The Assessment of Sperm DNA Integrity: Implications for Assisted Reproductive Technology Fertility Outcomes across Livestock Species
Abstract
Simple Summary
Abstract
1. Introduction
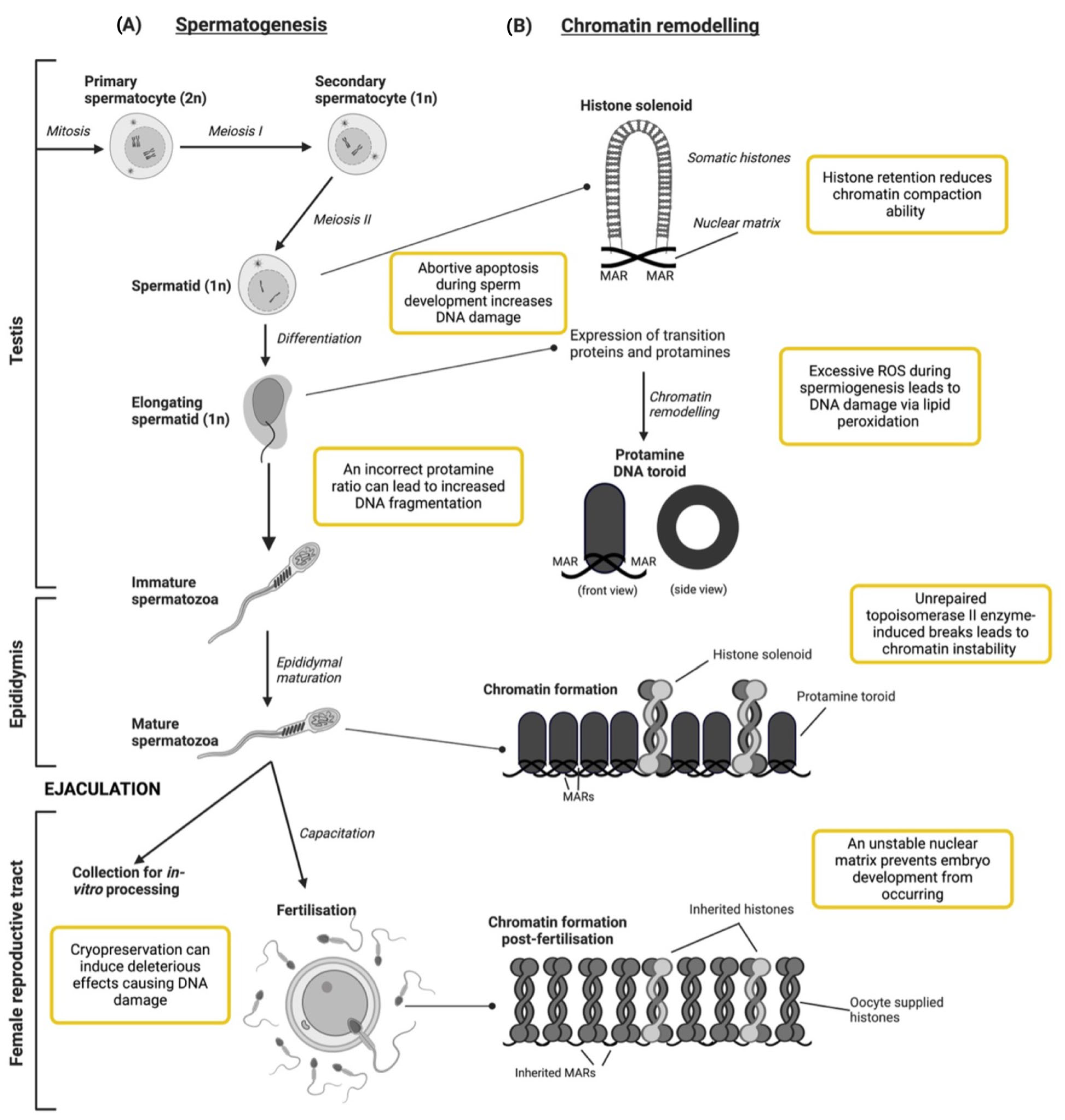
2. DNA Maturation and the Development of Spermatozoa
2.1. Testicular Development
Susceptibility of Chromatin to Damage during Testicular Development
2.2. Epididymal Maturation and Fertilisation within the Female Reproductive Tract
3. Current Measures of DNA Integrity Used across Species
3.1. Sperm Chromatin Structure Assay (SCSA)
3.2. Single-Cell Gel Electrophoresis (COMET)
3.3. Transferase dUTP Nick End Labelling (TUNEL)
3.4. Chromomycin A3 (CMA3)
3.5. Toluidine Blue (TB)
3.6. Sperm Chromatin Dispersion (SCD) Test
3.7. 8-Hydroxyguanine
4. Implications for the Livestock Artificial Breeding Industry
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Agarwal, A.; Mulgund, A.; Hamada, A.; Chyatte, M.R. A unique view on male infertility around the globe. Reprod. Biol. Endocrinol. 2015, 13, 37. (In English) [Google Scholar] [CrossRef] [PubMed]
- Comhaire, F.; Vermeulen, L. Human semen analysis. Hum. Reprod. Update 1995, 1, 343–362. [Google Scholar] [CrossRef] [PubMed]
- Evans, G.; Maxwell, W.M.C. Salamon’s Artificial Insemination of Sheep and Goats; Butterworths: Sydney, Australia, 1987; p. 194. [Google Scholar]
- Mortimer, D.; Mortimer, S.T. Computer-Aided Sperm Analysis (CASA) of Sperm Motility and Hyperactivation. In Spermatogenesis: Methods and Protocols; Carrell, D.T., Aston, K.I., Eds.; Humana Press: Totowa, NJ, USA, 2013; pp. 77–87. [Google Scholar]
- López-Fernández, C.; Fernández, J.L.; Gosálbez, A.; Arroyo, F.; Vázquez, J.M.; Holt, W.V.; Gosálvez, J. Dynamics of sperm DNA fragmentation in domestic animals: III. Ram. Theriogenology 2008, 70, 898–908. [Google Scholar] [CrossRef] [PubMed]
- Wouters-Tyrou, D.; Martinage, A.; Chevaillier, P.; Sautière, P. Nuclear basic proteins in spermiogenesis. Biochimie 1998, 80, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Shamsi, M.B.; Venkatesh, S.; Tanwar, M.; Singh, G.; Mukherjee, S.; Malhotra, N.; Kumar, R.; Gupta, N.P.; Mittal, S.; Dada, R. Comet assay: A prognostic tool for DNA integrity assessment in infertile men opting for assisted reproduction. Indian J. Med. Res. 2010, 131, 675–681. Available online: https://journals.lww.com/ijmr/Fulltext/2010/31050/Comet_assay__A_prognostic_tool_for_DNA_integrity.14.aspx (accessed on 13 December 2023). [PubMed]
- David, I.; Kohnke, P.; Lagriffoul, G.; Praud, O.; Plouarboué, F.; Degond, P.; Druart, X. Mass sperm motility is associated with fertility in sheep. Anim. Reprod. Sci. 2015, 161, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Kruger, T.F.; Acosta, A.A.; Simmons, K.F.; Swanson, R.J.; Matta, J.F.; Oehninger, S. Predictive value of abnormal sperm morphology in in vitro fertilization. Fertil. Steril. 1988, 49, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Söderquist, L.; Janson, L.; Larsson, K.; Einarsson, S. Sperm Morphology and Fertility in A. I. Bulls. J. Vet. Med. Ser. A 1991, 38, 534–543. [Google Scholar] [CrossRef]
- Kumaresan, A.; Johannisson, A.; Al-Essawe, E.M.; Morrell, J.M. Sperm viability, reactive oxygen species, and DNA fragmentation index combined can discriminate between above- and below-average fertility bulls. J. Dairy Sci. 2017, 100, 5824–5836. [Google Scholar] [CrossRef]
- Nagy, S.; Johannisson, A.; Wahlsten, T.; Ijäs, R.; Andersson, M.; Rodriguez-Martinez, H. Sperm chromatin structure and sperm morphology: Their association with fertility in AI-dairy Ayrshire sires. Theriogenology 2013, 79, 1153–1161. [Google Scholar] [CrossRef]
- Neuhauser, S.; Bollwein, H.; Siuda, M.; Handler, J. Effects of Different Freezing Protocols on Motility, Viability, Mitochondrial Membrane Potential, Intracellular Calcium Level, and DNA Integrity of Cryopreserved Equine Epididymal Sperm. J. Equine Vet. Sci. 2019, 82, 102801. [Google Scholar] [CrossRef]
- Aitken, R.J.; De Iuliis, G.N. On the possible origins of DNA damage in human spermatozoa. Mol. Hum. Reprod. 2009, 16, 3–13. [Google Scholar] [CrossRef]
- Didion, B.A.; Kasperson, K.M.; Wixon, R.L.; Evenson, D.P. Boar Fertility and Sperm Chromatin Structure Status: A Retrospective Report. J. Androl. 2009, 30, 655–660. [Google Scholar] [CrossRef]
- Nordstoga, A.; Krogenæs, A.; Nødtvedt, A.; Farstad, W.; Waterhouse, K. The Relationship Between Post-Thaw Sperm DNA Integrity and Non-Return Rate Among Norwegian Cross-Bred Rams. Reprod. Domest. Anim. 2013, 48, 207–212. [Google Scholar] [CrossRef]
- Peris-Frau, P.; Álvarez-Rodríguez, M.; Martín-Maestro, A.; Iniesta-Cuerda, M.; Sánchez-Ajofrín, I.; Garde, J.J.; Rodriguez-Martinez, H.; Soler, A.J. Comparative evaluation of DNA integrity using sperm chromatin structure assay and Sperm-Ovis-Halomax during in vitro capacitation of cryopreserved ram spermatozoa. Reprod. Domest. Anim. 2019, 54, 46–49. [Google Scholar] [CrossRef]
- Chi, H.-J.; Kim, S.-G.; Kim, Y.-Y.; Park, J.-Y.; Yoo, C.-S.; Park, I.-H.; Sun, H.-G.; Kim, J.-W.; Lee, K.-H.; Park, H.-D. ICSI significantly improved the pregnancy rate of patients with a high sperm DNA fragmentation index. Clin. Exp. Reprod. Med. 2017, 44, 132–140. [Google Scholar] [CrossRef]
- Santolaria, P.; Vicente-Fiel, S.; Palacín, I.; Fantova, E.; Blasco, M.E.; Silvestre, M.A.; Yániz, J.L. Predictive capacity of sperm quality parameters and sperm subpopulations on field fertility after artificial insemination in sheep. Anim. Reprod. Sci. 2015, 163, 82–88. [Google Scholar] [CrossRef]
- Waberski, D.; Schapmann, E.; Henning, H.; Riesenbeck, A.; Brandt, H. Sperm chromatin structural integrity in normospermic boars is not related to semen storage and fertility after routine AI. Theriogenology 2011, 75, 337–345. [Google Scholar] [CrossRef]
- de Kretser, D.M.; Loveland, K.L.; Meinhardt, A.; Simorangkir, D.; Wreford, N. Spermatogenesis. Hum. Reprod. 1998, 13 (Suppl. S1), 1–8. [Google Scholar] [CrossRef]
- Clermont, Y.; Leblond, C.P. Spermiogenesis of man, monkey, ram and other mammals as shown by the “periodic acid-schiff” technique. Am. J. Anat. 1955, 96, 229–253. [Google Scholar] [CrossRef]
- Ward, W.S. Function of sperm chromatin structural elements in fertilization and development. Mol. Hum. Reprod. 2009, 16, 30–36. [Google Scholar] [CrossRef]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Meistrich, M. Histone and basic nuclear protein transitions in mammalian spermatogenesis. Histones Other Basic Nucl. Proteins 1989, 166, 165. [Google Scholar]
