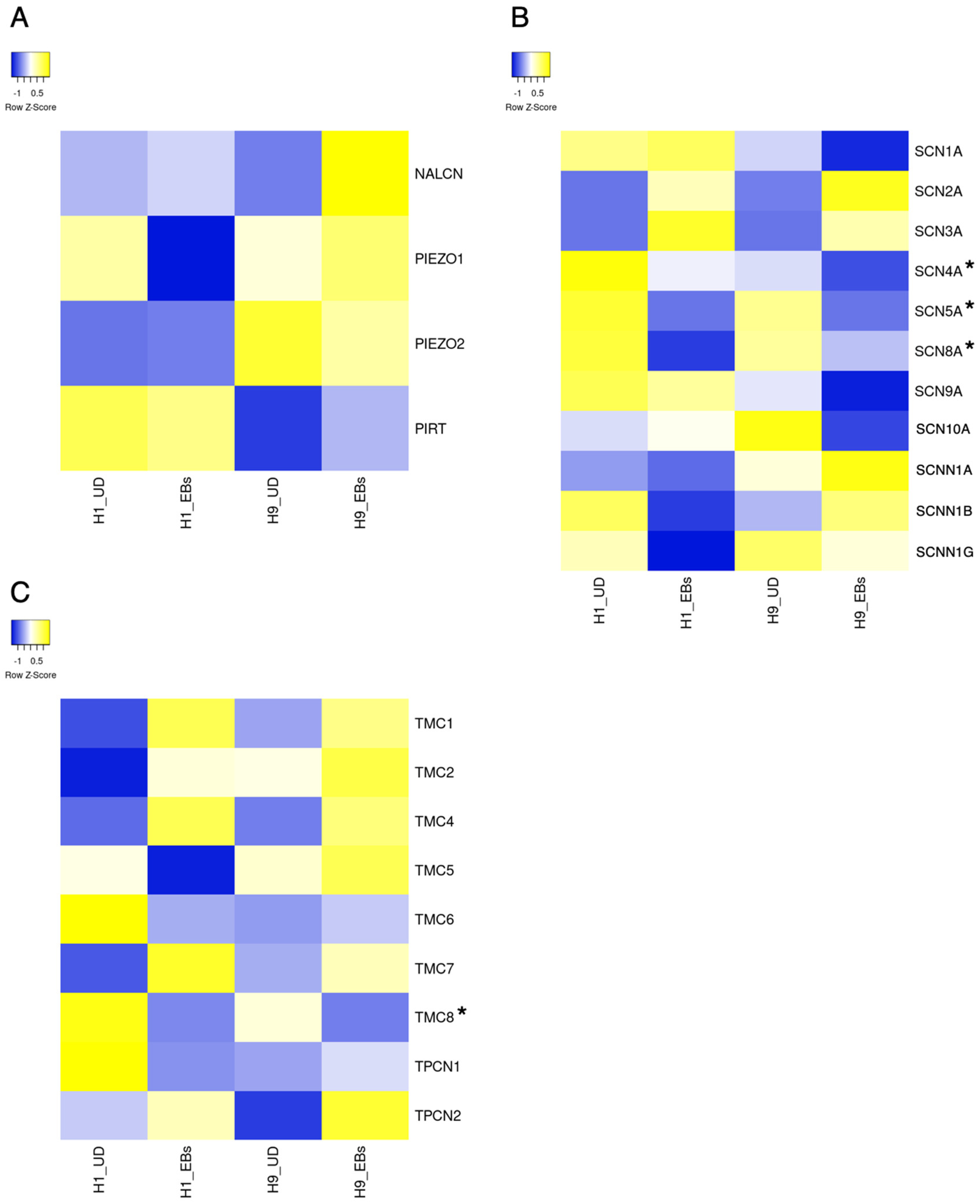
4. Sodium Channels
The Role of Sodium Channels in Pluripotent Stem Cells
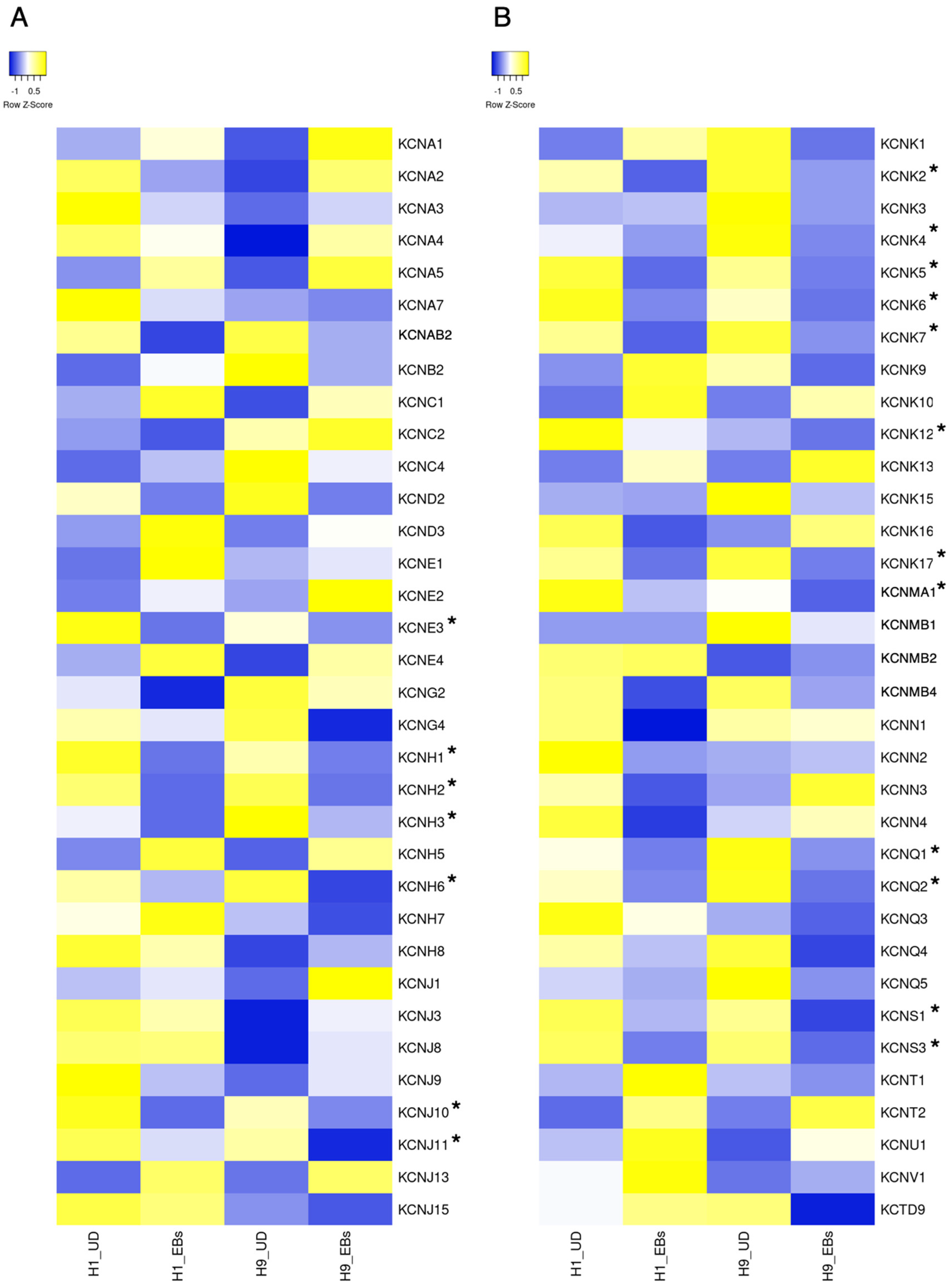
5. Potassium Channels
The Role of Potassium Channels in Pluripotent Stem Cells
6. Calcium Channels
The Role of Calcium Channels in Pluripotent Stem Cells
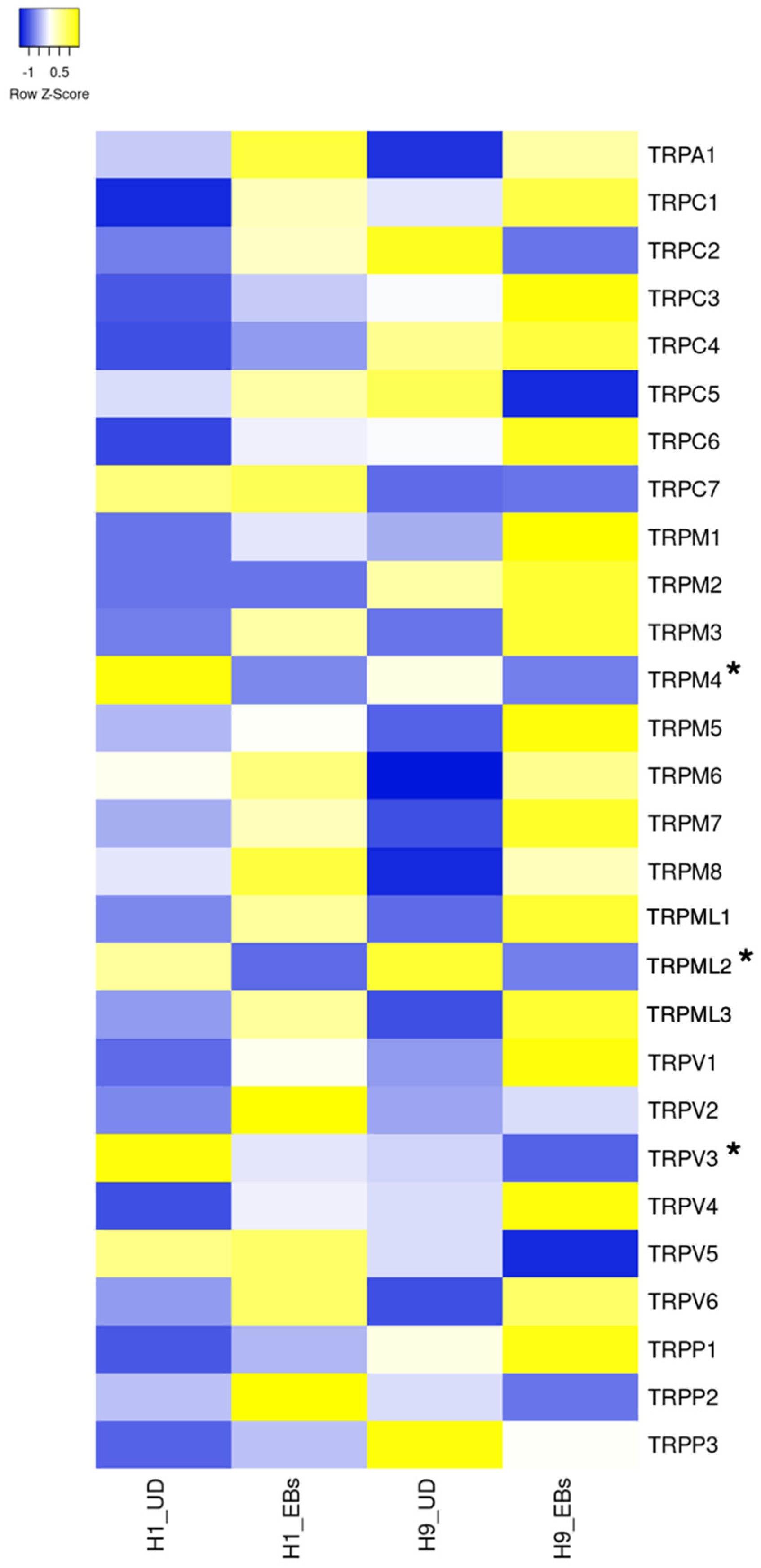
7. Transient Receptor Potential Channels
7.1. The Role of TRPC Channels in Pluripotent Stem Cells
7.2. The Role of TRPM Channels in Pluripotent Stem Cells
7.3. The Role of TRPV Channels in Pluripotent Stem Cells
8. Piezo Channels
The Role of Piezo Channels in Pluripotent Stem Cells
9. Cyclic Nucleotide-Gated and Hyperpolarization-Activated Channels
The Role of HCN Channels in Pluripotent Stem Cells
10. Acid-Sensing Ion Channels
11. Chloride Channels
12. Transmembrane Channel-like Channels
13. Two-Pore Channels
The Role of TPC Channels in Pluripotent Stem Cells
14. Summary of Channels Expressed in PSCs
15. Candidates of Channels Which Have Some Roles in PSCs
16. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Niwa, H.; Ogawa, K.; Shimosato, D.; Adachi, K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 2009, 460, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuka, S.; Nakai-Futatsugi, Y.; Niwa, H. LIF signal in mouse embryonic stem cells. JAK-STAT 2015, 4, e1086520. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vallier, L.; Mendjan, S.; Brown, S.; Chng, Z.; Teo, A.; Smithers, L.E.; Trotter, M.W.; Cho, C.H.; Martinez, A.; Rugg-Gunn, P.; et al. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development 2009, 136, 1339–1349. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Romorini, L.; Garate, X.; Neiman, G.; Luzzani, C.; Furmento, V.A.; Guberman, A.S.; Sevlever, G.E.; Scassa, M.E.; Miriuka, S.G. AKT/GSK3β signaling pathway is critically involved in human pluripotent stem cell survival. Sci. Rep. 2016, 6, 35660. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xu, C.; Rosler, E.; Jiang, J.; Lebkowski, J.S.; Gold, J.D.; O’Sullivan, C.; Delavan-Boorsma, K.; Mok, M.; Bronstein, A.; Carpenter, M.K. Basic fibroblast growth factor supports undifferentiated human embryonic stem cell growth without conditioned medium. Stem. Cells 2005, 23, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Levenstein, M.E.; Ludwig, T.E.; Xu, R.H.; Llanas, R.A.; VanDenHeuvel-Kramer, K.; Manning, D.; Thomson, J.A. Basic fibroblast growth factor support of human embryonic stem cell self-renewal. Stem Cells 2006, 24, 568–574. