The MCM2-7 Complex: Roles beyond DNA Unwinding
Abstract
Simple Summary
Abstract
1. Introduction
2. The MCM2-7 Complex and Cohesin Regulation
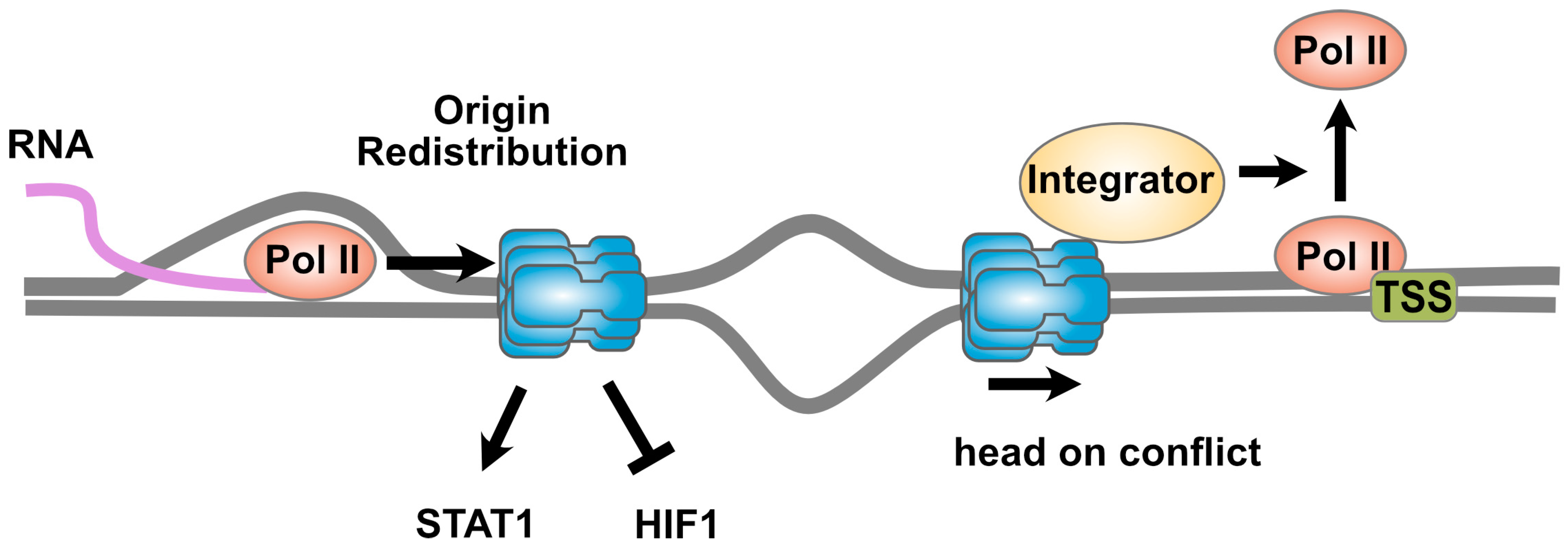
3. Sharing the Highway: The MCM2-7 Complex and Transcription
4. MCM2-7 Complexes and the Intra-S Checkpoint
5. The MCM2-7 Complex, Histone Inheritance, and Epigenetic Memory
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Remus, D.; Beuron, F.; Tolun, G.; Griffith, J.D.; Morris, E.P.; Diffley, J.F. Concerted Loading of Mcm2–7 Double Hexamers around DNA during DNA Replication Origin Licensing. Cell 2009, 139, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.J.; Indiani, C.; O’Donnell, M. Reconstitution of the Mcm2-7p Heterohexamer, Subunit Arrangement, and ATP Site Architecture. J. Biol. Chem. 2003, 278, 4491–4499. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhai, Y.; Zhang, Y.; Li, W.; Yang, M.; Lei, J.; Tye, B.-K.; Gao, N. Structure of the eukaryotic MCM complex at 3.8 Å. Nature 2015, 524, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Schwacha, A. The Mcm2-7 Complex Has In Vitro Helicase Activity. Mol. Cell 2008, 31, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Ishimi, Y. A DNA Helicase Activity Is Associated with an MCM4, -6, and -7 Protein Complex. J. Biol. Chem. 1997, 272, 24508–24513. [Google Scholar] [CrossRef] [PubMed]
- Evrin, C.; Clarke, P.; Zech, J.; Lurz, R.; Sun, J.; Uhle, S.; Li, H.; Stillman, B.; Speck, C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl. Acad. Sci. USA 2009, 106, 20240–20245. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.P.; Stillman, B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 1992, 357, 128–134. [Google Scholar] [CrossRef]
- Li, N.; Lam, W.H.; Zhai, Y.; Cheng, J.; Cheng, E.; Zhao, Y.; Gao, N.; Tye, B.-K. Structure of the origin recognition complex bound to DNA replication origin. Nature 2018, 559, 217–222. [Google Scholar] [CrossRef] [PubMed]
- McGarry, T.J.; Kirschner, M.W. Geminin, an Inhibitor of DNA Replication, Is Degraded during Mitosis. Cell 1998, 93, 1043–1053. [Google Scholar] [CrossRef]
- Méndez, J.; Stillman, B. Chromatin Association of Human Origin Recognition Complex, Cdc6, and Minichromosome Maintenance Proteins during the Cell Cycle: Assembly of Prereplication Complexes in Late Mitosis. Mol. Cell. Biol. 2000, 20, 8602–8612. [Google Scholar] [CrossRef]
- Donovan, S.; Harwood, J.; Drury, L.S.; Diffley, J.F.X. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. USA 1997, 94, 5611–5616. [Google Scholar] [CrossRef] [PubMed]
- Maiorano, D.; Moreau, J.; Méchali, M. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature 2000, 404, 622–625. [Google Scholar] [CrossRef] [PubMed]
- Nishitani, H.; Lygerou, Z.; Nishimoto, T.; Nurse, P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature 2000, 404, 625–628. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.; Tye, B.K. Initiating DNA synthesis: From recruiting to activating the MCM complex. J. Cell Sci. 2001, 114, 1447–1454. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Li, N.; Huo, Y.; Dang, S.; Tye, B.-K.; Gao, N.; Zhai, Y. Structural Insight into the MCM double hexamer activation by Dbf4-Cdc7 kinase. Nat. Commun. 2022, 13, 1396. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Gao, N.; Zhai, Y. DDK promotes DNA replication initiation: Mechanistic and structural insights. Curr. Opin. Struct. Biol. 2023, 78, 102504. [Google Scholar] [CrossRef]
- Sheu, Y.-J.; Stillman, B. Cdc7-Dbf4 Phosphorylates MCM Proteins via a Docking Site-Mediated Mechanism to Promote S Phase Progression. Mol. Cell 2006, 24, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Douglas, M.E.; Ali, F.A.; Costa, A.; Diffley, J.F.X. The mechanism of eukaryotic CMG helicase activation. Nature 2018, 555, 265–268. [Google Scholar] [CrossRef]