- Barral, S.; Morozumi, Y.; Tanaka, H.; Montellier, E.; Govin, J.; de Dieuleveult, M.; Charbonnier, G.; Couté, Y.; Puthier, D.; Buchou, T.; et al. Histone Variant H2A.L.2 Guides Transition Protein-Dependent Protamine Assembly in Male Germ Cells. Mol. Cell 2017, 66, 89–101.e8. [Google Scholar] [CrossRef]
- Akama, K.; Sato, H.; Furihata-Yamauchi, M.; Komatsu, Y.; Tobita, T.; Nakano, M. Interaction of nucleosome core DNA with transition proteins 1 and 3 from boar late spermatid nuclei. J. Biochem. 1996, 119, 448–455. [Google Scholar] [CrossRef]
- Chevaillier, P.; Chirat, F.; Sautiere, P. The amino acid sequence of the ram spermatidal protein 3. Eur. J. Biochem. 1998, 258, 460–464. [Google Scholar] [CrossRef]
- Steger, K.; Klonisch, T.; Gavenis, K.; Drabent, B.; Doenecke, D.; Bergmann, M. Expression of mRNA and protein of nucleoproteins during human spermiogenesis. Mol. Hum. Reprod. 1998, 4, 939–945. [Google Scholar] [CrossRef]
- Yu, Y.E.; Zhang, Y.; Unni, E.; Shirley, C.R.; Deng, J.M.; Russell, L.D.; Weil, M.M.; Behringer, R.R.; Meistrich, M.L. Abnormal spermatogenesis and reduced fertility in transition nuclear protein 1-deficient mice. Proc. Natl. Acad. Sci. USA 2000, 97, 4683–4688. [Google Scholar] [CrossRef]
- Shirley, C.R.; Hayashi, S.; Mounsey, S.; Yanagimachi, R.; Meistrich, M.L. Abnormalities and reduced reproductive potential of sperm from Tnp1- and Tnp2-null double mutant mice. Biol. Reprod. 2004, 71, 1220–1229. [Google Scholar] [CrossRef]
- Hud, N.V.; Milanovich, F.P.; Balhorn, R. Evidence of Novel Secondary Structure in DNA-Bound Protamine Is Revealed by Raman Spectroscopy. Biochemistry 1994, 33, 7528–7535. [Google Scholar] [CrossRef]
- Castillo, J.; Simon, L.; de Mateo, S.; Lewis, S.; Oliva, R. Protamine/DNA Ratios and DNA Damage in Native and Density Gradient Centrifuged Sperm From Infertile Patients. J. Androl. 2011, 32, 324–332. [Google Scholar] [CrossRef]
- Cho, C.; Willis, W.D.; Goulding, E.H.; Jung-Ha, H.; Choi, Y.-C.; Hecht, N.B.; Eddy, E.M. Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nat. Genet. 2001, 28, 82–86. [Google Scholar] [CrossRef]
- Moritz, L.; Schon, S.B.; Rabbani, M.; Sheng, Y.; Pendlebury, D.F.; Agrawal, R.; Sultan, C.; Jorgensen, K.; Zheng, X.; Diehl, A.; et al. Single residue substitution in protamine 1 disrupts sperm genome packaging and embryonic development in mice. bioRxiv 2021, 460631. [Google Scholar] [CrossRef]
- Bench, G.S.; Friz, A.M.; Corzett, M.H.; Morse, D.H.; Balhorn, R. DNA and total protamine masses in individual sperm from fertile mammalian subjects. Cytometry 1996, 23, 263–271. (In English) [Google Scholar] [CrossRef]
- Erkek, S.; Hisano, M.; Liang, C.-Y.; Gill, M.; Murr, R.; Dieker, J.; Schübeler, D.; Vlag, J.v.d.; Stadler, M.B.; Peters, A.H.F.M. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat. Struct. Mol. Biol. 2013, 20, 868–875. [Google Scholar] [CrossRef]
- Tanphaichitr, N.; Sobhon, P.; Taluppeth, N.; Chalermisarachai, P. Basic nuclear proteins in testicular cells and ejaculated spermatozoa in man. Exp. Cell Res. 1978, 117, 347–356. [Google Scholar] [CrossRef]
- Arpanahi, A.; Brinkworth, M.; Iles, D.; Krawetz, S.A.; Paradowska, A.; Platts, A.E.; Saida, M.; Steger, K.; Tedder, P.; Miller, D. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res. 2009, 19, 1338–1349. (In English) [Google Scholar] [CrossRef]
- Ukogu, O.A.; Smith, A.D.; Devenica, L.M.; Bediako, H.; McMillan, R.B.; Ma, Y.; Balaji, A.; Schwab, R.D.; Anwar, S.; Dasgupta, M.; et al. Protamine loops DNA in multiple steps. Nucleic Acids Res. 2020, 48, 6108–6119. [Google Scholar] [CrossRef]
- Sotolongo, B.; Huang, T.T.F.; Isenberger, E.; Ward, W.S. An Endogenous Nuclease in Hamster, Mouse, and Human Spermatozoa Cleaves DNA into Loop-Sized Fragments. J. Androl. 2005, 26, 272–280. [Google Scholar] [CrossRef]
- Suzuki, M.; Crozatier, C.; Yoshikawa, K.; Mori, T.; Yoshikawa, Y. Protamine-induced DNA compaction but not aggregation shows effective radioprotection against double-strand breaks. Chem. Phys. Lett. 2009, 480, 113–117. [Google Scholar] [CrossRef]
- Takata, H.; Hanafusa, T.; Mori, T.; Shimura, M.; Iida, Y.; Ishikawa, K.; Yoshikawa, K.; Yoshikawa, Y.; Maeshima, K. Chromatin compaction protects genomic DNA from radiation damage. PLoS ONE 2013, 8, e75622. (In English) [Google Scholar] [CrossRef]
- Ward, W.S.; Kimura, Y.; Yanagimachi, R. An intact sperm nuclear matrix may be necessary for the mouse paternal genome to participate in embryonic development. Biol. Reprod. 1999, 60, 702–706. (In English) [Google Scholar] [CrossRef]
- Balhorn, R.; Brewer, L.; Corzett, M. DNA condensation by protamine and arginine-rich peptides: Analysis of toroid stability using single DNA molecules. Mol. Reprod. Dev. 2000, 56, 230–234. [Google Scholar] [CrossRef]
- Hess, R.A.; de Franca, L.R. Spermatogenesis and Cycle of the Seminiferous Epithelium. In Molecular Mechanisms in Spermatogenesis; Cheng, C.Y., Ed.; Springer: New York, NY, USA, 2008; pp. 1–15. [Google Scholar]
- Amann, R.P. The Cycle of the Seminiferous Epithelium in Humans: A Need to Revisit? J. Androl. 2008, 29, 469–487. [Google Scholar] [CrossRef]
- Graham, J.K. Analysis of Stallion Semen and its Relation to Fertility. Vet. Clin. N. Am. Equine Pract. 1996, 12, 119–130. [Google Scholar] [CrossRef]
- Rousseaux, J.; Rousseaux-Prevost, R. Molecular Localization of Free Thiols in Human Sperm Chromatin1. Biol. Reprod. 1995, 52, 1066–1072. [Google Scholar] [CrossRef]
- Gilbert, D.L. Perspective on the history of oxygen and life. In Oxygen and Living Processes: An Interdisciplinary Approach; Springer: Berlin/Heidelberg, Germany, 1981; pp. 1–43. [Google Scholar]
- Aitken, R.J.; Irvine, D.S.; Wu, F.C. Prospective analysis of sperm-oocyte fusion and reactive oxygen species generation as criteria for the diagnosis of infertility. Am. J. Obstet. Gynecol. 1991, 164, 542–551. [Google Scholar] [CrossRef]
- de Lamirande, E.; Gagnon, C. Human sperm hyperactivation and capacitation as parts of an oxidative process. Free Radic. Biol. Med. 1993, 14, 157–166. (In English) [Google Scholar] [CrossRef]
- Sánchez, R.; Sepúlveda, C.; Risopatrón, J.; Villegas, J.; Giojalas, L.C. Human sperm chemotaxis depends on critical levels of reactive oxygen species. Fertil. Steril. 2010, 93, 150–153. [Google Scholar] [CrossRef]
- Lopes, S.; Jurisicova, A.; Sun, J.G.; Casper, R.F. Reactive oxygen species: Potential cause for DNA fragmentation in human spermatozoa. Hum. Reprod. 1998, 13, 896–900. [Google Scholar] [CrossRef]
- Agarwal, A.; Saleh, R.A.; Bedaiwy, M.A. Role of reactive oxygen species in the pathophysiology of human reproduction. Fertil. Steril. 2003, 79, 829–843. [Google Scholar] [CrossRef] [PubMed]
