Trait Variation and Spatiotemporal Dynamics across Avian Secondary Contact Zones
Abstract
Simple Summary
Abstract
1. Introduction
2. Trait Divergence in Avian SCZs
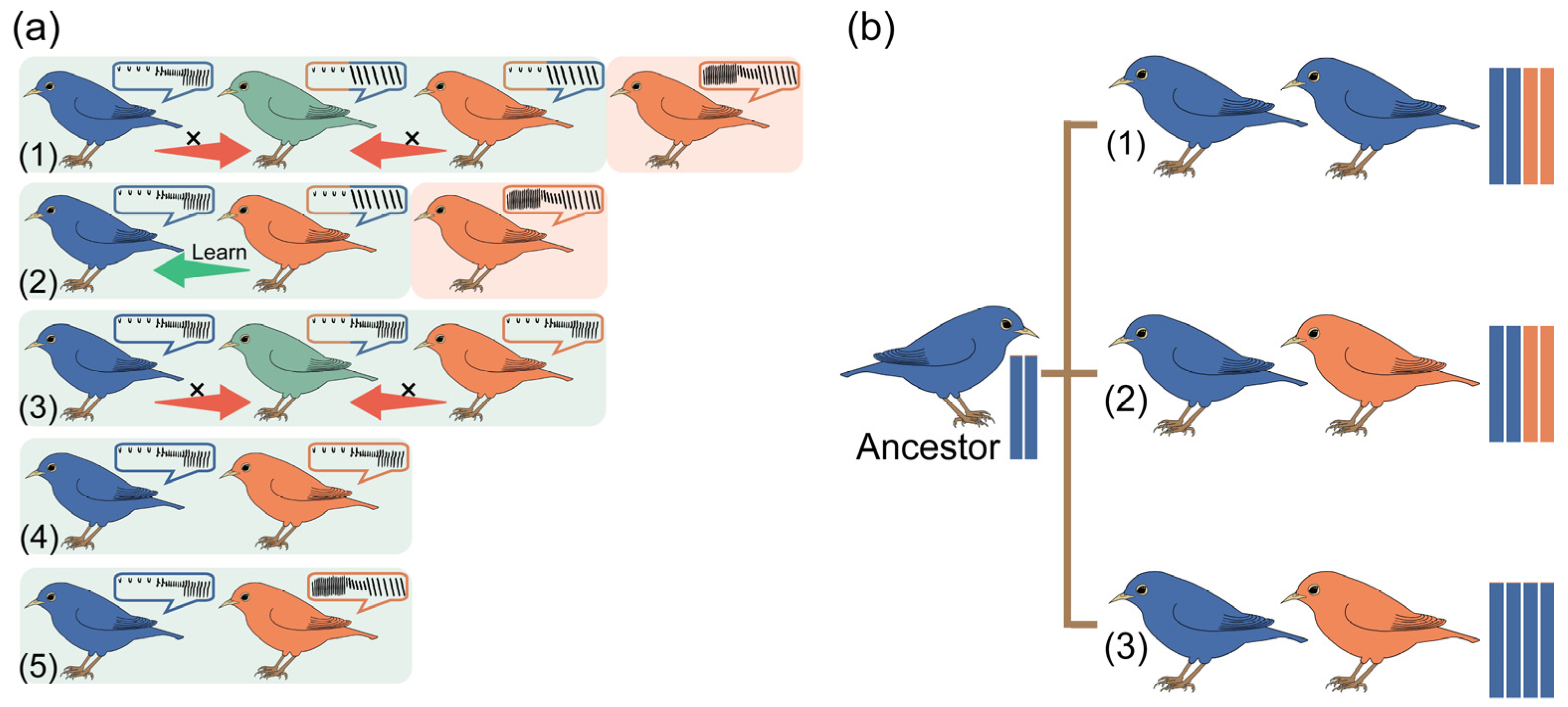
2.1. Vocalization
2.2. Plumage
2.3. Beak Morphology
2.4. Migratory Behavior
3. Spatiotemporal Dynamics
4. Perspectives
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Etges, W.J.; Strand, A.E.; Williams, L.M.; Oleksiak, M.F.; Sotka, E.E. Can Diversifying Selection Be Distinguished from History in Geographic Clines? A Population Genomic Study of Killifish (Fundulus heteroclitus). PLoS ONE 2012, 7, e45138. [Google Scholar]
- Hewitt, G.M. Hybrid zones—Natural laboratories for evolutionary studies. Trends Ecol. Evol. 1988, 3, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, G. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef]
- Hewitt, G.M. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 1996, 58, 247–276. [Google Scholar] [CrossRef]
- Shafer, A.B.A.; Cullingham, C.I.; Côté, S.D.; Coltman, D.W. Of glaciers and refugia: A decade of study sheds new light on the phylogeography of northwestern North America. Mol. Ecol. 2010, 19, 4589–4621. [Google Scholar] [CrossRef]
- Galbreath, K.E.; Hafner, D.J.; Zamudio, K.R. When cold is better: Climate-driven elevation shifts yield complex patterns of diversification and demography in an alpine specialist (American pika, Ochotona princeps). Evolution 2009, 63, 2848–2863. [Google Scholar] [CrossRef]
- Hewitt, G.M. Quaternary phylogeography: The roots of hybrid zones. Genetica 2011, 139, 617–638. [Google Scholar] [CrossRef]
- Kaya, S.; Kabasakal, B.; Erdogan, A. Geographic Genetic Structure of Alectoris chukar in Turkiye: Post-LGM-Induced Hybridization and Human-Mediated Contaminations. Biology 2023, 12, 401. [Google Scholar] [CrossRef] [PubMed]
- Machado, A.P.; Cumer, T.; Iseli, C.; Beaudoing, E.; Ducrest, A.-L.; Dupasquier, M.; Guex, N.; Dichmann, K.; Lourenco, R.; Lusby, J.; et al. Unexpected post-glacial colonisation route explains the white colour of barn owls (Tyto alba) from the British Isles. Mol. Ecol. 2022, 31, 482–497. [Google Scholar] [CrossRef]
- Marques, V.; Hinojosa, J.C.; Dapporto, L.; Talavera, G.; Stefanescu, C.; Gutierrez, D.; Vila, R. The opposed forces of differentiation and admixture across glacial cycles in the butterfly Aglais Urticae. Mol. Ecol. 2024, 33, e17304. [Google Scholar] [CrossRef]
- Nolte, A.W.; Freyhof, J.; Tautz, D. When invaders meet locally adapted types:: Rapid moulding of hybrid zones between sculpins (Cottus, Pisces) in the Rhine system. Mol. Ecol. 2006, 15, 1983–1993. [Google Scholar] [CrossRef] [PubMed]
- Carantón-Ayala, D.; Avendaño, J.E.; Cadena, C.D. Hybridization in brushfinches (Atlapetes, Emberizidae) from the southeast Andes of Colombia: A consequence of habitat disturbance? J. Ornithol. 2018, 159, 713–722. [Google Scholar] [CrossRef]