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Varzideh, F.; Gambardella, J.; Kansakar, U.; Jankauskas, S.S.; Santulli, G. Molecular Mechanisms Underlying Pluripotency and Self-Renewal of Embryonic Stem Cells. Int. J. Mol. Sci. 2023, 24, 8386. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Greber, B.; Lehrach, H.; Adjaye, J. Fibroblast growth factor 2 modulates transforming growth factor beta signaling in mouse embryonic fibroblasts and human ESCs (hESCs) to support hESC self-renewal. Stem Cells 2007, 25, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Ryazanova, L.V.; Rondon, L.J.; Zierler, S.; Hu, Z.; Galli, J.; Yamaguchi, T.P.; Mazur, A.; Fleig, A.; Ryazanov, A.G. TRPM7 is essential for Mg2+ homeostasis in mammals. Nat. Commun. 2010, 1, 109. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ng, S.Y.; Chin, C.H.; Lau, Y.T.; Luo, J.; Wong, C.K.; Bian, Z.X.; Tsang, S.Y. Role of voltage-gated potassium channels in the fate determination of embryonic stem cells. J. Cell. Physiol. 2010, 224, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Che, C.; Cheng, J.; Li, P.; Yang, Y. Ion channels in stem cells and their roles in stem cell biology and vascular diseases. J. Mol. Cell Cardiol. 2022, 166, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Khalid, O.; Namazi, A.; Tu, T.G.; Elie, O.; Lee, C.; Kim, Y. Discovery of consensus gene signature and intermodular connectivity defining self-renewal of human embryonic stem cells. Stem Cells 2014, 32, 1468–1479. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic. Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dormeyer, W.; van Hoof, D.; Braam, S.R.; Heck, A.J.; Mummery, C.L.; Krijgsveld, J. Plasma membrane proteomics of human embryonic stem cells and human embryonal carcinoma cells. J. Proteome. Res. 2008, 7, 2936–2951. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Rushing, S.N.; Kong, C.W.; Fu, J.; Lieu, D.K.; Chan, C.W.; Deng, W.; Li, R.A. Electrophysiological properties of human induced pluripotent stem cells. Am. J. Physiol. Cell Physiol. 2010, 298, C486–C495. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Meisler, M.H.; O’Brien, J.E.; Sharkey, L.M. Sodium channel gene family: Epilepsy mutations, gene interactions and modifier effects. J. Physiol. 