- Sakkas, D.; Mariethoz, E.; St. John, J.C. Abnormal Sperm Parameters in Humans Are Indicative of an Abortive Apoptotic Mechanism Linked to the Fas-Mediated Pathway. Exp. Cell Res. 1999, 251, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.; Haaf, T. DNA nicks and increased sensitivity of DNA to fluorescence in situ end labeling during functional spermiogenesis. Biotechniques 1998, 25, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Balhorn, R. A model for the structure of chromatin in mammalian sperm. J. Cell Biol. 1982, 93, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Meyer-Ficca, M.L.; Lonchar, J.D.; Ihara, M.; Meistrich, M.L.; Austin, C.A.; Meyer, R.G. Poly(ADP-Ribose) Polymerases PARP1 and PARP2 Modulate Topoisomerase II Beta (TOP2B) Function During Chromatin Condensation in Mouse Spermiogenesis1. Biol. Reprod. 2011, 84, 900–909. [Google Scholar] [CrossRef] [PubMed]
- Manicardi, G.C.; Bianchi, P.; Pantano, S.; Azzoni, P.; Bizzaro, D.; Bianchi, U.; Sakkas, D. Presence of endogenous nicks in DNA of ejaculated human spermatozoa and its relationship to chromomycin A3 accessibility. Biol. Reprod. 1995, 52, 864–867. [Google Scholar] [CrossRef] [PubMed]
- Henkel, R.; Kierspel, E.; Stalf, T.; Mehnert, C.; Menkveld, R.; Tinneberg, H.R.; Schill, W.B.; Kruger, T.F. Effect of reactive oxygen species produced by spermatozoa and leukocytes on sperm functions in non-leukocytospermic patients. Fertil. Steril. 2005, 83, 635–642. [Google Scholar] [CrossRef]
- Dacheux, J.-L.; Dacheux, F. New insights into epididymal function in relation to sperm maturation. Reproduction 2014, 147, R27–R42. (In English) [Google Scholar] [CrossRef]
- Calvin, H.I.; Bedford, J.M. Formation of disulphide bonds in the nucleus and accessory structures of mammalian spermatozoa during maturation in the epididymis. J. Reprod. Fertil. Suppl. 1971, 13, 65–75. [Google Scholar]
- Marushige, Y.; Marushige, K. Properties of chromatin isolated from bull spermatozoa. Biochim. Biophys. Acta (BBA)–Nucleic Acids Protein Synth. 1974, 340, 498–508. [Google Scholar] [CrossRef]
- Leclerc, P.; De Lamirande, E.; Gagnon, C. Regulation of Protein-Tyrosine Phosphorylation and Human Sperm Capacitation by Reactive Oxygen Derivatives. Free Radic. Biol. Med. 1997, 22, 643–656. [Google Scholar] [CrossRef] [PubMed]
- van der Heijden, G.W.; Ramos, L.; Baart, E.B.; van den Berg, I.M.; Derijck, A.A.H.A.; van der Vlag, J.; Martini, E.; de Boer, P. Sperm-derived histones contribute to zygotic chromatin in humans. BMC Dev. Biol. 2008, 8, 34. [Google Scholar] [CrossRef]
- Kopečný, V.; Pavlok, A. Autoradiographic study of mouse spermatozoan arginine-rich nuclear protein in fertilization. J. Exp. Zool. 1975, 191, 85–95. [Google Scholar] [CrossRef] [PubMed]
- McLay, D.W.; Clarke, H.J. Remodelling the paternal chromatin at fertilization in mammals. Reproduction 2003, 125, 625–633. [Google Scholar] [CrossRef]
- Ogura, A.; Matsuda, J.; Yanagimachi, R. Birth of normal young after electrofusion of mouse oocytes with round spermatids. Proc. Natl. Acad. Sci. USA 1994, 91, 7460–7462. (In English) [Google Scholar] [CrossRef]
- Hammoud, S.S.; Nix, D.A.; Zhang, H.; Purwar, J.; Carrell, D.T.; Cairns, B.R. Distinctive chromatin in human sperm packages genes for embryo development. Nature 2009, 460, 473–478. (In English) [Google Scholar] [CrossRef]
- Williams, C.J. Signalling mechanisms of mammalian oocyte activation. Hum. Reprod. Update 2002, 8, 313–321. [Google Scholar] [CrossRef]
- Maxwell, W.M.C.; Watson, P.F. Recent progress in the preservation of ram semen. Anim. Reprod. Sci. 1996, 42, 55–65. [Google Scholar] [CrossRef]
- O’connell, M.; Mcclure, N.; Lewis, S. The effects of cryopreservation on sperm morphology, motility and mitochondrial function. Hum. Reprod. 2002, 17, 704–709. [Google Scholar] [CrossRef]
- Peris, S.I.; Morrier, A.; Dufour, M.; Bailey, J.L. Cryopreservation of Ram Semen Facilitates Sperm DNA Damage: Relationship between Sperm Andrological Parameters and the Sperm Chromatin Structure Assay. J. Androl. 2004, 25, 224–233. [Google Scholar] [CrossRef]
- Bucak, M.N.; Ateşşahin, A.; Yüce, A. Effect of anti-oxidants and oxidative stress parameters on ram semen after the freeze–thawing process. Small Rumin. Res. 2008, 75, 128–134. [Google Scholar] [CrossRef]
- Salamon, S.; Maxwell, W.M.C. Storage of ram semen. Anim. Reprod. Sci. 2000, 62, 77–111. [Google Scholar] [CrossRef] [PubMed]
- Evenson, D.P.; Darzynkiewicz, Z.; Melamed, M.R. Relation of Mammalian Sperm Chromatin Heterogeneity to Fertility. Science 1980, 210, 1131–1133. [Google Scholar] [CrossRef] [PubMed]
- Ostling, O.; Johanson, K.J. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem. Biophys. Res. Commun. 1984, 123, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.P.; McCoy, M.T.; Tice, R.R.; Schneider, E.L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988, 175, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Floyd, R.A.; Watson, J.J.; Wong, P.K.; Altmiller, D.H.; Rickard, R.C. Hydroxyl free radical adduct of deoxyguanosine: Sensitive detection and mechanisms of formation. Free Radic. Res. Commun. 1986, 1, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Gorczyca, W.; Gong, J.; Darzynkiewcz, Z. Detection of DNA strandsbreaks in individual apoptotic cells by in situ terminal deoxynucleotidyl transferase and nick translation assays. Cancer Res. 1993, 53, 1945–1951. [Google Scholar] [PubMed]
- Erenpreiss, J.; Bars, J.; Lipatnikova, V.; Erenpreisa, J.; Zalkalns, J. Comparative Study of Cytochemical Tests for Sperm Chromatin Integrity. J. Androl. 2001, 22, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Roti, J.L.R.; Wright, W.D. Visualization of DNA loops in nucleoids from HeLa cells: Assays for DNA damage and repair. Cytometry 1987, 8, 461–467. [Google Scholar] [CrossRef]
- Vogelstein, B.; Pardoll, D.M.; Coffey, D.S. Supercoiled loops and eucaryotic DNA replicaton. Cell 1980, 22, 79–85. (In English) [Google Scholar] [CrossRef]
- Fernández, J.L.; Muriel, L.; Rivero, M.T.; Goyanes, V.; Vazquez, R.; Alvarez, J.G. The sperm chromatin dispersion test: A simple method for the determination of sperm DNA fragmentation. J. Androl. 2003, 24, 59–66. (In English) [Google Scholar] [CrossRef]