- Shipilina, D.; Serbyn, M.; Ivanitskii, V.; Marova, I.; Backström, N. Patterns of genetic, phenotypic, and acoustic variation across a chiffchaff (Phylloscopus collybita abietinus/tristis) hybrid zone. Ecol. Evol. 2017, 7, 2169–2180. [Google Scholar] [CrossRef] [PubMed]
- Farrington, H.L.; Lawson, L.P.; Clark, C.M.; Petren, K. The evolutionary history of Darwin’s finches: Speciation, gene flow, and introgression in a fragmented landscape. Evolution 2014, 68, 2932–2944. [Google Scholar] [CrossRef]
- Abbott, R.; Albach, D.; Ansell, S.; Arntzen, J.W.; Baird, S.J.E.; Bierne, N.; Boughman, J.; Brelsford, A.; Buerkle, C.A.; Buggs, R.; et al. Hybridization and speciation. J. Evol. Biol. 2013, 26, 229–246. [Google Scholar] [CrossRef] [PubMed]
- Barrera-Guzmán, A.O.; Aleixo, A.; Shawkey, M.D.; Weir, J.T. Hybrid speciation leads to novel male secondary sexual ornamentation of an Amazonian bird. Proc. Natl. Acad. Sci. USA 2018, 115, E218–E225. [Google Scholar] [CrossRef]
- Seehausen, O. Conservation: Losing biodiversity by reverse speciation. Curr. Biol. 2006, 16, R334–R337. [Google Scholar] [CrossRef]
- Todesco, M.; Pascual, M.A.; Owens, G.L.; Ostevik, K.L.; Moyers, B.T.; Hübner, S.; Heredia, S.M.; Hahn, M.A.; Caseys, C.; Bock, D.G.; et al. Hybridization and extinction. Evol. Appl. 2016, 9, 892–908. [Google Scholar] [CrossRef]
- Vali, U.; Dombrovski, V.; Treinys, R.; Bergmanis, U.; Daroczi, S.J.; Dravecky, M.; Ivanovski, V.; Lontkowski, J.; Maciorowski, G.; Meyburg, B.-U.; et al. Widespread hybridization between the Greater Spotted Eagle Aquila clanga and the Lesser Spotted Eagle Aquila pomarina (Aves: Accipitriformes) in Europe. Biol. J. Linn. Soc. 2010, 100, 725–736. [Google Scholar] [CrossRef]
- Beysard, M.; Heckel, G. Structure and dynamics of hybrid zones at different stages of speciation in the common vole (Microtus arvalis). Mol. Ecol. 2014, 23, 673–687. [Google Scholar] [CrossRef]
- Buggs, R.J. Empirical study of hybrid zone movement. Heredity 2007, 99, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Menon, M.; Landguth, E.; Leal-Saenz, A.; Bagley, J.C.; Schoettle, A.W.; Wehenkel, C.; Flores-Renteria, L.; Cushman, S.A.; Waring, K.M.; Eckert, A.J. Tracing the footprints of a moving hybrid zone under a demographic history of speciation with gene flow. Evol. Appl. 2020, 13, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Milá, B.; McCormack, J.E.; Castañeda, G.; Wayne, R.K.; Smith, T.B. Recent postglacial range expansion drives the rapid diversification of a songbird lineage in the genus Junco. Proc Biol Sci. 2007, 274, 2653–2660. [Google Scholar]
- Rising, J.D. Morphological variation and evolution in some North-American orioles. Syst. Zool. 1970, 19, 315. [Google Scholar] [CrossRef]
- Sibley, C.G.; Short, L.L., Jr. Hybridization in some Indian Bulbuls Pycnonotus cafer × P. Leucogenys. Ibis 1959, 101, 177–182. [Google Scholar] [CrossRef]
- Heaney, L.R.; Timm, R.M. Morphology, genetics, and ecology of pocket gophers (genus Geomys) in a narrow hybrid zone. Biol. J. Linn. Soc. 1985, 25, 301–317. [Google Scholar] [CrossRef]