- Gambus, A.; Jones, R.C.; Sanchez-Diaz, A.; Kanemaki, M.; van Deursen, F.; Edmondson, R.D.; Labib, K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006, 8, 358–366. [Google Scholar] [CrossRef]
- Yuan, Z.; Georgescu, R.; Bai, L.; Zhang, D.; Li, H.; O’donnell, M.E. DNA unwinding mechanism of a eukaryotic replicative CMG helicase. Nat. Commun. 2020, 11, 688. [Google Scholar] [CrossRef]
- Bailey, R.; Moreno, S.P.; Gambus, A. Termination of DNA replication forks: “Breaking up is hard to do”. Nucleus 2015, 6, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Dewar, J.M.; Low, E.; Mann, M.; Räschle, M.; Walter, J.C. CRL2Lrr1 promotes unloading of the vertebrate replisome from chromatin during replication termination. Genes Dev. 2017, 31, 275–290. [Google Scholar] [CrossRef] [PubMed]
- Maric, M.; Maculins, T.; De Piccoli, G.; Labib, K. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science 2014, 346, 440. [Google Scholar] [CrossRef] [PubMed]
- Moreno, S.P.; Bailey, R.; Campion, N.; Herron, S.; Gambus, A. Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science 2014, 346, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan, M.; Nguyen, T.; Gallo, D.; Luthra, N.; Brown, G.W.; Saridakis, V.; Frappier, L. A Role for USP7 in DNA Replication. Mol. Cell. Biol. 2014, 34, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, A.; Frappier, L.; Méchali, M. MCM-BP regulates unloading of the MCM2–7 helicase in late S phase. Genes Dev. 2011, 25, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Kusunoki, S.; Ishimi, Y. Interaction of human minichromosome maintenance protein-binding protein with minichromosome maintenance 2–7. FEBS J. 2014, 281, 1057–1067. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.Q.; Jackson, D.A.; Blow, J.J. Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev. 2007, 21, 3331–3341. [Google Scholar] [CrossRef] [PubMed]
- Ibarra, A.; Schwob, E.; Méndez, J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc. Natl. Acad. Sci. USA 2008, 105, 8956–8961. [Google Scholar] [CrossRef]
- Courtot, L.; Hoffmann, J.-S.; Bergoglio, V. The Protective Role of Dormant Origins in Response to Replicative Stress. Int. J. Mol. Sci. 2018, 19, 3569. [Google Scholar] [CrossRef]
- Agarwal, A.; Korsak, S.; Choudhury, A.; Plewczynski, D. The dynamic role of cohesin in maintaining human genome architecture. BioEssays 2023, 45, e2200240. [Google Scholar] [CrossRef] [PubMed]
- Sumara, I.; Vorlaufer, E.; Stukenberg, P.; Kelm, O.; Redemann, N.; Nigg, E.A.; Peters, J.-M. The Dissociation of Cohesin from Chromosomes in Prophase Is Regulated by Polo-like Kinase. Mol. Cell 2002, 9, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Sumara, I.; Vorlaufer, E.; Gieffers, C.; Peters, B.H.; Peters, J.-M. Characterization of Vertebrate Cohesin Complexes and Their Regulation in Prophase. J. Cell Biol. 2000, 151, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.S.; Yiu, P.; Chou, M.F.; Gygi, S.; Walter, J.C. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat. Cell Biol. 2004, 6, 991–996. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, P.J.; Hirano, T. Scc2 Couples Replication Licensing to Sister Chromatid Cohesion in Xenopus Egg Extracts. Curr. Biol. 2004, 14, 1598–1603. [Google Scholar] [CrossRef]
- Ciosk, R.; Shirayama, M.; Shevchenko, A.; Tanaka, T.; Toth, A.; Shevchenko, A.; Nasmyth, K. Cohesin’s Binding to Chromosomes Depends on a Separate Complex Consisting of Scc2 and Scc4 Proteins. Mol. Cell 2000, 5, 243–254. [Google Scholar] [CrossRef]
- Watrin, E.; Schleiffer, A.; Tanaka, K.; Eisenhaber, F.; Nasmyth, K.; Peters, J.-M. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr. Biol. 2006, 16, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.S.; Basu, A.; Bermudez, V.; Hurwitz, J.; Walter, J.C. Cdc7–Drf1 kinase links chromosome cohesion to the initiation of DNA replication in Xenopus egg extracts. Genes Dev. 2008, 22, 1894–1905. [Google Scholar] [CrossRef]