- Wdowiak, A.; Bakalczuk, S.; Bakalczuk, G. The effect of sperm DNA fragmentation on the dynamics of the embryonic development in intracytoplasmatic sperm injection. Reprod. Biol. 2015, 15, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Meseguer, M.; Santiso, R.; Garrido, N.; García-Herrero, S.; Remohí, J.; Fernandez, J.L. Effect of sperm DNA fragmentation on pregnancy outcome depends on oocyte quality. Fertil. Steril. 2011, 95, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Sivanarayana, T.; Ravi Krishna, C.; Jaya Prakash, G.; Krishna, K.M.; Madan, K.; Sudhakar, G.; Rama Raju, G.A. Sperm DNA fragmentation assay by sperm chromatin dispersion (SCD): Correlation between DNA fragmentation and outcome of intracytoplasmic sperm injection. Reprod. Med. Biol. 2014, 13, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.-T.; Wang, R.-X.; He, B.; Mo, W.-Y.; Huang, L.; Wang, S.-K.; Mao, X.-B.; Cheng, J.-P.; Huang, Y.-Y.; Liu, R. Effect of sperm DNA fragmentation on clinical outcomes for Chinese couples undergoing in vitro fertilization or intracytoplasmic sperm injection. J. Int. Med. Res. 2016, 44, 1283–1291. [Google Scholar] [CrossRef] [PubMed]
- Al Omrani, B.; Al Eisa, N.; Javed, M.; Al Ghedan, M.; Al Matrafi, H.; Al Sufyan, H. Associations of sperm DNA fragmentation with lifestyle factors and semen parameters of Saudi men and its impact on ICSI outcome. Reprod. Biol. Endocrinol. 2018, 16, 49. [Google Scholar] [CrossRef]
- Muriel, L.; Garrido, N.; Fernández, J.L.; Remohí, J.; Pellicer, A.; de los Santos, M.J.; Meseguer, M. Value of the sperm deoxyribonucleic acid fragmentation level, as measured by the sperm chromatin dispersion test, in the outcome of in vitro fertilization and intracytoplasmic sperm injection. Fertil. Steril. 2006, 85, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Vicente-Fiel, S.; Palacín, I.; Santolaria, P.; Fantova, E.; Quintín-Casorrán, F.J.; Sevilla-Mur, E.; Yániz, J.L. In vitro assessment of sperm quality from rams of high and low field fertility. Anim. Reprod. Sci. 2014, 146, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Karoui, S.; Díaz, C.; González-Marín, C.; Amenabar, M.E.; Serrano, M.; Ugarte, E.; Gosálvez, J.; Roy, R.; López-Fernández, C.; Carabaño, M.J. Is sperm DNA fragmentation a good marker for field AI bull fertility? J. Anim. Sci. 2012, 90, 2437–2449. [Google Scholar] [CrossRef]
- Crespo, F.; Quiñones-Pérez, C.; Ortiz, I.; Diaz-Jimenez, M.; Consuegra, C.; Pereira, B.; Dorado, J.; Hidalgo, M. Seasonal variations in sperm DNA fragmentation and pregnancy rates obtained after artificial insemination with cooled-stored stallion sperm throughout the breeding season (spring and summer). Theriogenology 2020, 148, 89–94. [Google Scholar] [CrossRef]
- Siddhartha, N.; Reddy, N.S.; Pandurangi, M.; Muthusamy, T.; Vembu, R.; Kasinathan, K. The Effect of Sperm DNA Fragmentation Index on the Outcome of Intrauterine Insemination and Intracytoplasmic Sperm Injection. J. Hum. Reprod. Sci. 2019, 12, 189–198. Available online: https://journals.lww.com/jhrs/fulltext/2019/12030/the_effect_of_sperm_dna_fragmentation_index_on_the.4.aspx (accessed on 22 January 2024). [CrossRef] [PubMed]
- Avendaño, C.; Franchi, A.; Duran, H.; Oehninger, S. DNA fragmentation of normal spermatozoa negatively impacts embryo quality and intracytoplasmic sperm injection outcome. Fertil. Steril. 2010, 94, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-C.; Lin, D.P.-C.; Tsao, H.-M.; Cheng, T.-C.; Liu, C.-H.; Lee, M.-S. Sperm DNA fragmentation negatively correlates with velocity and fertilization rates but might not affect pregnancy rates. Fertil. Steril. 2005, 84, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Bakos, H.W.; Thompson, J.G.; Feil, D.; Lane, M. Sperm DNA damage is associated with assisted reproductive technology pregnancy. Int. J. Androl. 2008, 31, 518–526. [Google Scholar] [CrossRef] [PubMed]
- Anzar, M.; He, L.; Buhr, M.M.; Kroetsch, T.G.; Pauls, K.P. Sperm Apoptosis in Fresh and Cryopreserved Bull Semen Detected by Flow Cytometry and Its Relationship with Fertility. Biol. Reprod. 2002, 66, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, K.E.; Haugan, T.; Kommisrud, E.; Tverdal, A.; Flatberg, G.; Farstad, W.; Evenson, D.P.; De Angelis, P.M. Sperm DNA damage is related to field fertility of semen from young Norwegian Red bulls. Reprod. Fertil. Dev. 2006, 18, 781–788. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Li, G.; Jin, H.; Guo, Y.; Sun, Y. The effect of sperm DNA fragmentation index on assisted reproductive technology outcomes and its relationship with semen parameters and lifestyle. Transl. Androl. Urol. 2019, 8, 356–365. Available online: https://tau.amegroups.org/article/view/27211 (accessed on 22 January 2024). [CrossRef]
- Zhang, Z.; Zhu, L.-L.; Jiang, H.-S.; Chen, H.; Chen, Y.; Dai, Y.-T. Predictors of pregnancy outcome for infertile couples attending IVF and ICSI programmes. Andrologia 2016, 48, 962–969. [Google Scholar] [CrossRef]
- Speyer, B.E.; Pizzey, A.R.; Ranieri, M.; Joshi, R.; Delhanty, J.D.A.; Serhal, P. Fall in implantation rates following ICSI with sperm with high DNA fragmentation. Hum. Reprod. 2010, 25, 1609–1618. [Google Scholar] [CrossRef]
- Narud, B.; Klinkenberg, G.; Khezri, A.; Zeremichael, T.T.; Stenseth, E.-B.; Nordborg, A.; Haukaas, T.H.; Morrell, J.M.; Heringstad, B.; Myromslien, F.D.; et al. Differences in sperm functionality and intracellular metabolites in Norwegian Red bulls of contrasting fertility. Theriogenology 2020, 157, 24–32. [Google Scholar] [CrossRef]
- Karabinus, D.S.; Evenson, D.P.; Jost, L.K.; Baer, R.K.; Kaproth, M.T. Comparison of Semen Quality in Young and Mature Holstein Bulls Measured by Light Microscopy and Flow Cytometry. J. Dairy Sci. 1990, 73, 2364–2371. [Google Scholar] [CrossRef]
- Sellem, E.; Broekhuijse, M.L.W.J.; Chevrier, L.; Camugli, S.; Schmitt, E.; Schibler, L.; Koenen, E.P.C. Use of combinations of in vitro quality assessments to predict fertility of bovine semen. Theriogenology 2015, 84, 1447–1454. [Google Scholar] [CrossRef] [PubMed]
- Christensen, P.; Labouriau, R.; Birck, A.; Boe-Hansen, G.B.; Pedersen, J.; Borchersen, S. Relationship among seminal quality measures and field fertility of young dairy bulls using low-dose inseminations. J. Dairy Sci. 2011, 94, 1744–1754. [Google Scholar] [CrossRef] [PubMed]
- Boe-Hansen, G.B.; Christensen, P.; Vibjerg, D.; Nielsen, M.B.F.; Hedeboe, A.M. Sperm chromatin structure integrity in liquid stored boar semen and its relationships with field fertility. Theriogenology 2008, 69, 728–736. [Google Scholar] [CrossRef]
- Morrell, J.M.; Johannisson, A.; Dalin, A.-M.; Hammar, L.; Sandebert, T.; Rodriguez-Martinez, H. Sperm morphology and chromatin integrity in Swedish warmblood stallions and their relationship to pregnancy rates. Acta Vet. Scand. 2008, 50, 2. [Google Scholar] [CrossRef]
