Activation of Cryptochrome 4 from Atlantic Herring
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
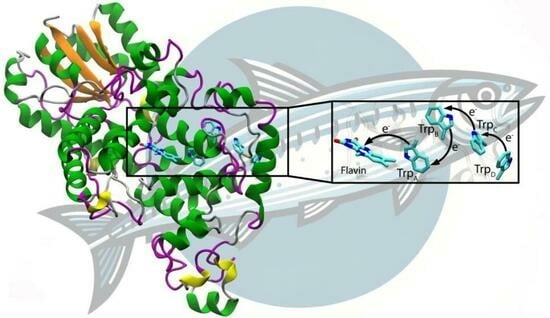
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| Trp | Tryptophan |
| ET | Electron transfer |
| RP | Radical pair |
| RPM | Radical pair mechanism |
| FAD | Flavin adenine dinucleotide |
| Cry1/4 | Cryptochrome 1/4 |
| Ch | Clupea harengus |
| Er | Erithacus rubecula |
| At | Arabidopsis thaliana |
| MD | Molecular dynamics |
| QM/MM | Quantum mechanics/molecular mechanics |
| Å | Ångström |
| NVT | Canonical ensemble |
| NPT | Isothermal–isobaric ensemble |
| DFTB | Density functional based tight-binding |
References
- Wiltschko, W. On the effect of static magnetic fields on the migratory orientation of the robin (Erithacus rubecula). Z. Tierpsychol. 1968, 25, 537–558. [Google Scholar] [CrossRef] [PubMed]
- Wiltschko, W.; Wiltschko, R. Interrelation of magnetic compass and star orientation in night-migrating birds. J. Fish Biol. 1976, 109, 91–99. [Google Scholar] [CrossRef]
- Kirschvink, J.L. Biogenic magnetite as a basis for magnetic field sensitivity in animals. Biosystems 1981, 13, 181–201. [Google Scholar] [CrossRef] [PubMed]
- Spiecker, L.; Laurien, M.; Dammann, W.; Franke, A.; Clemmesen, C.; Gerlach, G. Juvenile Atlantic herring (Clupea harengus) use a time-compensated sun compass for orientation. J. Exp. Biol. 2022, 225, jeb244607. [Google Scholar] [CrossRef] [PubMed]
- Myklatun, A.; Lauri, A.; Eder, S.H.K.; Cappetta, M.; Shcherbakov, D.; Wurst, W.; Winklhofer, M.; Westmeyer, G.G. Zebrafish and medaka offer insights into the neurobehavioral correlates of vertebrate magnetoreception. Nat. Commun. 2018, 9, 802. [Google Scholar] [CrossRef] [PubMed]
- Takebe, A.; Furutani, T.; Wada, T.; Koinuma, M.; Kubo, Y.; Okano, K.; Okano, T. Zebrafish respond to the geomagnetic field by bimodal and group-dependent orientation. Sci. Rep. 2012, 2, 727. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.B. Use of the earth’s magnetic field by orienting cave salamanders (Eurycea lucifuga). J. Comp. Physiol. A 1977, 121, 273–288. [Google Scholar] [CrossRef]
- Mouritsen, H. Long-distance navigation and magnetoreception in migratory animals. Nature 2018, 558, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Formicki, K.; Korzelecka-Orkisz, A.; Tański, A. Magnetoreception in fish. J. Fish Biol. 2019, 95, 73–91. [Google Scholar] [CrossRef]
- Naisbett-Jones, L.C.; Lohmann, K.J. Magnetoreception and magnetic navigation in fishes: A half century of discovery. J. Comp. Physiol. A 2022, 208, 19–40. [Google Scholar] [CrossRef]
- Wan, G.; Hayden, A.N.; Iiams, S.E.; Merlin, C. Cryptochrome 1 mediates light-dependent inclination magnetosensing in monarch butterflies. Nat. Commun. 2021, 12, 771. [Google Scholar] [CrossRef] [PubMed]
- Schulten, K.; Swenberg, C.E.; Weller, A. A biomagnetic sensory mechanism based on magnetic field modulated coherent electron spin motion. Z. Phys. Chem. 1978, 111, 1–5. [Google Scholar] [CrossRef]
- Hore, P.J.; Mouritsen, H. The radical-pair mechanism of magnetoreception. Annu. Rev. Biophys. 2016, 45, 299–344. [Google Scholar] [CrossRef]
- Ritz, T.; Adem, S.; Schulten, K. A model for photoreceptor-based magnetoreception in birds. Biophys. J. 2000, 78, 707–718. [Google Scholar] [CrossRef]
- Shaw, J.; Boyd, A.; House, M.; Woodward, R.; Mathes, F.; Cowin, G.; Saunders, M.; Baer, B. Magnetic particle-mediated magnetoreception. J. R. Soc. Interface 2015, 12, 20150499. [Google Scholar] [CrossRef]