- Takahashi, T.S.; Walter, J.C. Cdc7–Drf1 is a developmentally regulated protein kinase required for the initiation of vertebrate DNA replication. Gene Dev. 2005, 19, 2295–2300. [Google Scholar] [CrossRef]
- Zheng, G.; Kanchwala, M.; Xing, C.; Yu, H.; University of Texas Southwestern Medical Center; States, U. MCM2–7-dependent cohesin loading during S phase promotes sister-chromatid cohesion. eLife 2018, 7, e33920. [Google Scholar] [CrossRef]
- Guillou, E.; Ibarra, A.; Coulon, V.; Casado-Vela, J.; Rico, D.; Casal, I.; Schwob, E.; Losada, A.; Méndez, J. Cohesin organizes chromatin loops at DNA replication factories. Genes Dev. 2010, 24, 2812–2822. [Google Scholar] [CrossRef] [PubMed]
- Gerlich, D.; Koch, B.; Dupeux, F.; Peters, J.-M.; Ellenberg, J. Live-Cell Imaging Reveals a Stable Cohesin-Chromatin Interaction after but Not before DNA Replication. Curr. Biol. 2006, 16, 1571–1578. [Google Scholar] [CrossRef] [PubMed]
- Skibbens, R.V.; Corson, L.B.; Koshland, D.; Hieter, P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999, 13, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Tóth, A.; Ciosk, R.; Uhlmann, F.; Galova, M.; Schleiffer, A.; Nasmyth, K. Yeast Cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999, 13, 320–333. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shi, X.; Li, Y.; Kim, B.-J.; Jia, J.; Huang, Z.; Yang, T.; Fu, X.; Jung, S.Y.; Wang, Y.; et al. Acetylation of Smc3 by Eco1 Is Required for S Phase Sister Chromatid Cohesion in Both Human and Yeast. Mol. Cell 2008, 31, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Ünal, E.; Heidinger-Pauli, J.M.; Kim, W.; Guacci, V.; Onn, I.; Gygi, S.P.; Koshland, D.E. A Molecular Determinant for the Establishment of Sister Chromatid Cohesion. Science 2008, 321, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shahar, T.R.; Heeger, S.; Lehane, C.; East, P.; Flynn, H.; Skehel, M.; Uhlmann, F. Eco1-Dependent Cohesin Acetylation During Establishment of Sister Chromatid Cohesion. Science 2008, 321, 563–566. [Google Scholar] [CrossRef]
- Uhlmann, F.; Nasmyth, K. Cohesion between sister chromatids must be established during DNA replication. Curr. Biol. 1998, 8, 1095–1102. [Google Scholar] [CrossRef]
- Moldovan, G.-L.; Pfander, B.; Jentsch, S. PCNA Controls Establishment of Sister Chromatid Cohesion during S Phase. Mol. Cell 2006, 23, 723–732. [Google Scholar] [CrossRef]
- Hou, F.; Zou, H. Two Human Orthologues of Eco1/Ctf7 Acetyltransferases Are Both Required for Proper Sister-Chromatid Cohesion. Mol. Biol. Cell 2005, 16, 3908–3918. [Google Scholar] [CrossRef]
- Bellows, A.M.; Kenna, M.A.; Cassimeris, L.; Skibbens, R.V. Human EFO1p exhibits acetyltransferase activity and is a unique combination of linker histone and Ctf7p/Eco1p chromatid cohesion establishment domains. Nucleic Acids Res. 2003, 31, 6334–6343. [Google Scholar] [CrossRef] [PubMed]
- Alomer, R.M.; da Silva, E.M.L.; Chen, J.; Piekarz, K.M.; McDonald, K.; Sansam, C.G.; Sansam, C.L.; Rankin, S. Esco1 and Esco2 regulate distinct cohesin functions during cell cycle progression. Proc. Natl. Acad. Sci. USA 2017, 114, 9906–9911. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Jones, M.J.K.; Jallepalli, P.V. Cohesin recruits the Esco1 acetyltransferase genome wide to repress transcription and promote cohesion in somatic cells. Proc. Natl. Acad. Sci. USA 2015, 112, 11270–11275. [Google Scholar] [CrossRef] [PubMed]
- Lafont, A.L.; Song, J.; Rankin, S. Sororin cooperates with the acetyltransferase Eco2 to ensure DNA replication-dependent sister chromatid cohesion. Proc. Natl. Acad. Sci. USA 2010, 107, 20364–20369. [Google Scholar] [CrossRef]