- Simon, L.; Proutski, I.; Stevenson, M.; Jennings, D.; McManus, J.; Lutton, D.; Lewis, S.E. Sperm DNA damage has a negative association with live-birth rates after IVF. Reprod. Biomed. Online 2013, 26, 68–78. (In English) [Google Scholar] [CrossRef]
- Simões, R.; Feitosa, W.B.; Siqueira, A.F.P.; Nichi, M.; Paula-Lopes, F.F.; Marques, M.G.; Peres, M.A.; Barnabe, V.H.; Visintin, J.A.; Assumpção, M.E.O. Influence of bovine sperm DNA fragmentation and oxidative stress on early embryo in vitro development outcome. Reproduction 2013, 146, 433–441. [Google Scholar] [CrossRef]
- Serafini, R.; Love, C.C.; Coletta, A.; Mari, G.; Mislei, B.; Caso, C.; Di Palo, R. Sperm DNA integrity in frozen-thawed semen from Italian Mediterranean Buffalo bulls and its relationship to in vivo fertility. Anim. Reprod. Sci. 2016, 172, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.; Andrabi, S.M.H.; Jahan, S. Semen quality parameters as fertility predictors of water buffalo bull spermatozoa during low-breeding season. Theriogenology 2016, 86, 1516–1522. [Google Scholar] [CrossRef]
- Eid, L.N.; Shamiah, S.M.; El-Regalaty, H.A.M.; El-Keraby, F.E. Sperm DNA damage and embryonic development as related to fertility potential of buffalo bulls. J. Anim. Poult. Prod. 2011, 2, 65–74. [Google Scholar] [CrossRef]
- Pourmasumi, S.; Khoradmehr, A.; Rahiminia, T.; Sabeti, P.; Talebi, A.R.; Ghasemzadeh, J. Evaluation of sperm chromatin integrity using aniline blue and toluidine blue staining in infertile and normozoospermic men. J. Reprod. Infertil. 2019, 20, 95. [Google Scholar] [PubMed]
- Tsarev, I.; Bungum, M.; Giwercman, A.; Erenpreisa, J.; Ebessen, T.; Ernst, E.; Erenpreiss, J. Evaluation of male fertility potential by Toluidine Blue test for sperm chromatin structure assessment. Hum. Reprod. 2009, 24, 1569–1574. [Google Scholar] [CrossRef] [PubMed]
- Souza, E.T.; Silva, C.V.; Travençolo, B.A.N.; Alves, B.G.; Beletti, M.E. Sperm chromatin alterations in fertile and subfertile bulls. Reprod. Biol. 2018, 18, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Iranpour, F.G.; Nasr-Esfahani, M.H.; Valojerdi, M.R.; Taki Al-Taraihi, T.M. Chromomycin A3 staining as a useful tool for evaluation of male fertility. J. Assist. Reprod. Genet. 2000, 17, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Iranpour, F.G. Impact of sperm chromatin evaluation on fertilization rate in intracytoplasmic sperm injection. Adv. BioMed Res. 2014, 3, 229. (In English) [Google Scholar] [CrossRef] [PubMed]
- Castro, L.S.; Siqueira, A.F.P.; Hamilton, T.R.S.; Mendes, C.M.; Visintin, J.A.; Assumpção, M.E.O.A. Effect of bovine sperm chromatin integrity evaluated using three different methods on in vitro fertility. Theriogenology 2018, 107, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Evenson, D.P. The Sperm Chromatin Structure Assay (SCSA®) and other sperm DNA fragmentation tests for evaluation of sperm nuclear DNA integrity as related to fertility. Anim. Reprod. Sci. 2016, 169, 56–75. [Google Scholar] [CrossRef] [PubMed]
- Evenson, D.P.; Larson, K.L.; Jost, L.K. Sperm Chromatin Structure Assay: Its Clinical Use for Detecting Sperm DNA Fragmentation in Male Infertility and Comparisons With Other Techniques. J. Androl. 2002, 23, 25–43. [Google Scholar] [CrossRef]
- Rybar, R.; Kopecka, V.; Prinosilova, P.; Kubickova, S.; Veznik, Z.; Rubes, J. Fertile bull sperm aneuploidy and chromatin integrity in relationship to fertility. Int. J. Androl. 2010, 33, 613–622. [Google Scholar] [CrossRef]
- Varghese, A.C.; Fischer-Hammadeh, C.; Hammadeh, M.E. Acridine Orange Test for Assessment of Human Sperm DNA Integrity. In Sperm Chromatin: Biological and Clinical Applications in Male Infertility and Assisted Reproduction; Zini, A., Agarwal, A., Eds.; Springer: New York, NY, USA, 2011; pp. 189–199. [Google Scholar]
- Evenson, D.P.; Jost, L.K.; Baer, R.K.; Turner, T.W.; Schrader, S.M. Individuality of DNA denaturation patterns in human sperm as measured by the sperm chromatin structure assay. Reprod. Toxicol. 1991, 5, 115–125. [Google Scholar] [CrossRef]
- Sailer, B.L.; Jost, L.K.; Evenson, D.P. Mammalian Sperm DNA Susceptibility to In Situ Denaturation Associated with the Presence of DNA Strand Breaks as Measured by the Terminal Deoxynucleotidyl Transferase Assay. J. Androl. 1995, 16, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Evenson, D.P. Loss of livestock breeding efficiency due to uncompensable sperm nuclear defects. Reprod. Fertil. Dev. 1999, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Januskauskas, A.; Johannisson, A.; Rodriguez-Martinez, H. Subtle membrane changes in cryopreserved bull semen in relation with sperm viability, chromatin structure, and field fertility. Theriogenology 2003, 60, 743–758. [Google Scholar] [CrossRef] [PubMed]
- Langendam, M.W.; Magnuson, K.; Williams, A.R.; Walker, V.R.; Howdeshell, K.L.; Rooney, A.A.; Hooijmans, C.R. Developing a database of systematic reviews of animal studies. Regul. Toxicol. Pharmacol. 2021, 123, 104940. (In English) [Google Scholar] [CrossRef] [PubMed]
- Larson-Cook, K.L.; Brannian, J.D.; Hansen, K.A.; Kasperson, K.M.; Aamold, E.T.; Evenson, D.P. Relationship between the outcomes of assisted reproductive techniques and sperm DNA fragmentation as measured by the sperm chromatin structure assay. Fertil. Steril. 2003, 80, 895–902. [Google Scholar] [CrossRef]
- Sharma, R.; Iovine, C.; Agarwal, A.; Henkel, R. TUNEL assay—Standardized method for testing sperm DNA fragmentation. Andrologia 2021, 53, e13738. [Google Scholar] [CrossRef] [PubMed]
- Olive, P.L.; Banáth, J.P. The comet assay: A method to measure DNA damage in individual cells. Nat. Protoc. 2006, 1, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.R. The comet assay for DNA damage and repair. Mol. Biotechnol. 2004, 26, 249–261. [Google Scholar] [CrossRef]
- Ribas-Maynou, J.; García-Peiró, A.; Fernández-Encinas, A.; Abad, C.; Amengual, M.J.; Prada, E.; Navarro, J.; Benet, J. Comprehensive analysis of sperm DNA fragmentation by five different assays: TUNEL assay, SCSA, SCD test and alkaline and neutral Comet assay. Andrology 2013, 1, 715–722. [Google Scholar] [CrossRef]
- de Freitas Pacheco, J.A.; Landaberry, S.J.C.; Singh, P.D. Spectrophotometric observations of the comet Halley during the 1985–86 apparition. Mon. Not. R. Astron. Soc. 1988, 235, 457–464. [Google Scholar] [CrossRef][Green Version]