- Willett, C.S.; Ford, M.J.; Harrison, R.G. Inferences about the origin of a field cricket hybrid zone from a mitochondrial DNA phylogeny. Heredity 1997, 79, 484–494. [Google Scholar] [CrossRef][Green Version]
- Bernatchez, L.; Dodson, J.J. Allopatric origin of sympatric populations of lake whitefish (Coregonus clupeaformis) as revealed by mitochondrial-dna restriction analysis. Evolution 1990, 44, 1263–1271. [Google Scholar] [PubMed]
- Latta, R.G.; Mitton, J.B. Historical separation and present gene flow through a zone of secondary contact in ponderosa pine. Evolution 1999, 53, 769–776. [Google Scholar] [CrossRef]
- Vijverberg, K.; Kuperus, P.; Breeuwer, J.A.J.; Bachmann, K. Incipient adaptive radiation of New Zealand and Australian Microseris (Asteraceae): An amplified fragment length polymorphism (AFLP) study. J. Evol. Biol. 2000, 13, 997–1008. [Google Scholar] [CrossRef]
- Arnegard, M.E.; Markert, J.A.; Danley, P.D.; Stauffer, J.R.; Ambali, A.J.; Kocher, T.D. Population structure and colour variation of the cichlid fish Labeotropheus fuelleborni Ahl along a recently formed archipelago of rocky habitat patches in southern Lake Malawi. Proc. R. Soc. B Biol. Sci. 1999, 266, 119–130. [Google Scholar] [CrossRef]
- Hogner, S.; Riera, A.B.; Wold, M.; Lifjeld, J.T.; Johnsen, A. Intergeneric hybridization between Common Redstart Phoenicurus phoenicurus and Whinchat Saxicola rubetra revealed by molecular analyses. J. Ornithol. 2015, 156, 829–836. [Google Scholar] [CrossRef]
- da Silva Coelho, F.A.; Gill, S.; Tomlin, C.M.; Papavassiliou, M.; Farley, S.D.; Cook, J.A.; Sonsthagen, S.A.; Sage, G.K.; Heaton, T.H.; Talbot, S.L.; et al. Ancient bears provide insights into Pleistocene ice age refugia in Southeast Alaska. Mol. Ecol. 2023, 32, 3641–3656. [Google Scholar] [CrossRef]
- Bensch, S.; Andersson, T.; Åkesson, S. Morphological and molecular variation across a migratory divide in willow warblers, Phylloscopus Trochilus. Evolution 1999, 53, 1925–1935. [Google Scholar] [CrossRef]
- Avise, J.C.; Avise, J.C. Molecular Markers, Natural History and Evolution; Springer Science & Business Media: Berlin/Heidelberg, Germany, 1994; Volume i–xiv, pp. 1–511. [Google Scholar]
- Eliásová, K.; Lledó, J.I.L.; Grau, J.H.; Loudová, M.; Bannikova, A.A.; Zolotareva, K.; Benes, V.; Hulva, P.; Bolfíková, B.C. Contrasting levels of hybridization across the two contact zones between two hedgehog species revealed by genome-wide SNP data. Heredity 2022, 129, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Stemshorn, K.C.; Reed, F.A.; Nolte, A.W.; Tautz, D. Rapid formation of distinct hybrid lineages after secondary contact of two fish species (Cottus sp.). Mol. Ecol. 2011, 20, 1475–1491. [Google Scholar] [CrossRef]