- Higashi, T.L.; Ikeda, M.; Tanaka, H.; Nakagawa, T.; Bando, M.; Shirahige, K.; Kubota, Y.; Takisawa, H.; Masukata, H.; Takahashi, T.S. The Prereplication Complex Recruits XEco2 to Chromatin to Promote Cohesin Acetylation in Xenopus Egg Extracts. Curr. Biol. 2012, 22, 977–988. [Google Scholar] [CrossRef]
- Ivanov, M.P.; Ladurner, R.; Poser, I.; Beveridge, R.; Rampler, E.; Hudecz, O.; Novatchkova, M.; Hériché, J.; Wutz, G.; van der Lelij, P.; et al. The replicative helicase MCM recruits cohesin acetyltransferase ESCO2 to mediate centromeric sister chromatid cohesion. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Minamino, M.; Tei, S.; Negishi, L.; Kanemaki, M.T.; Yoshimura, A.; Sutani, T.; Bando, M.; Shirahige, K. Temporal Regulation of ESCO2 Degradation by the MCM Complex, the CUL4-DDB1-VPRBP Complex, and the Anaphase-Promoting Complex. Curr. Biol. 2018, 28, 2665–2672.e5. [Google Scholar] [CrossRef] [PubMed]
- Bender, D.; Da Silva, E.M.L.; Chen, J.; Poss, A.; Gawey, L.; Rulon, Z.; Rankin, S. Multivalent interaction of ESCO2 with the replication machinery is required for sister chromatid cohesion in vertebrates. Proc. Natl. Acad. Sci. USA 2020, 117, 1081–1089. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Lafont, A.; Chen, J.; Wu, F.M.; Shirahige, K.; Rankin, S. Cohesin acetylation promotes sister chromatid cohesion only in association with the replication machin-ery. J. Biol. Chem. 2012, 287, 34325–34336. [Google Scholar] [CrossRef]
- Yoshimura, A.; Sutani, T.; Shirahige, K. Functional control of Eco1 through the MCM complex in sister chromatid cohesion. Gene 2021, 784, 145584. [Google Scholar] [CrossRef]
- Rao, S.S.P.; Huang, S.-C.; St Hilaire, B.G.; Engreitz, J.M.; Perez, E.M.; Kieffer-Kwon, K.-R.; Sanborn, A.L.; Johnstone, S.E.; Bascom, G.D.; Bochkov, I.D.; et al. Cohesin Loss Eliminates All Loop Domains. Cell 2017, 171, 305–320.e24. [Google Scholar] [CrossRef] [PubMed]
- Davidson, I.F.; Bauer, B.; Goetz, D.; Tang, W.; Wutz, G.; Peters, J.-M. DNA loop extrusion by human cohesin. Science 2019, 366, 1338–1345. [Google Scholar] [CrossRef]
- Dequeker, B.J.H.; Scherr, M.J.; Brandão, H.B.; Gassler, J.; Powell, S.; Gaspar, I.; Flyamer, I.M.; Lalic, A.; Tang, W.; Stocsits, R.; et al. MCM complexes are barriers that restrict cohesin-mediated loop extrusion. Nature 2022, 606, 197–203. [Google Scholar] [CrossRef]
- Li, Y.; Haarhuis, J.H.I.; Cacciatore, Á.S.; Oldenkamp, R.; Van Ruiten, M.S.; Willems, L.; Teunissen, H.; Muir, K.W.; De Wit, E.; Rowland, B.D.; et al. The structural basis for cohesin–CTCF-anchored loops. Nature 2020, 578, 472–476. [Google Scholar] [CrossRef]
- García-Muse, T.; Aguilera, A. Transcription–replication conflicts: How they occur and how they are resolved. Nat. Rev. Mol. Cell Biol. 2016, 17, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Hamperl, S.; Cimprich, K.A. Conflict Resolution in the Genome: How Transcription and Replication Make It Work. Cell 2016, 167, 1455–1467. [Google Scholar] [CrossRef] [PubMed]
- Yankulov, K.; Todorov, I.; Romanowski, P.; Licatalosi, D.; Cilli, K.; McCracken, S.; Laskey, R.; Bentley, D.L. MCM Proteins Are Associated with RNA Polymerase II Holoenzyme. Mol. Cell. Biol. 1999, 19, 6154–6163. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Snyder, M.; Huang, X.-Y.; Zhang, J.J. The minichromosome maintenance proteins 2-7 (MCM2-7) are necessary for RNA polymerase II (Pol II)-mediated transcription. J. Biol. Chem. 2009, 284, 13466–13472. [Google Scholar] [CrossRef]
- Gros, J.; Kumar, C.; Lynch, G.; Yadav, T.; Whitehouse, I.; Remus, D. Post-licensing Specification of Eukaryotic Replication Origins by Facilitated Mcm2-7 Sliding along DNA. Mol. Cell 2015, 60, 797–807. [Google Scholar] [CrossRef]