- McKelvey-Martin, V.J.; Green, M.H.L.; Schmezer, P.; Pool-Zobel, B.L.; De Méo, M.P.; Collins, A. The single cell gel electrophoresis assay (comet assay): A European review. Mutat. Res. Fundam. Mol. Mech. Mutagen. 1993, 288, 47–63. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Gupta, S.; Sharma, R. Andrological Evaluation of Male Infertility; Springer International Publishing: Cham, Switzerland, 2016; pp. 113–133. [Google Scholar]
- Bianchi, P.G.; Manicardi, G.C.; Urner, F.; Campana, A.; Sakkas, D. Chromatin packaging and morphology in ejaculated human spermatozoa: Evidence of hidden anomalies in normal spermatozoa. Mol. Hum. Reprod. 1996, 2, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Lolis, D.; Georgiou, I.; Syrrou, M.; Zikopoulos, K.; Konstantelli, M.; Messinis, I. Chromomycin A3-staining as an indicator of protamine deficiency and fertilization. Int. J. Androl. 1996, 19, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Hosseinifar, H.; Yazdanikhah, S.; Modarresi, T.; Totonchi, M.; Sadighi Gilani, M.; Sabbaghian, M. Correlation between sperm DNA fragmentation index and CMA 3 positive spermatozoa in globozoospermic patients. Andrology 2015, 3, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Sridharan, G.; Shankar, A.A. Toluidine blue: A review of its chemistry and clinical utility. J. Oral Maxillofac. Pathol. 2012, 16, 251–255. (In English) [Google Scholar] [CrossRef] [PubMed]
- Alfred, Y.; Simon, A.; Bruno, A.; Patrice, D.; Boula, S.S.; Jean-Paul, D.; Gangbo, F.; Anatole, L. Assessment of Spermatic Chromatin Decondensation by the Toluidine Blue Assay in Infertile Patients in Cotonou. Adv. Reprod. Sci. 2022, 10, 37–48. [Google Scholar] [CrossRef]
- Kipper, B.H.; Trevizan, J.T.; Carreira, J.T.; Carvalho, I.R.; Mingoti, G.Z.; Beletti, M.E.; Perri, S.H.V.; Franciscato, D.A.; Pierucci, J.C.; Koivisto, M.B. Sperm morphometry and chromatin condensation in Nelore bulls of different ages and their effects on IVF. Theriogenology 2017, 87, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Dogan, S.; Vargovic, P.; Oliveira, R.; Belser, L.E.; Kaya, A.; Moura, A.; Sutovsky, P.; Parrish, J.; Topper, E.; Memili, E. Sperm Protamine-Status Correlates to the Fertility of Breeding Bulls. Biol. Reprod. 2015, 92, 92. [Google Scholar] [CrossRef] [PubMed]
- Gaggini, T.S.; Rocha, L.O.; Souza, E.T.; de Rezende, F.M.; Antunes, R.C.; Beletti, M.E. Head morphometry and chromatin instability in normal boar spermatozoa and in spermatozoa with cytoplasmic droplets. Anim. Reprod. 2018, 14, 1253–1258. [Google Scholar] [CrossRef]
- Xu, L.R.; Carr, M.M.; Bland, A.P.; Hall, G.A. Histochemistry and morphology of porcine mast cells. J. Histochem. 1993, 25, 516–522. (In English) [Google Scholar] [CrossRef]
- Fernández, J.L.; Johnston, S.; Gosálvez, J. Sperm Chromatin Dispersion (SCD) Assay. In A Clinician’s Guide to Sperm DNA and Chromatin Damage; Zini, A., Agarwal, A., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 137–152. [Google Scholar]
- Kim, S.M.; Kim, S.K.; Jee, B.C.; Kim, S.H. Effect of Sperm DNA Fragmentation on Embryo Quality in Normal Responder Women in In Vitro Fertilization and Intracytoplasmic Sperm Injection. Yonsei Med. J. 2019, 60, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Herbert, K.E.; Evans, M.D.; Finnegan, M.T.V.; Farooq, S.; Mistry, N.; Podmore, I.D.; Farmer, P.; Lunec, J. A novel HPLC procedure for the analysis of 8-oxoguanine in DNA. Free Radic. Biol. Med. 1996, 20, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Shigenaga, M.K.; Aboujaoude, E.N.; Chen, Q.; Ames, B.N. [2] Assays of oxidative DNA damage biomarkers 8-oxo-2′-deoxyguanosine and 8-oxoguanine in nuclear DNA and biological fluids by high-performance liquid chromatography with electrochemical detection. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1994; Volume 234, pp. 16–33. [Google Scholar]
- Svoboda, P.; Maekawa, M.; Kawai, K.; Tominaga, T.; Savela, K.; Kasai, H. Urinary 8-hydroxyguanine may be a better marker of oxidative stress than 8-hydroxydeoxyguanosine in relation to the life spans of various species. Antioxid. Redox Signal. 2006, 8, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Kawai, K.; Kasai, H.; Li, Y.S.; Kawasaki, Y.; Watanabe, S.; Ohta, M.; Honda, T.; Yamato, H. Measurement of 8-hydroxyguanine as an oxidative stress biomarker in saliva by HPLC-ECD. Genes Environ. 2018, 40, 5. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Schipper, H.M.; Velly, A.M.; Mohit, S.; Gornitsky, M. Salivary biomarkers of oxidative stress: A critical review. Free Radic. Biol. Med. 2015, 85, 95–104. (In English) [Google Scholar] [CrossRef] [PubMed]
- Guz, J.; Gackowski, D.; Foksinski, M.; Rozalski, R.; Zarakowska, E.; Siomek, A.; Szpila, A.; Kotzbach, M.; Kotzbach, R.; Olinski, R. Comparison of Oxidative Stress/DNA Damage in Semen and Blood of Fertile and Infertile Men. PLoS ONE 2013, 8, e68490. [Google Scholar] [CrossRef]
- Kim, S.W.; Kim, B.; Mok, J.; Kim, E.S.; Park, J. Dysregulation of the Acrosome Formation Network by 8-oxoguanine (8-oxoG) in Infertile Sperm: A Case Report with Advanced Techniques. Int. J. Mol. Sci. 2021, 22, 5857. Available online: https://www.mdpi.com/1422-0067/22/11/5857 (accessed on 22 January 2024). [CrossRef]
| Assay | Predominant Species | Equipment | Technique | Advantages | Limitations |
|---|---|---|---|---|---|
| SCSA | Human, bull, ram, boar, and stallion | Flow cytometry | Measures susceptibility of sperm nuclear DNA to acid-induced denaturation. | Efficient, quantitative assessment. Has shown consistency in results. | Expensive equipment (flow cytometry) and skilled interpretation and training needed. |
| TUNEL | Human, bull, stallion | Flow cytometry or microscopy | Labels free 3′-OH termini on ss-DNA or ds-DNA to detect DNA strand breaks and apoptosis. | High predictive value. Direct, easy to use and efficient. | Expensive equipment if flow cytometry used. Skilled equipment training needed. |
| SCD | Human, ram, bull | Microscopy | Sperm subject to protein depletion treatment and assay relies on response of fragmented or unfragmented sperm DNA by lysis. | Simultaneously preserves sperm morphology. Simple and efficient. No expensive equipment cost. | Observer subjectivity |
| COMET | Human, bull | Microscopy | Assesses level of SSB or DSB in a cell following agarose cell suspension, cell lysis, DNA denaturation, electrophoresis and microscopy. | Can be quantified to determine cells’ degree of DNA fragmentation. Can be used on a smaller population of cells. | Time-consuming. High level of training required to use specific computer programmes. No strong conclusions regarding fertility. |