- McKinney, G.J.; Larson, W.A.; Seeb, L.W.; Seeb, J.E. RADseq provides unprecedented insights into molecular ecology and evolutionary genetics: Comment on Breaking RAD by Lowry et al. (2016). Mol. Ecol. Resour. 2017, 17, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Stiller, J.; Feng, S.; Chowdhury, A.-A.; Rivas-Gonzalez, I.; Duchene, D.A.; Fang, Q.; Deng, Y.; Kozlov, A.; Stamatakis, A.; Claramunt, S.; et al. Complexity of avian evolution revealed by family-level genomes. Nature 2024, 629, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.; Wu, L.; Zhang, D.; Wang, H.; Dong, F.; Yang, L.; Wang, S.; Amano, H.E.; Zhang, W.; Jia, C.; et al. Landscape Heterogeneity Explains the Genetic Differentiation of a Forest Bird across the Sino-Himalayan Mountains. Mol. Biol. Evol. 2024, 41, msae027. [Google Scholar] [CrossRef]
- Zhang, D.; Rheindt, F.E.; She, H.; Cheng, Y.; Song, G.; Jia, C.; Qu, Y.; Alstrom, P.; Lei, F. Most Genomic Loci Misrepresent the Phylogeny of an Avian Radiation Because of Ancient Gene Flow. Syst. Biol. 2021, 70, 961–975. [Google Scholar] [CrossRef]
- Xiong, X.; Liu, J.; Rao, Y. Whole Genome Resequencing Helps Study Important Traits in Chickens. Genes 2023, 14, 1198. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; She, H.; Rheindt, F.E.; Wu, L.; Wang, H.; Zhang, K.; Cheng, Y.; Song, G.; Jia, C.; Qu, Y.; et al. Genomic and phenotypic changes associated with alterations of migratory behaviour in a songbird. Mol. Ecol. 2023, 32, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Burri, R.; Nater, A.; Kawakami, T.; Mugal, C.F.; Olason, P.I.; Smeds, L.; Suh, A.; Dutoit, L.; Bures, S.; Garamszegi, L.Z.; et al. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Res. 2015, 25, 1656–1665. [Google Scholar] [CrossRef]
- Ottenburghs, J.; Honka, J.; Müsken, G.; Ellegren, H. Recent introgression between Taiga Bean Goose and Tundra Bean Goose results in a largely homogeneous landscape of genetic differentiation. Heredity 2020, 125, 73–84. [Google Scholar] [CrossRef]
- Jeffery, N.W.; Bradbury, I.R.; Stanley, R.R.E.; Wringe, B.F.; Van Wyngaarden, M.; Ben Lowen, J.; McKenzie, C.H.; Matheson, K.; Sargent, P.S.; DiBacco, C. Genomewide evidence of environmentally mediated secondary contact of European green crab (Carcinus maenas) lineages in eastern North America. Evol. Appl. 2018, 11, 869–882. [Google Scholar] [CrossRef]
- Morales, H.E.; Sunnucks, P.; Joseph, L.; Pavlova, A. Perpendicular axes of differentiation generated by mitochondrial introgression. Mol. Ecol. 2017, 26, 3241–3255. [Google Scholar] [CrossRef]
- Jetz, W.; Thomas, G.H.; Joy, J.B.; Hartmann, K.; Mooers, A.O. The global diversity of birds in space and time. Nature 2012, 491, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Zhang, R.Y.; Machado-Stredel, F.; Alstrom, P.; Johansson, U.S.; Irestedt, M.; Mays, H.L.; McKay, B.D.; Nishiumi, I.; Cheng, Y.L.; et al. Great journey of Great Tits (Parus major group): Origin, diversification and historical demographics of a broadly distributed bird lineage. J. Biogeogr. 2020, 47, 1585–1598. [Google Scholar] [CrossRef]
- Jiang, Z.Y.; Song, G.; Luo, X.; Zhang, D.Z.; Lei, F.M.; Qu, Y.H. Recurrent selection and reduction in recombination shape the genomic landscape of divergence across multiple population pairs of Green-backed Tit. Evol. Lett. 2023, 7, 99–111. [Google Scholar] [CrossRef]