- Liu, Y.; Ai, C.; Gan, T.; Wu, J.; Jiang, Y.; Liu, X.; Lu, R.; Gao, N.; Li, Q.; Ji, X.; et al. Transcription shapes DNA replication initiation to preserve genome integrity. Genome Biol. 2021, 22, 1–27. [Google Scholar] [CrossRef]
- Scherr, M.J.; Wahab, S.A.; Remus, D.; Duderstadt, K.E. Mobile origin-licensing factors confer resistance to conflicts with RNA polymerase. Cell Rep. 2022, 38, 110531. [Google Scholar] [CrossRef] [PubMed]
- Petermann, E.; Lan, L.; Zou, L. Sources, resolution and physiological relevance of R-loops and RNA–DNA hybrids. Nat. Rev. Mol. Cell Biol. 2022, 23, 521–540. [Google Scholar] [CrossRef] [PubMed]
- Gan, W.; Guan, Z.; Liu, J.; Gui, T.; Shen, K.; Manley, J.L.; Li, X. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011, 25, 2041–2056. [Google Scholar] [CrossRef] [PubMed]
- Hamperl, S.; Bocek, M.J.; Saldivar, J.C.; Swigut, T.; Cimprich, K.A. Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell 2017, 170, 774–786.e19. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; Gómez-González, B. DNA–RNA hybrids: The risks of DNA breakage during transcription. Nat. Struct. Mol. Biol. 2017, 24, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.-H.; Kelman, Z. The Replicative Helicases of Bacteria, Archaea, and Eukarya Can Unwind RNA-DNA Hybrid Substrates. J. Biol. Chem. 2006, 281, 26914–26921. [Google Scholar] [CrossRef]
- Baillat, D.; Hakimi, M.-A.; Näär, A.M.; Shilatifard, A.; Cooch, N.; Shiekhattar, R. Integrator, a Multiprotein Mediator of Small Nuclear RNA Processing, Associates with the C-Terminal Repeat of RNA Polymerase II. Cell 2005, 123, 265–276. [Google Scholar] [CrossRef]
- Stein, C.B.; Field, A.R.; Mimoso, C.A.; Zhao, C.; Huang, K.-L.; Wagner, E.J.; Adelman, K. Integrator endonuclease drives promoter-proximal termination at all RNA polymerase II-transcribed loci. Mol. Cell 2022, 82, 4232–4245.e11. [Google Scholar] [CrossRef]
- Bhowmick, R.; Mehta, K.P.; Lerdrup, M.; Cortez, D. Integrator facilitates RNAPII removal to prevent transcription-replication collisions and genome instability. Mol. Cell 2023, 83, 2357–2366.e8. [Google Scholar] [CrossRef]
- Zhang, J.J.; Zhao, Y.; Chait, B.T.; Lathem, W.W.; Ritzi, M.; Knippers, R.; Darnell, J.E. Ser727-dependent recruitment of MCM5 by Stat1α in IFN-γ-induced transcriptional activation. EMBO J. 1998, 17, 6963–6971. [Google Scholar] [CrossRef]
- DaFonseca, C.J.; Shu, F.; Zhang, J.J. Identification of two residues in MCM5 critical for the assembly of MCM complexes and Stat1-mediated transcription activation in response to IFN-γ. Proc. Natl. Acad. Sci. USA 2001, 98, 3034–3039. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Zhong, Z.; Darnell, J.E. Maximal activation of transcription by statl and stat3 requires both tyrosine and serine phosphorylation. Cell 1995, 82, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Snyder, M.; He, W.; Zhang, J.J. The DNA replication factor MCM5 is essential for Stat1-mediated transcriptional activation. Proc. Natl. Acad. Sci. USA 2005, 102, 14539–14544. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.L.; Jiang, B.-H.; Rue, E.A.; Semenza, G.L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 1995, 92, 5510–5514. [Google Scholar] [CrossRef] [PubMed]
- Iyer, N.V.; Kotch, L.E.; Agani, F.; Leung, S.W.; Laughner, E.; Wenger, R.H.; Gassmann, M.; Gearhart, J.D.; Lawler, A.M.; Yu, A.Y.; et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 1998, 12, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Hubbi, M.E.; Luo, W.; Baek, J.H.; Semenza, G.L. MCM Proteins Are Negative Regulators of Hypoxia-Inducible Factor 1. Mol. Cell 2011, 42, 700–712. [Google Scholar] [CrossRef] [PubMed]