| TB | Human, bull | Microscopy | Metachromatic dye binds to damaged dense chromatin and phosphate groups of DNA strands. | Simple and inexpensive. Highly predictive of human fertility. | Labour intensive. Can only access a limited number of sperm cells. Observer subjectivity. |
| CMA3 | Human, bull | Flow cytometry or microscopy | CMA3 compete for protamine binding sites to identify endogenous nicks in decondensed abnormal sperm. | Directly related to degree of protamination. Can identify abnormalities in histone–protamine displacement. | Observer subjectivity with microscopy. Expensive equipment if flow cytometry used. |
| Species | ART Method | Sperm DNA Integrity Measure | Correlation/Outcome | Reference |
|---|---|---|---|---|
| SCA | ||||
| Human (n = 165) | ICSI | DFI | DFI lower when pregnancy was achieved (14.86%) than when no embryonic heartbeat detected (17.37%); p = 0.031 Low DFI in spermatozoa corresponded with faster embryo development to reach the blastocyst stage DFI positively correlated with a delay in 8 out of 13 embryonic development periods | [86] |
| Human (n = 420) | ICSI (OO or DO) | SDF before and after SU of fresh and frozen–thawed sperm | SDF increase of 10%; probability of negative pregnancy outcome increased by 1.31 SDF affects pregnancy outcome (OR 0.973, 95% CI 0.948–0.999, R2 0.069; p = 0.037) | [87] |
| Human (n = 377) | ICSI | DFI >30% (mean 39.25) | DFI > 30% decreased number of 8-cell embryos on day 3 (3.97) Number of blastocysts formed on day 5 (1.6), CP (23.13%) and LB (13.43%) from <30% DFI (5.96, 2.44, 35.83%, 28.75%; n = 237); (p = 0.001, p = 0.001, p = 0.05, p = 0.005) Miscarriage rate (9.7%) increased from <30% DFI (7.08%); p = 0.005 | [88] |
| Human (n = 135) | ICSI | SDF > 22.3% | SDF > 22.3% FR (55.1%) lower than sperm > 22.3% SDF (74.9%); p < 0.001. SDF negatively correlates with FR (r = −0.433; p < 0.001). | [89] |
| Human (n = 94) | ICSI | Low DFI (<15%); (n = 50) Mod DFI (15–30%); (n = 31) High DFI (>30%); (n = 13) | High-DFI group was unable to achieve pregnancy following ICSI. | [90] |
| Human (n = 85) | IVF/ICSI | SDF | SDF negatively correlated with FR (r = −0.241; p = 0.045) and implantation rate (r = −0.25; p = 0.042) of samples assessed post-capacitation Asymmetrical nuclei more frequent with increased SDF in capacitated sperm; p < 0.05 Higher SDF lowered ability to develop expanded blastocyts on d6; p < 0.05 | [91] |
| Human (n = 867) | IVF (n = 379) | Low DFI (<30%); (n = 343) High DFI (>30%); (n = 36) | FR higher in High-DFI group (86.9%) than Low DFI (78.4%); p < 0.05 CP lower in High-DFI group (25%) than Low DFI (48.6%) p < 0.05 | [18] |
| Rasa Aragonesa ram (n = 8) | Field fertility odds ratio 1.4–1.7 (HF; n = 4) 0.6–0.9 (LF; n = 4) | SDF | SDF following 6 and 24 h incubation at 37 °C higher in LF (20.27%, 31.24%) than HF (14.42%, 22.32%), p < 0.05, p < 0.01 | [92] |
| Holstein bull (n = 201) | AI (≈533/bull) | SDF | SDF following 0 h incubation negatively correlates with ESBV and EGV (r = −0.45, r = −0.36; p < 0.0001) SDF following 6 h incubation negatively correlates with ESBV and EGV (r = −0.49, r = −0.38; p < 0.0001) | [93] |
| Stallion (n = 11) Moderate fertility = PR <50% (n = 8) Good fertility = PR >50% (n = 3) | Uterine AI (catheter) | SDF in semen cool-stored in spring vs. summer | Negative correlation between SDF and PR (r = −0.619; p < 0.001) Moderate-fertility group has higher SDF at 0 (7.9) and 6 h (15.4) cooled storage compared to good-fertility group (3.83%, 9.58%); p < 0.05 SDF rate higher in sperm cool-stored in summer than in spring and PR lower in summer than spring; p < 0.05 | [94] |
| TUNEL | ||||
| Human (n = 105) | ICSI | DFI > 20% (% TUNEL positive) | DFI > 20% decreased the number of good-quality embryos (6.63), implantation rate (4.9%), and number of pregnancies (3) from <20% DFI (11, 15.79%, 9); p = 0.018, p = 0.002, p = 0.046 | [95] |
| Human (n = 36) | ICSI | SDF | SDF negatively correlated with mean total embryo score (r = −0.64, p < 0.001) and mean transferred embryo score (r = −0.63, p < 0.001) SDF TH 17.6% is predictive of pregnancy (p < 0.021) | [96] |
| Human (n = 303) | IVF/ICSI | SDF | SDF higher in ICSI (6.8%) than IVF (1.9%) group (p < 0.05) SDF negatively correlated with IVF and ICSI FR (r = −0.357; p < 0.001, r = −0.222; p = 0.04) Good embryo rate (p < 0.05) SDF <4% (46.4%) higher than 10–15% SDF (31.6%) in IVF group SDF <4% (45.6%) higher than 10–15% SDF (33.0%) in all samples | [97] |
| Human (n = 45) | IVF | SDF (%TUNEL positive) | SDF negatively correlated with FR; p < 0.05 >55% SDF resulted in lower FR than <35% SDF; p < 0.05 | [98] |
| Human (n = 68) | ICSI | SDF (%TUNEL positive) | SDF higher in non-pregnant group than pregnant group | [98] |
| Holstein bulls (n = 5) | AI | SDF (% TUNEL positive) | SDF higher in LF (below-average fertility) bulls (20–25%) than average or HF (above-average fertility) bulls (<15%); p < 0.05 | [99] |
| Norwegian red bulls (n = 30) | AI | SDF (% TUNEL positive) 4.8–9.4% 9.4–21.2% | 2.2–4.8% SDF 10% significantly higher odds of AI success (p = 0.006) | [100] |
| SCSA | ||||
| Human (n = 2262) | ART (AIH-IUI n = 1185, IVF n = 1221, ICSI n = 216) | High DFI (≥30%) Med DFI (15–30%) Low DFI (≤15%) | AIH-IUI: Early abortion rate increased in High-DFI (27.3%) and Med-DFI (14.6%) groups compared to Low DFI (4.9%); p < 0.05 | [101] |
| Human (n = 1316) | IVF | DFI >11.3% | DFI higher in non-pregnant (17%) than pregnant group (14.9%); p = 0.001 DFI is a predictor of pregnancy outcome (p = 0.023) DFI TH > 11.3% is predictive of pregnancy outcome (AUCROC 0.574, 95% CI 0.541–0.607, SE 56.1%, SP 60%, PPV 77.9%, NPV 35.1%) | [102] |
| Human (n = 266) | ICSI | DFI >30.3% | DFI higher in non-pregnant (31.5%) than pregnant group (26.3%); p = 0.01 DFI is a predictor of pregnancy outcome (p = 0.004) DFI TH > 30.3% is predictive of pregnancy outcome (AUCROC 0.567, 95% CI 0.487–0.647, SE 50.6%, SP 68.8%, PPV 79.3%, NPV 37.0%) | [102] |
| Human (n = 96) | ICSI (n = 155) | 19% DFI TH | DFI TH ≥ 18–19% predicts the outcome of ICSI (p < 0.005) DFI negatively correlates with continuing pregnancies (r = −0.184, p = 0.022), and positively correlates with non-pregnancy (r = 0.197, p = 0.014) Continuing pregnancy rate and implantation rate lower in ≥19% DFI group (14.9%, 12.1%) than <19% DFI (34.6%, 27.2%); p = 0.005, 0.001 Non-pregnancy rate significantly higher in ≥19% DFI group (75.7%) than <19% DFI (55.6%) p = 0.008 | [103] |
| Ram (n = 15) | Vaginal AI | Mean DFI and heterogeneity (SD DFI) of SDF in the total sperm population | Mean DFI negatively associated with 25 d NRR (OR 0.98, 95% CI 0.97–1, p = 0.039) SD DFI negatively associated with 25 d NRR (OR 0.98, 95% CI 0.97–0.99, p = 0.001 | [16] |