- Jarvis, E.D.; Mirarab, S.; Aberer, A.J.; Li, B.; Houde, P.; Li, C.; Ho, S.Y.W.; Faircloth, B.C.; Nabholz, B.; Howard, J.T.; et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 2014, 346, 1320–1331. [Google Scholar] [CrossRef]
- Prum, R.O.; Berv, J.S.; Dornburg, A.; Field, D.J.; Townsend, J.P.; Lemmon, E.M.; Lemmon, A.R. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 2015, 526, 569-U247. [Google Scholar] [CrossRef]
- Feng, S.H.; Stiller, J.; Deng, Y.; Armstrong, J.; Fang, Q.; Reeve, A.H.; Xie, D.; Chen, G.J.; Guo, C.X.; Faircloth, B.C.; et al. Dense sampling of bird diversity increases power of comparative genomics. Nature 2021, 592, E24. [Google Scholar] [CrossRef]
- Qu, Y.H.; Chen, C.H.; Xiong, Y.; She, H.S.; Zhang, Y.E.; Cheng, Y.L.; DuBay, S.; Li, D.M.; Ericson, P.G.P.; Hao, Y.; et al. Rapid phenotypic evolution with shallow genomic differentiation during early stages of high elevation adaptation in Eurasian Tree Sparrows. Natl. Sci. Rev. 2020, 7, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.H.; Chen, C.H.; Chen, X.M.; Hao, Y.; She, H.S.; Wang, M.X.; Ericson, P.G.P.; Lin, H.Y.; Cai, T.L.; Song, G.; et al. The evolution of ancestral and species-specific adaptations in snowfinches at the Qinghai-Tibet Plateau. Proc. Natl. Acad. Sci. USA 2021, 118, e2012398118. [Google Scholar] [CrossRef]
- Hao, Y.; Song, G.; Zhang, Y.E.; Zhai, W.W.; Jia, C.X.; Ji, Y.Z.; Tang, S.Y.; Lv, H.R.; Qu, Y.H.; Lei, F.M. Divergent contributions of coding and noncoding sequences to initial high-altitude adaptation in passerine birds endemic to the Qinghai-Tibet Plateau. Mol. Ecol. 2023, 32, 3524–3540. [Google Scholar] [CrossRef] [PubMed]
- Gwee, C.Y.; Garg, K.M.; Chattopadhyay, B.; Sadanandan, K.R.; Prawiradilaga, D.M.; Irestedt, M.; Lei, F.M.; Bloch, L.M.; Lee, J.G.H.; Irham, M.; et al. Phylogenomics of white-eyes, a ‘great speciator’, reveals Indonesian archipelago as the center of lineage diversity. eLife 2020, 9, e62765. [Google Scholar] [CrossRef]
- Dai, C.Y.; Feng, P. Multiple concordant cytonuclear divergences and potential hybrid speciation within a species complex in Asia. Mol. Phylogenetics Evol. 2023, 180, 107709. [Google Scholar] [CrossRef]
- Konishi, M.; Emlen, S.T.; Ricklefs, R.E.; Wingfield, J.C. Contributions of bird studies to biology. Science 1989, 246, 465–472. [Google Scholar] [CrossRef]
- Ottenburghs, J. Exploring the hybrid speciation continuum in birds. Ecol. Evol. 2018, 8, 13027–13034. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Jiao, X.L.; Zhang, D.Z.; Cheng, Y.L.; Song, G.; Qu, Y.H.; Lei, F.M. Comparative Genomics and Evolution of Avian Specialized Traits. Curr. Genom. 2021, 22, 496–511. [Google Scholar] [CrossRef]
- Kapusta, A.; Suh, A. Evolution of bird genomes-a transposon's-eye view. Ann. N. Y. Acad. Sci. 2017, 1389, 164–185. [Google Scholar] [CrossRef] [PubMed]
- Hillier, L.W.; Miller, W.; Birney, E.; Warren, W.; Hardison, R.C.; Ponting, C.P.; Bork, P.; Burt, D.W.; Groenen, M.A.M.; Delany, M.E.; et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 2004, 432, 695–716. [Google Scholar]