- Hubbi, M.E.; Kshitiz, K.; Gilkes, D.M.; Rey, S.; Wong, C.C.; Luo, W.; Kim, D.-H.; Dang, C.V.; Levchenko, A.; Semenza, G.L. A Nontranscriptional Role for HIF-1α as a Direct Inhibitor of DNA Replication. Sci. Signal. 2013, 6, ra10. [Google Scholar] [CrossRef]
- Katou, Y.; Kanoh, Y.; Bando, M.; Noguchi, H.; Tanaka, H.; Ashikari, T.; Sugimoto, K.; Shirahige, K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 2003, 424, 1078–1083. [Google Scholar] [CrossRef] [PubMed]
- Waterman, D.P.; Haber, J.E.; Smolka, M.B. Checkpoint Responses to DNA Double-Strand Breaks. Annu. Rev. Biochem. 2020, 89, 103–133. [Google Scholar] [CrossRef]
- Galanti, L.; Pfander, B. Right time, right place— DNA damage and DNA replication checkpoints collectively safeguard S phase. EMBO J. 2018, 37, e100681. [Google Scholar] [CrossRef]
- Branzei, D.; Foiani, M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair 2007, 6, 994–1003. [Google Scholar] [CrossRef] [PubMed]
- Numata, Y.; Ishihara, S.; Hasegawa, N.; Nozaki, N.; Ishimi, Y. Interaction of human MCM2-7 proteins with TIM, TIPIN and Rb. J. Biochem. 2010, 147, 917–927. [Google Scholar] [CrossRef]
- Chao, W.C.; Murayama, Y.; Muñoz, S.; Costa, A.; Uhlmann, F.; Singleton, M.R. Structural Studies Reveal the Functional Modularity of the Scc2-Scc4 Cohesin Loader. Cell Rep. 2015, 12, 719–725. [Google Scholar] [CrossRef]
- Bando, M.; Katou, Y.; Komata, M.; Tanaka, H.; Itoh, T.; Sutani, T.; Shirahige, K. Csm3, Tof1, and Mrc1 Form a Heterotrimeric Mediator Complex That Associates with DNA Replication Forks. J. Biol. Chem. 2009, 284, 34355–34365. [Google Scholar] [CrossRef] [PubMed]
- Bastia, D.; Srivastava, P.; Zaman, S.; Choudhury, M.; Mohanty, B.K.; Bacal, J.; Langston, L.D.; Pasero, P.; O’donnell, M.E. Phosphorylation of CMG helicase and Tof1 is required for programmed fork arrest. Proc. Natl. Acad. Sci. USA 2016, 113, E3639–E3648. [Google Scholar] [CrossRef]
- Baretić, D.; Jenkyn-Bedford, M.; Aria, V.; Cannone, G.; Skehel, M.; Yeeles, J.T. Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork. Mol. Cell 2020, 78, 926–940.e13. [Google Scholar] [CrossRef] [PubMed]
- Gambus, A.; van Deursen, F.; Polychronopoulos, D.; Foltman, M.; Jones, R.C.; Edmondson, R.D.; Calzada, A.; Labib, K. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase α within the eukaryotic replisome. EMBO J. 2009, 28, 2992–3004. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Bekker-Jensen, S.; Mailand, N.; Lukas, C.; Bartek, J.; Lukas, J. Claspin Operates Downstream of TopBP1 To Direct ATR Signaling towards Chk1 Activation. Mol. Cell. Biol. 2006, 26, 6056–6064. [Google Scholar] [CrossRef] [PubMed]
- Tourrière, H.; Versini, G.; Cordón-Preciado, V.; Alabert, C.; Pasero, P. Mrc1 and Tof1 Promote Replication Fork Progression and Recovery Independently of Rad53. Mol. Cell 2005, 19, 699–706. [Google Scholar] [CrossRef]
- Ünsal-Kaçmaz, K.; Chastain, P.D.; Qu, P.-P.; Minoo, P.; Cordeiro-Stone, M.; Sancar, A.; Kaufmann, W.K. The Human Tim/Tipin Complex Coordinates an Intra-S Checkpoint Response to UV That Slows Replication Fork Displacement. Mol. Cell. Biol. 2007, 27, 3131–3142. [Google Scholar] [CrossRef]
- Leman, A.R.; Noguchi, C.; Lee, C.Y.; Noguchi, E. Human Timeless and Tipin stabilize replication forks and facilitate sister-chromatid cohesion. J. Cell Sci. 2010, 123, 660–670. [Google Scholar] [CrossRef] [PubMed]
- Calzada, A.; Hodgson, B.; Kanemaki, M.; Bueno, A.; Labib, K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 2005, 19, 1905–1919. [Google Scholar] [CrossRef] [PubMed]
- Byun, T.S.; Pacek, M.; Yee, M.-C.; Walter, J.C.; Cimprich, K.A. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005, 19, 1040–1052. [Google Scholar] [CrossRef] [PubMed]