| Finnish Ayrshire bull (n = 43) >55% 60 d NRR (F; n = 21) <55% 60 d NRR (SF; n = 22) | AI (n ≈ 5964/bull) | DFI SD-DFI HDS | HDS higher in F (0.61%) than SF bulls (0.48%); p < 0.05 HDS positively correlated with calving rate (r = 0.31; p < 0.05) | [12] |
| Norwegian red bull (HF; n = 19) (LF; n = 18) | AI | DFI HDS | HF bulls had lower DFI and HDS (1.84%, 2.93%) than LF bulls (3.5%, 4.31%) p < 0.01 DFI and HDS negatively correlate with 56 d NRR56 (r = −0.57, p = 0.0003, r = −0.37, p = 0.026) DFI significantly predicts 56 d NRR (p < 0.01) | [104] |
| Norwegian red bulls (n = 30) | AI | DFI | 7.5–21.6% DFI reduced odds of AI success from average (6%; p = 0.011) 1.6–3.8% DFI increased odds of AI success from average (7%; p = 0.010) | [100] |
| Swedish red bull (n = 14) Holstein bull (n = 6) | AI | DFI 3.31% TH Below-average 56 d NRR (BAB, n = 5) Average 56 d NRR (AB, n = 9) Above-average 56 d NRR (AAB, n = 6) | DFI decreased in AAB (2.88%) compared to BAB and AB (6.23%, 4.65%); p < 0.05 DFI negatively correlated with adjusted 56 d NRR (r = −0.61; p = 0.01) DFI can differentiate between BAB and AAB (R2 = 0.56; p = 0.02) DFI TH 3.31% accurately predicts 56 d NRR (SE 66.7%, SP 100%, AUCROC 0.8) | [11] |
| Holstein bulls (n = 20) | AI | DFI (COMP αt) | DFI negatively correlated to NRR (r = −0.60 p < 0.01) DFI lower in mature bulls than in young bulls (p < 0.01) | [105] |
| Holstein bulls (n = 19) | AI (n = 192) | SDF | SDF negatively correlated with 56 d NRR (r = −0.287, r2 = 0.082; p < 0.05) | [106] |
| Bulls: Holstein (n = 156) Jersey (n = 39) | AI (n = 75,610) | DFI SD-DFI HDS | Sperm without DFI (97.5%) predicts 56 d NRR (p < 0.0001) Sperm with moderate DFI (2.4%) predicts 56 d NRR (p < 0.0001) Sperm with High DFI (0.2%) predicts 56 d NRR (p < 0.0003) SD-DFI (33.3%) predicts 56 d NRR (p < 0.0001) HDS (2.8%) predicts 56 d NRR (p < 0.0004) | [107] |
| Boar (n = 18) | AI | Mean DFI SD DFI of SDF in the total sperm population | DFI negatively correlated with farrowing rate (r = −0.55, p < 0.01) and ANB (r = −0.54, p < 0.01) SD DFI negatively correlated with farrowing rate (r = −0.67, p < 0.002) and ANB (r = −0.54, p < 0.02) DFI TH 6% is predictive of farrowing rate and ANB (OR 1.5, 95% CI 1.21–1.94, p = 0.0003, SE 83%) SD DFI TH 40 is predictive of farrowing rate and ANB (OR 2.5, 95% CI 1.87–3.32, p = 0.001, SE 92%) | [15] |
| Boar (n = 160) | AI | DFI > 3% DFI > 2.1% | DFI > 3% (0 h storage) reduced ANB/litter (13.9) from DFI < 3% (14.87–14.94); p < 0.01 For Landrace and Danish Large White boars, DFI > 2.1% after 24 h 18 °C storage had lower litter size (14.4, 14.2) than DFI < 2.1% (15.1, 15.1); p < 0.01 | [108] |
| Stallion (n = 41) | AI | DFI SD DFI Mean DFI | DFI negatively correlated with PR (r = −0.63, p < 0.05) | [109] |
| COMET | ||||
| Human (n = 339) | IVF (n = 203) ICSI (n = 136) | >50% SDF (Comet Score) TH | SDF higher in non-pregnant group than LB or miscarriage groups (p < 0.05) following IVF SDF TH > 50% reduced pregnancy (16.2, p = 0.005) and LB rates (13.1%, p = 0.007) following IVF | [110] |
| Bulls (n = 45) | IVP (-ET) | Mean-DNA Mean H-DNA T-DNA | T-DNA higher in group 4 (8.53%) than group 3 (4.31%); p < 0.05 Mean-DNA and Mean H-DNA negatively correlated with blastocyst rate (r = −0.375, r = 0.389; p = 0.02, p = 0.016) | [111] |
| Italian Mediterranean Buffalo bulls (n = 3) | AI (n = 528) | % H-DNA % T-DNA TM OTM | % H-DNA TH ≥ 86% (and its relative % T-DNA < 14%) predicts successful d30/45 pregnancy (AUCROC 0.56, SE 81%, SP 26%; p < 0.05) Tail area TH ≤ 58 μm2 predicts successful d30/45 pregnancy (AUCROC 0.56, SE 80%, SP 26%; p < 0.05) | [112] |
| Nili–Ravi Water Buffalo bull (n = 5) | AI (n = 514) | Comet length, % H-DNA, %T-DNA, tail length, TM, OTM | Tail length negatively correlated with fertility rate (r = −0.7; p = 0.04) | [113] |
| Buffalo bulls (n = 6) (HF; n = 3) (LF; n = 3) | IVP (-ET) | SDF (% of cells comet tail+) | SDF higher in LF group (18.72%) than HF group (8.94%; p < 0.05). | [114] |
| TB | ||||
| Humans (n = 1386) | Infertile and normospermic | SDF% | Mean TB staining was higher in infertile group than normospermic (p = 0.005) | [115] |
| Human (n = 142) | Infertile and fertile men | %TB dark cells %TB light cells | TB dark cells and light cells had 92% and 90% specificity, respectively, for predicting infertility Both poor predictors of fertility (42 and 32% sensitivity, respectively) | [116] |
| Bull (n = 8) | Fertile and sub-fertile (subjects to scrotal insulation) | Chromatin alteration types (Base, Basal half, Central axis, Dispersed and Whole) | Greater (p < 0.01) chromatin decondensation and heterogeneity were recorded in sub-fertile bulls | [117] |
| CMA3 | ||||
| Human (n = 139) | IVF | %CMA3 positivity | %CMA3 positivity has negative correlation with farrowing rate %CMA3 positivity significant difference between fertilising and non-fertilising patients. | [118] |
| Human (n = 30) | ICSI | %CMA3 | %CMA3 positivity showed significant negative correlation with FR | [119] |
| Bulls (n = 12) (140 bulls ranked on embryo development rate, chose bottom 6 and top 6) | IVP | %CMA3 | Significant difference in %CMA3 between groups (p = 0.03), suggesting greater protamine deficiency in the high fertility group | [120] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robertson, M.J.; Chambers, C.; Spanner, E.A.; de Graaf, S.P.; Rickard, J.P. The Assessment of Sperm DNA Integrity: Implications for Assisted Reproductive Technology Fertility Outcomes across Livestock Species. Biology 2024, 13, 539. https://doi.org/10.3390/biology13070539
Robertson MJ, Chambers C, Spanner EA, de Graaf SP, Rickard JP. The Assessment of Sperm DNA Integrity: Implications for Assisted Reproductive Technology Fertility Outcomes across Livestock Species. Biology. 2024; 13(7):539. https://doi.org/10.3390/biology13070539
Chicago/Turabian StyleRobertson, Maya J., Caitlin Chambers, Eloise A. Spanner, Simon P. de Graaf, and Jessica P. Rickard. 2024. "The Assessment of Sperm DNA Integrity: Implications for Assisted Reproductive Technology Fertility Outcomes across Livestock Species" Biology 13, no. 7: 539. https://doi.org/10.3390/biology13070539
APA StyleRobertson, M. J., Chambers, C., Spanner, E. A., de Graaf, S. P., & Rickard, J. P. (2024). The Assessment of Sperm DNA Integrity: Implications for Assisted Reproductive Technology Fertility Outcomes across Livestock Species. Biology, 13(7), 539. https://doi.org/10.3390/biology13070539