- Warren, W.C.; Clayton, D.F.; Ellegren, H.; Arnold, A.P.; Hillier, L.W.; Künstner, A.; Searle, S.; White, S.; Vilella, A.J.; Fairley, S.; et al. The genome of a songbird. Nature 2010, 464, 757–762. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.H.; Zhao, H.W.; Han, N.J.; Zhou, G.Y.; Song, G.; Gao, B.; Tian, S.L.; Zhang, J.B.; Zhang, R.Y.; Meng, X.H.; et al. Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat. Commun. 2013, 4, 2071. [Google Scholar] [CrossRef]
- Leroy, H.; Bowie, R.C.K.; Rubacova, L.; Matysiokova, B.; Remes, V. A late burst of colour evolution in a radiation of songbirds (Passeriformes: Parulidae) suggests secondary contact drives signal divergence. J. Evol. Biol. 2024, 37, 401–413. [Google Scholar] [CrossRef]
- DeRaad, D.A.; Applewhite, E.E.; Tsai, W.L.E.; Terrill, R.S.; Kingston, S.E.; Braun, M.J.; McCormack, J.E. Hybrid zone or hybrid lineage: A genomic reevaluation of Sibley’s classic species conundrum in Pipilo towhees. Evolution 2023, 77, 852–869. [Google Scholar] [CrossRef]
- Pulido-Santacruz, P.; Aleixo, A.; Weir, J.T. Genomic data reveal a protracted window of introgression during the diversification of a neotropical woodcreeper radiation. Evolution 2020, 74, 842–858. [Google Scholar] [CrossRef]
- Ocampo, D.; Winker, K.; Miller, M.J.; Sandoval, L.; Uy, J.A.C. Replicate contact zones suggest a limited role of plumage in reproductive isolation among subspecies of the variable seedeater (Sporophila corvina). Mol. Ecol. 2023, 32, 3586–3604. [Google Scholar] [CrossRef]
- Nichols, P.; Genner, M.J.; van Oosterhout, C.; Smith, A.; Parsons, P.; Sungani, H.; Swanstrom, J.; Joyce, D.A. Secondary contact seeds phenotypic novelty in cichlid fishes. Proc. R. Soc. B Biol. Sci. 2015, 282, 20142272. [Google Scholar] [CrossRef]
- Parsons, K.J.; Son, Y.H.; Albertson, R.C. Hybridization Promotes Evolvability in African Cichlids: Connections between Transgressive Segregation and Phenotypic Integration. Evol. Biol. 2011, 38, 306–315. [Google Scholar] [CrossRef]
- Funk, E.R.; Taylor, S.A. High-throughput sequencing is revealing genetic associations with avian plumage color. Auk 2019, 136, ukz048. [Google Scholar] [CrossRef]
- Brenowitz, E.A.; Margoliash, D.; Nordeen, K.W. An introduction to birdsong and the avian song system. J. Neurobiol. 1997, 33, 495–500. [Google Scholar] [CrossRef]
- Nowicki, S.; Searcy, W.A. Song function and the evolution of female preferences—Why birds sing, why brains matter. In Behavioral Neurobiology of Birdsong; Zeigler, H.P., Marler, P., Eds.; The New York Academy of Sciences: New York, NY, USA, 2004; pp. 704–723. [Google Scholar]
- Alstrom, P.; Ranft, R. The use of sounds in avian systematics and the importance of bird sound archives. Bull. Br. Ornithol. Club 2003, 123A, 114–135. [Google Scholar]
- Wei, C.; Sangster, G.; Olsson, U.; Rasmussen, P.C.; Svensson, L.; Yao, C.-T.; Carey, G.J.; Leader, P.J.; Zhang, R.; Chen, G.; et al. Cryptic species in a colorful genus: Integrative taxonomy of the bush robins (Aves, Muscicapidae, Tarsiger) suggests two overlooked species. Mol. Phylogenetics Evol. 2022, 175, 107580. [Google Scholar] [CrossRef]