- Nedelcheva, M.N.; Roguev, A.; Dolapchiev, L.B.; Shevchenko, A.; Taskov, H.B.; Shevchenko, A.; Stewart, A.F.; Stoynov, S.S. Uncoupling of Unwinding from DNA Synthesis Implies Regulation of MCM Helicase by Tof1/Mrc1/Csm3 Checkpoint Complex. J. Mol. Biol. 2005, 347, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Elledge, S.J. Sensing DNA Damage Through ATRIP Recognition of RPA-ssDNA Complexes. Science 2003, 300, 1542–1548. [Google Scholar] [CrossRef] [PubMed]
- MacDougall, C.A.; Byun, T.S.; Van, C.; Yee, M.-C.; Cimprich, K.A. The structural determinants of checkpoint activation. Genes Dev. 2007, 21, 898–903. [Google Scholar] [CrossRef] [PubMed]
- Michael, W.M.; Ott, R.; Fanning, E.; Newport, J. Activation of the DNA Replication Checkpoint Through RNA Synthesis by Primase. Science 2000, 289, 2133–2137. [Google Scholar] [CrossRef] [PubMed]
- Van, C.; Yan, S.; Michael, W.M.; Waga, S.; Cimprich, K.A. Continued primer synthesis at stalled replication forks contributes to checkpoint activation. J. Cell Biol. 2010, 189, 233–246. [Google Scholar] [CrossRef]
- Simon, A.C.; Zhou, J.C.; Perera, R.L.; van Deursen, F.; Evrin, C.; Ivanova, M.E.; Kilkenny, M.L.; Renault, L.; Kjaer, S.; Matak-Vinković, D.; et al. A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. Nature 2014, 510, 293–297. [Google Scholar] [CrossRef]
- Castaneda, J.C.; Schrecker, M.; Remus, D.; Hite, R.K. Mechanisms of loading and release of the 9-1-1 checkpoint clamp. Nat. Struct. Mol. Biol. 2022, 29, 369–375. [Google Scholar] [CrossRef]
- Mordes, D.A.; Glick, G.G.; Zhao, R.; Cortez, D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008, 22, 1478–1489. [Google Scholar] [CrossRef] [PubMed]
- Kemp, M.G.; Akan, Z.; Yilmaz, S.; Grillo, M.; Smith-Roe, S.L.; Kang, T.-H.; Cordeiro-Stone, M.; Kaufmann, W.K.; Abraham, R.T.; Sancar, A.; et al. Tipin-Replication Protein A Interaction Mediates Chk1 Phosphorylation by ATR in Response to Genotoxic Stress. J. Biol. Chem. 2010, 285, 16562–16571. [Google Scholar] [CrossRef]
- Komata, M.; Bando, M.; Araki, H.; Shirahige, K. The Direct Binding of Mrc1, a Checkpoint Mediator, to Mcm6, a Replication Helicase, Is Essential for the Replication Checkpoint against Methyl Methanesulfonate-Induced Stress. Mol. Cell. Biol. 2009, 29, 5008–5019. [Google Scholar] [CrossRef] [PubMed]
- Cortez, D.; Glick, G.; Elledge, S.J. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. USA 2004, 101, 10078–10083. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.Y.; Shevchenko, A.; Shevchenko, A.; Dunphy, W.G. Mcm2 Is a Direct Substrate of ATM and ATR during DNA Damage and DNA Replication Checkpoint Responses. J. Biol. Chem. 2004, 279, 53353–53364. [Google Scholar] [CrossRef] [PubMed]
- Trenz, K.; Errico, A.; Costanzo, V. Plx1 is required for chromosomal DNA replication under stressful conditions. EMBO J. 2008, 27, 876–885. [Google Scholar] [CrossRef]
- Tsvetkov, L.; Stern, D.F. Interaction of Chromatin-associated Plk1 and Mcm7. J. Biol. Chem. 2005, 280, 11943–11947. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Aslanian, A.; Fu, K.; Tsuji, T.; Zhang, Y. The Interaction between Checkpoint Kinase 1 (Chk1) and the Minichromosome Maintenance (MCM) Complex Is Required for DNA Damage-induced Chk1 Phosphorylation. J. Biol. Chem. 2014, 289, 24716–24723. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Pozo, F.M.; Wisotsky, J.N.; Wang, B.; Jacobberger, J.W.; Zhang, Y. Phosphorylation of Minichromosome Maintenance 3 (MCM3) by Checkpoint Kinase 1 (Chk1) Negatively Regulates DNA Replication and Checkpoint Activation. J. Biol. Chem. 2015, 290, 12370–12378. [Google Scholar] [CrossRef]
- Escobar, T.M.; Loyola, A.; Reinberg, D. Parental nucleosome segregation and the inheritance of cellular identity. Nat. Rev. Genet. 2021, 22, 379–392. [Google Scholar] [CrossRef]
- Petryk, N.; Dalby, M.; Wenger, A.; Stromme, C.B.; Strandsby, A.; Andersson, R.; Groth, A. MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 2018, 361, 1389–1392. [Google Scholar] [CrossRef] [PubMed]