- Winklhofer, M.; Kirschvink, J.L. A quantitative assessment of torque-transducer models for magnetoreception. J. R. Soc. Interface 2010, 7, S273–S289. [Google Scholar] [CrossRef] [PubMed]
- Solov’yov, I.A.; Domratcheva, T.; Schulten, K. Separation of photo-induced radical pair in cryptochrome to a functionally critical distance. Sci. Rep. 2014, 4, 3845. [Google Scholar] [CrossRef]
- Solov’yov, I.A.; Mouritsen, H.; Schulten, K. Acuity of a cryptochrome and vision-based magnetoreception system in birds. Biophys. J. 2010, 99, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Solov’yov, I.A.; Greiner, W. Theoretical Analysis of an Iron Mineral-Based Magnetoreceptor Model in Birds. Biophys. J. 2007, 93, 1493–1509. [Google Scholar] [CrossRef]
- Wong, S.Y.; Frederiksen, A.; Hanić, M.; Schuhmann, F.; Grüning, G.; Hore, P.; Solov’yov, I.A. Navigation of migratory songbirds: A quantum magnetic compass sensor. Neuroforum 2021, 27, 141–150. [Google Scholar] [CrossRef]
- Solov’yov, I.A.; Schulten, K. Reaction Kinetics and Mechanism of Magnetic Field Effects in Cryptochrome. J. Phys. Chem. B 2012, 116, 1089–1099. [Google Scholar] [CrossRef] [PubMed]
- Wiltschko, W.; Wiltschko, R. Light-dependent magnetoreception in birds: The behaviour of European robins, Erithacus rubecula, under monochromatic light of various wavelengths and intensities. J. Exp. Biol. 2001, 204, 3295–3302. [Google Scholar] [CrossRef] [PubMed]
- Wiltschko, W.; Wiltschko, R. Migratory orientation of European robins is affected by the wavelength of light as well as by a magnetic pulse. J. Comp. Physiol. A 1995, 177, 363–369. [Google Scholar] [CrossRef]
- Xu, J.; Jarocha, L.E.; Zollitsch, T.; Konowalczyk, M.; Henbest, K.B.; Richert, S.; Golesworthy, M.J.; Schmidt, J.; Déjean, V.; Sowood, D.J.; et al. Magnetic sensitivity of cryptochrome 4 from a migratory songbird. Nature 2021, 594, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Günther, A.; Einwich, A.; Sjulstok, E.; Feederle, R.; Bolte, P.; Koch, K.W.; Solov’yov, I.A.; Mouritsen, H. Double-cone localization and seasonal expression pattern suggest a role in magnetoreception for European robin cryptochrome 4. Curr Biol. 2018, 28, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Zoltowski, B.D.; Chelliah, Y.; Wickramaratne, A.; Jarocha, L.; Karki, N.; Xu, W.; Mouritsen, H.; Hore, P.J.; Hibbs, R.E.; Green, C.B.; et al. Chemical and structural analysis of a photoactive vertebrate cryptochrome from pigeon. Proc. Natl. Acad. Sci. USA 2019, 116, 19449–19457. [Google Scholar] [CrossRef] [PubMed]
- Timmer, D.; Frederiksen, A.; Lünemann, D.C.; Thomas, A.R.; Xu, J.; Bartölke, R.; Schmidt, J.; Kubař, T.; De Sio, A.; Solov’yov, I.A.; et al. Tracking the Electron Transfer Cascade in European Robin Cryptochrome 4 Mutants. J. Am. Chem. Soc. 2023, 145, 11566–11578. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.Y.; Wei, Y.; Mouritsen, H.; Solov’yov, I.A.; Hore, P.J. Cryptochrome magnetoreception: Four tryptophans could be better than three. J. R. Soc. Interface 2021, 18, 20210601. [Google Scholar] [CrossRef] [PubMed]
- Bellinger, M.R.; Wei, J.; Hartmann, U.; Cadiou, H.; Winklhofer, M.; Banks, M.A. Conservation of magnetite biomineralization genes in all domains of life and implications for magnetic sensing. Proc. Natl. Acad. Sci. USA 2022, 119, e2108655119. [Google Scholar] [CrossRef]