- Liou, L.W.; Price, T.D. Speciation by reinforcement of premating isolation. Evolution 1994, 48, 1451–1459. [Google Scholar] [CrossRef]
- Servedio, M.R.; Noor, M.A.F. The role of reinforcement in speciation: Theory and data. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 339–364. [Google Scholar] [CrossRef]
- Irwin, D.E.; Price, T. Sexual imprinting, learning and speciation. Heredity 1999, 82, 347–354. [Google Scholar] [CrossRef]
- Vabishchevich, A.P.; Formozov, N.A. Song variability in Pied Flycatchers Ficedula hypoleuca: Impact of the sympatry with Collared Flycatchers F. Albicollis. Acta Ornithol. 2010, 45, 189–202. [Google Scholar] [CrossRef]
- Secondi, J.; Bordas, P.; Hipsley, C.A.; Bensch, S. Bilateral Song Convergence in a Passerine Hybrid Zone: Genetics Contribute in One Species Only. Evol. Biol. 2011, 38, 441–452. [Google Scholar] [CrossRef]
- Vokurková, J.; Petrusková, T.; Reifová, R.; Kozman, A.; Morkovsky, L.; Kipper, S.; Weiss, M.; Reif, J.; Dolata, P.T.; Petrusek, A. The Causes and Evolutionary Consequences of Mixed Singing in Two Hybridizing Songbird Species (Luscinia spp.). PLoS ONE 2013, 8, e60172. [Google Scholar] [CrossRef]
- Qvarnström, A.; Haavie, J.; Sæther, S.A.; Eriksson, D.; Pärt, T. Song similarity predicts hybridization in flycatchers. J. Evol. Biol. 2006, 19, 1202–1209. [Google Scholar] [CrossRef]
- Cros, E.; Rheindt, F.E. Massive bioacoustic analysis suggests introgression across Pleistocene land bridges in Mixornis tit-babblers. J. Ornithol. 2017, 158, 407–419. [Google Scholar] [CrossRef]
- Reif, J.; Jiran, M.; Reifová, R.; Vokurková, J.; Dolata, P.T.; Petrusek, A.; Petrusková, T. Interspecific territoriality in two songbird species: Potential role of song convergence in male aggressive interactions. Anim. Behav. 2015, 104, 131–136. [Google Scholar] [CrossRef]
- Souriau, A.; Kohoutová, H.; Reif, J.; Vokurková, J.; Petrusek, A.; Reifová, R.; Petrusková, T. Can mixed singing facilitate coexistence of closely related nightingale species? Behav. Ecol. 2018, 29, 925–932. [Google Scholar] [CrossRef]
- Alatalo, R.V.; Eriksson, D.; Gustafsson, L.; Lundberg, A. Hybridization between pied and collared flycatchers—Sexual selection and speciation theory. J. Evol. Biol. 1990, 3, 375–389. [Google Scholar] [CrossRef]
- Haavie, J.; Borge, T.; Bures, S.; Garamszegi, L.Z.; Lampe, H.M.; Moreno, J.; Qvarnström, A.; Török, J.; Sætre, G.P. Flycatcher song in allopatry and sympatry: Convergence, divergence and reinforcement. J. Evol. Biol. 2004, 17, 227–237. [Google Scholar] [CrossRef]
- Nazarenko, A.A.; Valchuk, O.P.; Martens, J. Secondary contact and overlap of Parus major and Parus minor populations in the middle Amur River basin. Zool. Zhurnal 1999, 78, 372–381. [Google Scholar]