- Wenger, A.; Biran, A.; Alcaraz, N.; Redó-Riveiro, A.; Sell, A.C.; Krautz, R.; Flury, V.; Reverón-Gómez, N.; Solis-Mezarino, V.; Völker-Albert, M.; et al. Symmetric inheritance of parental histones governs epigenome maintenance and embryonic stem cell identity. Nat. Genet. 2023, 55, 1567–1578. [Google Scholar] [CrossRef] [PubMed]
- Gan, H.; Serra-Cardona, A.; Hua, X.; Zhou, H.; Labib, K.; Yu, C.; Zhang, Z. The Mcm2-Ctf4-Polα Axis Facilitates Parental Histone H3-H4 Transfer to Lagging Strands. Mol. Cell 2018, 72, 140–151.e3. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hua, X.; Serra-Cardona, A.; Xu, X.; Gan, S.; Zhou, H.; Yang, W.-S.; Chen, C.-L.; Xu, R.-M.; Zhang, Z. DNA polymerase α interacts with H3-H4 and facilitates the transfer of parental histones to lagging strands. Sci. Adv. 2020, 6, eabb5820. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-H.; Farina, A.; Bermudez, V.P.; Tappin, I.; Du, F.; Galal, W.C.; Hurwitz, J. Interaction between human Ctf4 and the Cdc45/Mcm2-7/GINS (CMG) replicative helicase. Proc. Natl. Acad. Sci. USA 2013, 110, 19760–19765. [Google Scholar] [CrossRef] [PubMed]
- Foltman, M.; Evrin, C.; De Piccoli, G.; Jones, R.C.; Edmondson, R.D.; Katou, Y.; Nakato, R.; Shirahige, K.; Labib, K. Eukaryotic Replisome Components Cooperate to Process Histones During Chromosome Replication. Cell Rep. 2013, 3, 892–904. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tang, Y.; Xu, J.; Leng, H.; Shi, G.; Hu, Z.; Wu, J.; Xiu, Y.; Feng, J.; Li, Q. The N-terminus of Spt16 anchors FACT to MCM2–7 for parental histone recycling. Nucleic Acids Res. 2023, 51, 11549–11567. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, X.; Feng, J.; Leng, H.; Li, S.; Xiao, J.; Liu, S.; Xu, Z.; Xu, J.; Li, D.; et al. The Histone Chaperone FACT Contributes to DNA Replication-Coupled Nucleosome Assembly. Cell Rep. 2016, 14, 1128–1141. [Google Scholar] [CrossRef] [PubMed]
- Safaric, B.; Chacin, E.; Scherr, M.J.; Rajappa, L.; Gebhardt, C.; Kurat, C.F.; Cordes, T.E.; Duderstadt, K. The fork protection complex recruits FACT to reorganize nucleosomes during replication. Nucleic Acids Res. 2022, 50, 1317–1334. [Google Scholar] [CrossRef]
- Tan, B.C.; Liu, H.; Lin, C.-L.; Lee, S.-C. Functional cooperation between FACT and MCM is coordinated with cell cycle and differential complex formation. J. Biomed. Sci. 2010, 17, 11. [Google Scholar] [CrossRef]
- Tan, B.C.-M.; Chien, C.-T.; Hirose, S.; Lee, S.-C. Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J. 2006, 25, 3975–3985. [Google Scholar] [CrossRef] [PubMed]
- Nathanailidou, P.; Dhakshnamoorthy, J.; Xiao, H.; Zofall, M.; Holla, S.; O’neill, M.; Andresson, T.; Wheeler, D.; Grewal, S.I.S. Specialized replication of heterochromatin domains ensures self-templated chromatin assembly and epigenetic inheritance. Proc. Natl. Acad. Sci. USA 2024, 121, e2315596121. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.L.; Winston, F. The conserved histone chaperone Spt6 is strongly required for DNA replication and genome stability. Cell Rep. 2023, 42, 112264. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rankin, B.D.; Rankin, S. The MCM2-7 Complex: Roles beyond DNA Unwinding. Biology 2024, 13, 258. https://doi.org/10.3390/biology13040258
Rankin BD, Rankin S. The MCM2-7 Complex: Roles beyond DNA Unwinding. Biology. 2024; 13(4):258. https://doi.org/10.3390/biology13040258
Chicago/Turabian StyleRankin, Brooke D., and Susannah Rankin. 2024. "The MCM2-7 Complex: Roles beyond DNA Unwinding" Biology 13, no. 4: 258. https://doi.org/10.3390/biology13040258
APA StyleRankin, B. D., & Rankin, S. (2024). The MCM2-7 Complex: Roles beyond DNA Unwinding. Biology, 13(4), 258. https://doi.org/10.3390/biology13040258