- Laurien, M.; Mende, L.; Luhrmann, L.; Frederiksen, A.; Aldag, M.; Spiecker, L.; Clemmensen, C.; Solov’yol, I.A.; Gerlach, G. Magnetic orientation in juvenile Atlantic herring (Clupea harengus) could involve Cryptochrome 4 as a potential magnetoreceptor. J. R. Soc. Interface, 2024; submitted. [Google Scholar]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 2010, 78, 1950–1958. [Google Scholar] [CrossRef]
- Lüdemann, G.; Solov’yov, I.A.; Kubař, T.; Elstner, M. Solvent driving force ensures fast formation of a persistent and well-separated radical pair in plant cryptochrome. J. Am. Chem. Soc. 2015, 137, 1147–1156. [Google Scholar] [CrossRef]
- Sjulstok, E.; Lüdemann, G.; Kubař, T.; Elstner, M.; Solov’yov, I.A. Molecular insights into variable electron transfer in amphibian cryptochrome. Biophys. J. 2018, 114, 2563–2572. [Google Scholar] [CrossRef]
- Fliege, J.; Svaiter, B.F. Steepest descent methods for multicriteria optimization. Math. Oper. Res. 2000, 51, 479–494. [Google Scholar] [CrossRef]
- Nosé, S.; Klein, M. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983, 50, 1055–1076. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed]
- Lacombat, F.; Espagne, A.; Dozova, N.; Plaza, P.; Müller, P.; Brettel, K.; Franz-Badur, S.; Essen, L.O. Ultrafast Oxidation of a Tyrosine by Proton-Coupled Electron Transfer Promotes Light Activation of an Animal-like Cryptochrome. J. Am. Chem. Soc. 2019, 141, 13394–13409. [Google Scholar] [CrossRef]
- Biskup, T.; Paulus, B.; Okafuji, A.; Hitomi, K.; Getzoff, E.D.; Weber, S.; Schleicher, E. Variable Electron Transfer Pathways in an Amphibian Cryptochrome. J. Biol. Chem. 2013, 288, 9249–9260. [Google Scholar] [CrossRef] [PubMed]
- Moser, C.C.; Keske, J.M.; Warncke, K.; Farid, R.S.; Dutton, P.L. Nature of biological electron transfer. Nature 1992, 355, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Page, C.C.; Moser, C.C.; Chen, X.; Dutton, P.L. Natural engineering principles of electron tunnelling in biological oxidation–reduction. Nature 1999, 402, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Schuhmann, F.; Kattnig, D.R.; Solov’yov, I.A. Exploring post-activation conformational changes in pigeon cryptochrome 4. J. Phys. Chem. B 2021, 125, 9652–9659. [Google Scholar] [CrossRef] [PubMed]
- Schuhmann, F.; Ryvkin, L.; McLaren, J.D.; Gerhards, L.; Solov’yov, I.A. Across atoms to crossing continents: Application of similarity measures to biological location data. PLoS ONE 2023, 18, e0284736. [Google Scholar] [CrossRef] [PubMed]
- Marcus, R.A.; Sutin, N. Electron transfers in chemistry and biology. Biochim. Biophys. Acta 1985, 811, 265–322. [Google Scholar] [CrossRef]
- Newton, M.D.; Sutin, N. Electron Transfer Reactions in Condensed Phases. Annu. Rev. Phys. Chem. 1984, 35, 437–480. [Google Scholar] [CrossRef]
- Warshel, A.; Parson, W.W. Computer simulations of electron-transfer reactions in solution and in photosynthetic reaction centers. Annu. Rev. Phys. Chem. 1991, 42, 279–309. [Google Scholar] [CrossRef]





| 1/k (ps) | TrpA–TrpB | TrpB–TrpC | TrpC–TrpD |
|---|---|---|---|
| 23 | 149 | 57 | |
| 141 | 82 | 240 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frederiksen, A.; Aldag, M.; Solov’yov, I.A.; Gerhards, L. Activation of Cryptochrome 4 from Atlantic Herring. Biology 2024, 13, 262. https://doi.org/10.3390/biology13040262
Frederiksen A, Aldag M, Solov’yov IA, Gerhards L. Activation of Cryptochrome 4 from Atlantic Herring. Biology. 2024; 13(4):262. https://doi.org/10.3390/biology13040262
Chicago/Turabian StyleFrederiksen, Anders, Mandus Aldag, Ilia A. Solov’yov, and Luca Gerhards. 2024. "Activation of Cryptochrome 4 from Atlantic Herring" Biology 13, no. 4: 262. https://doi.org/10.3390/biology13040262