2010, 588, 1841–1848. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, K.; Xue, T.; Tsang, S.Y.; Van Huizen, R.; Wong, C.W.; Lai, K.W.; Ye, Z.; Cheng, L.; Au, K.W.; Zhang, J.; et al. Electrophysiological properties of pluripotent human and mouse embryonic stem cells. Stem Cells 2005, 23, 1526–1534. [Google Scholar] [CrossRef] [PubMed]
- MacKinnon, R. Potassium channels. FEBS Lett. 2003, 555, 62–65. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Q.; Purhonen, P.; Hebert, H. Structure of potassium channels. Cell Mol. Life Sci. 2015, 72, 3677–3693. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sempou, E.; Kostiuk, V.; Zhu, J.; Cecilia Guerra, M.; Tyan, L.; Hwang, W.; Camacho-Aguilar, E.; Caplan, M.J.; Zenisek, D.; Warmflash, A.; et al. Membrane potential drives the exit from pluripotency and cell fate commitment via calcium and mTOR. Nat. Commun. 2022, 13, 6681, Erratum in Nat. Commun. 2023, 14, 3264. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Apáti, Á.; Berecz, T.; Sarkadi, B. Calcium signaling in human pluripotent stem cells. Cell Calcium. 2016, 59, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Hao, B.; Webb, S.E.; Miller, A.L.; Yue, J. The role of Ca2+ signaling on the self-renewal and neural differentiation of embryonic stem cells (ESCs). Cell Calcium. 2016, 59, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Forostyak, O.; Forostyak, S.; Kortus, S.; Sykova, E.; Verkhratsky, A.; Dayanithi, G. Physiology of Ca2+ signalling in stem cells of different origins and differentiation stages. Cell Calcium. 2016, 59, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Ermakov, A.; Daks, A.; Fedorova, O.; Shuvalov, O.; Barlev, N.A. Ca2+-depended signaling pathways regulate self-renewal and pluripotency of stem cells. Cell Biol. Int. 2018, 42, 1086–1096. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.J.; Wang, Y.J.; Zhang, M.; Zhang, P.; Liang, H.; Bai, H.J.; Yu, X.J.; Yang, H.T. Functional expression of the Ca2+ signaling machinery in human embryonic stem cells. Acta Pharmacol. Sin. 2017, 38, 1663–1672. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ermakov, A.; Pells, S.; Freile, P.; Ganeva, V.V.; Wildenhain, J.; Bradley, M.; Pawson, A.; Millar, R.; De Sousa, P.A. A role for intracellular calcium downstream of G-protein signaling in undifferentiated human embryonic stem cell culture. Stem Cell Res. 2012, 9, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.D.; Poole, K.; Martinac, B. Re-evaluating TRP channel mechanosensitivity. Trends Biochem. Sci. 2024, 49, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.B.; Webb, S.E.; Yue, J.; Moreau, M.; Leclerc, C.; Miller, A.L. TRPC3 is required for the survival, pluripotency and neural differentiation of mouse embryonic stem cells (mESCs). Sci. China Life Sci. 2018, 61, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Meier, A.B.; Lipp, P.; Laugwitz, K.L.; Dorn, T.; Moretti, A. Generation of heterozygous (MRli003-A-3) and homozygous (MRli003-A-4) TRPM4 knockout human iPSC lines. Stem Cell Res. 2022, 60, 102731. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Zhang, J.; Fu, W.; Li, S.; Tian, X.; Li, X.; Zhao, X.; Dong, J. Generation of a TRPM8 knockout hESC line (WAe009-A-A) derived from H9 using CRISPR/Cas9. Stem Cell Res. 2023, 67, 103040. [Google Scholar] [CrossRef] [PubMed]
- Runnels, L.W.; Yue, L.; Clapham, D.E. TRP-PLIK a bifunctional protein with kinase and ion channel activities. Science 2001, 291, 1043–1047. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, H.; Matsushita, M.; Nairn, A.C.; Kuriyan, J. Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity. Mol. Cell 2001, 7, 1047–1057. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Park, S.; Kwon, W.; Kim, D.; Park, J.K.; Han, J.E.; Cho, G.J.; Han, S.H.; Sung, Y.; Yi, J.K.; et al. Suppression of transient receptor potential melastatin 7 regulates pluripotency, proliferation, and differentiation of mouse embryonic stem cells via mechanistic target of rapamycin-extracellular signal-regulated kinase activation. J. Cell Biochem. 2022, 123, 547–567. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Tang, L.; Su, J.; Wang, H.; Liang, P.; Rong, K.; Gong, T. Generation of a TRPV1 knockout human pluripotent stem cell line (WAe009-A-U) using CRISPR/Cas9. Stem Cell Res. 2023, 72, 103202. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K.; Seta, H.; Haraguchi, Y.; Alsayegh, K.; Sekine, H.; Shimizu, T.; Hagiwara, N.; Yamazaki, K.; Okano, T. TRPV-1-mediated elimination of residual iPS cells in bioengineered cardiac cell sheet tissues. Sci. Rep. 2016, 6, 21747. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lo, I.C.; Chan, H.C.; Qi, Z.; Ng, K.L.; So, C.; Tsang, S.Y. TRPV3 Channel Negatively Regulates Cell Cycle Progression and Safeguards the Pluripotency of Embryonic Stem Cells. J. Cell Physiol. 2016, 231, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Murthy, S.E.; Dubin, A.E.; Patapoutian, A. Piezos thrive under pressure: Mechanically activated ion channels in health and disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Del Mármol, J.I.; Touhara, K.K.; Croft, G.; MacKinnon, R. Piezo1 forms a slowly-inactivating mechanosensory channel in mouse embryonic stem cells. Elife 2018, 7, e33149. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Biel, M.; Michalakis, S. Cyclic nucleotide-gated channels. Handb. Exp. Pharmacol. 2009, 191, 111–136. [Google Scholar] [CrossRef] [PubMed]
- Mazzolini, M.; Arcangeletti, M.; Marchesi, A.; Napolitano, L.M.R.; Grosa, D.; Maity, S.; Anselmi, C.; Torre, V. The gating mechanism in cyclic nucleotide-gated ion channels. Sci. Rep. 2018, 8, 45. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pan, Y.; Pohjolainen, E.; Schmidpeter, P.A.M.; Vaiana, A.C.; Nimigean, C.M.; Grubmüller, H.; Scheuring, S. Discrimination between cyclic nucleotides in a cyclic nucleotide-gated ion channel. Nat. Struct. Mol. Biol. 2023, 30, 512–520. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Benarroch, E.E. HCN channels: Function and clinical implications. Neurology 2013, 80, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Lau, Y.T.; Wong, C.K.; Luo, J.; Leung, L.H.; Tsang, P.F.; Bian, Z.X.; Tsang, S.Y. Effects of hyperpolarization-activated cyclic nucleotide-gated (HCN) channel blockers on the proliferation and cell cycle progression of embryonic stem cells. Pflug. Arch. 2011, 461, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Omelyanenko, A.; Sekyrova, P.; Andäng, M. ZD7288, a blocker of the HCN channel family, increases doubling time of mouse embryonic stem cells and modulates differentiation outcomes in a context-dependent manner. Springerplus 2016, 5, 41. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ruan, N.; Tribble, J.; Peterson, A.M.; Jiang, Q.; Wang, J.Q.; Chu, X.P. Acid-Sensing Ion Channels and Mechanosensation. Int. J. Mol. Sci. 2021, 22, 4810. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Verkman, A.S.; Galietta, L.J. Chloride channels as drug targets. Nat. Rev. Drug Discov. 2009, 8, 153–171. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zheng, W.; Holt, J.R. The Mechanosensory Transduction Machinery in Inner Ear Hair Cells. Annu. Rev. Biophys. 2021, 50, 31–51. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Patel, S.; Kilpatrick, B.S. Two-pore channels and disease. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 1678–1686. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gerndt, S.; Krogsaeter, E.; Patel, S.; Bracher, F.; Grimm, C. Discovery of lipophilic two-pore channel agonists. FEBS J. 2020, 287, 5284–5293. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.H.; Lu, Y.Y.; Yue, J. Two pore channel 2 differentially modulates neural differentiation of mouse embryonic stem cells. PLoS ONE 2013, 8, e66077. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gururaja, R.S.; Patel, N.J.; Singh, H. Intracellular Chloride Channels: Novel Biomarkers in Diseases. Front. Physiol. 2020, 11, 96. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ponsioen, B.; van Zeijl, L.; Langeslag, M.; Berryman, M.; Littler, D.; Jalink, K.; Moolenaar, W.H. Spatiotemporal regulation of chloride intracellular channel protein CLIC4 by RhoA. Mol. Biol. Cell 2009, 20, 4664–4672. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Suh, K.S.; Yuspa, S.H. Intracellular chloride channels: Critical mediators of cell viability and potential targets for cancer therapy. Curr. Pharm. Des. 2005, 11, 2753–2764. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, V.C.; Yang, H.H.; Craig-Lucas, A.; Dubois, W.; Carofino, B.L.; Lack, J.; Dwyer, J.E.; Simpson, R.M.; Cataisson, C.; Lee, M.P.; et al. Host CLIC4 expression in the tumor microenvironment is essential for breast cancer metastatic competence. PLoS Genet. 2022, 18, e1010271. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shukla, A.; Malik, M.; Cataisson, C.; Ho, Y.; Friesen, T.; Suh, K.S.; Yuspa, S.H. TGF-beta signalling is regulated by Schnurri-2-dependent nuclear translocation of CLIC4 and consequent stabilization of phospho-Smad2 and 3. Nat. Cell Biol. 2009, 11, 777–784. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mullen, A.C.; Wrana, J.L. TGF-β Family Signaling in Embryonic and Somatic Stem-Cell Renewal and Differentiation. Cold Spring Harb. Perspect. Biol. 2017, 9, a022186. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Beyer, T.A.; Weiss, A.; Khomchuk, Y.; Huang, K.; Ogunjimi, A.A.; Varelas, X.; Wrana, J.L. Switch enhancers interpret TGF-β and Hippo signaling to control cell fate in human embryonic stem cells. Cell Rep. 2013, 5, 1611–1624. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.; Teo, A.; Pauklin, S.; Hannan, N.; Cho, C.H.; Lim, B.; Vardy, L.; Dunn, N.R.; Trotter, M.; Pedersen, R.; et al. Activin/Nodal signaling controls divergent transcriptional networks in human embryonic stem cells and in endoderm progenitors. Stem Cells 2011, 29, 1176–1185. [Google Scholar] [CrossRef] [PubMed]
- Mullen, A.C.; Orlando, D.A.; Newman, J.J.; Lovén, J.; Kumar, R.M.; Bilodeau, S.; Reddy, J.; Guenther, M.G.; DeKoter, R.P.; Young, R.A. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell 2011, 147, 565–576. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, C.C.; Krogsaeter, E.; Kuo, C.Y.; Huang, M.C.; Chang, S.Y.; Biel, M. Endolysosomal cation channels point the way towards precision medicine of cancer and infectious diseases. Biomed. Pharmacother. 2022, 148, 112751. [Google Scholar] [CrossRef] [PubMed]
- Morgan, A.J.; Davis, L.C.; Galione, A. Choreographing endo-lysosomal Ca2+ throughout the life of a phagosome. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 119040. [Google Scholar] [CrossRef] [PubMed]
- Cuajungco, M.P.; Silva, J.; Habibi, A.; Valadez, J.A. The mucolipin-2 (TRPML2) ion channel: A tissue-specific protein crucial to normal cell function. Pflug. Arch. 2016, 468, 177–192. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yu, H.; Xie, M.; Meng, Z.; Lo, C.Y.; Chan, F.L.; Jiang, L.; Meng, X.; Yao, X. Endolysosomal ion channel MCOLN2 (Mucolipin-2) promotes prostate cancer progression via IL-1β/NF-κB pathway. Br. J. Cancer 2021, 125, 1420–1431. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Takase, O.; Yoshikawa, M.; Idei, M.; Hirahashi, J.; Fujita, T.; Takato, T.; Isagawa, T.; Nagae, G.; Suemori, H.; Aburatani, H.; et al. The role of NF-κB signaling in the maintenance of pluripotency of human induced pluripotent stem cells. PLoS ONE 2013, 8, e56399. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Armstrong, L.; Hughes, O.; Yung, S.; Hyslop, L.; Stewart, R.; Wappler, I.; Peters, H.; Walter, T.; Stojkovic, P.; Evans, J.; et al. The role of PI3K/AKT MAPK/ERK and NFkappabeta signalling in the maintenance of human embryonic stem cell pluripotency and viability highlighted by transcriptional profiling and functional analysis. Hum. Mol. Genet. 2006, 15, 1894–1913. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Zhao, Y.; Kusakizako, T.; Wang, Y.; Pan, C.; Zhang, Y.; Nureki, O.; Hattori, M.; Yan, Z. TMC1 and TMC2 Proteins Are Pore-Forming Subunits of Mechanosensitive Ion Channels. Neuron 2020, 105, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Tang, Y.; Luo, X.; Shi, X.; Song, F.; Ran, L. Pan-Cancer Analysis Reveals the Signature of TMC Family of Genes as a Promising Biomarker for Prognosis and Immunotherapeutic Response. Front. Immunol. 2021, 12, 715508. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]

| Ion Channels | Gene | Cell Type | Involved Signaling | Functional Role | Refs. |
|---|---|---|---|---|---|
| K+ channels | KCNs | Human ESCs and iPSCs | Cell cycle | Proliferation | [15,17] |
| Kv11.2 | KCNH6 | Human ESCs | mTOR pathway | Differentiation | [20] |
| TRPC3 | Trpc3 | Mouse ESCs | Ca2+ signaling | Pluripotency and survival | [28] |
| TRPM7 | Trpm7 | Mouse ESCs | mTOR and ERK pathway | Pluripotency and self-renewal | [33] |
| TRPM7 | Trpm7 | Mouse ESCs | Mg2+ homeostasis | Proliferation | [9] |
| Piezo1 | Piezo1 | Mouse ESCs | Mechanotransduction | Proliferation | [38] |
| HCN channels | HCNs | Mouse ESCs | Cell cycle | Proliferation | [43,44] |
| TPC2 | Tpc2 | Mouse ESCs | Ca2+ signaling | Pluripotency | [50] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaitsuka, T. The Unique Roles of Ion Channels in Pluripotent Stem Cells in Response to Biological Stimuli. Biology 2024, 13, 1043. https://doi.org/10.3390/biology13121043
Kaitsuka T. The Unique Roles of Ion Channels in Pluripotent Stem Cells in Response to Biological Stimuli. Biology. 2024; 13(12):1043. https://doi.org/10.3390/biology13121043
Chicago/Turabian StyleKaitsuka, Taku. 2024. "The Unique Roles of Ion Channels in Pluripotent Stem Cells in Response to Biological Stimuli" Biology 13, no. 12: 1043. https://doi.org/10.3390/biology13121043
APA StyleKaitsuka, T. (2024). The Unique Roles of Ion Channels in Pluripotent Stem Cells in Response to Biological Stimuli. Biology, 13(12), 1043. https://doi.org/10.3390/biology13121043