- Päckert, M.; Martens, J.; Eck, S.; Nazarenko, A.A.; Valchuk, O.P.; Petri, B.; Veith, M. The great tit (Parus major): A misclassified ring species. Biol. J. Linn. Soc. 2005, 86, 153–174. [Google Scholar] [CrossRef]
- Clausen, P.; Toft, S. Mixed singers and imitation singers among short-toed treecreepers. Br. Birds 1988, 81, 496–503. [Google Scholar]
- Martens, J.; Meincke, C. Territorial song on the siberian chiffchaff (Phylloscopus-collybita-tristis) and playback experiments within a central-european population (Ph-c-collybita). J. Fur Ornithol. 1989, 130, 455–473. [Google Scholar] [CrossRef]
- Marova, I.; Shipilina, D.; Fedorov, V.; Alekseev, V.; Lvanitskii, V. Interaction between Common and Siberian Chiffchaff in a contact zone. Ornis Fenn. 2017, 94, 66–81. [Google Scholar] [CrossRef]
- Reifova, R.; Reif, J.; Antczak, M.; Nachman, M.W. Ecological character displacement in the face of gene flow: Evidence from two species of nightingales. BMC Evol. Biol. 2011, 11, 138. [Google Scholar] [CrossRef]
- McGregor, P.K.; Krebs, J.R. Mating and song types in the great tit. Nature 1982, 297, 60–61. [Google Scholar] [CrossRef]
- Baker, M.C.; Bjerke, T.K.; Lampe, H.; Espmark, Y. Sexual-responses of female great tits to variation in size of males song repertoires. Am. Nat. 1986, 128, 491–498. [Google Scholar] [CrossRef]
- Paeckert, M. Song: The learned language of three major bird clades. In Bird Species: How They Arise, Modify and Vanish; Tietze, D.T., Ed.; Fascinating Life Sciences: Cambridge, UK, 2018; pp. 75–94. [Google Scholar]
- Hasselquist, D.; Bensch, S.; vonSchantz, T. Correlation between male song repertoire, extra-pair paternity and offspring survival in the great reed warbler. Nature 1996, 381, 229–232. [Google Scholar] [CrossRef]
- Catchpole, C.K. Sexual selection and the evolution of complex songs among european warblers of the genus acrocephalus. Behaviour 1980, 74, 149–166. [Google Scholar] [CrossRef]
- Mason, N.A.; Burns, K.J.; Tobias, J.A.; Claramunt, S.; Seddon, N.; Derryberry, E.P. Song evolution, speciation, and vocal learning in passerine birds. Evolution 2017, 71, 786–796. [Google Scholar] [CrossRef]
- Robinson, C.M.; Snyder, K.T.; Creanza, N. Correlated evolution between repertoire size and song plasticity predicts that sexual selection on song promotes open-ended learning. eLife 2019, 8, e44454. [Google Scholar] [CrossRef] [PubMed]
- Arato, J.; Fitch, W.T. Phylogenetic signal in the vocalizations of vocal learning and vocal non-learning birds. Philos. Trans. R. Soc. B-Biol. Sci. 2021, 376, 20200241. [Google Scholar] [CrossRef]
- Salomon, M.; Hemim, Y. Song variation in the chiffchaffs (Phylloscopus-collybita) of the western pyrenees—The contact zone between the collybita and brehmii forms. Ethology 1992, 92, 265–282. [Google Scholar] [CrossRef]
- Bensch, S.; Helbig, A.J.; Salomon, M.; Siebold, I. Amplified fragment length polymorphism analysis identifies hybrids between two subspecies of warblers. Mol. Ecol. 2002, 11, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Irwin, D.E. Song variation in an avian ring species. Evolution 2000, 54, 998–1010. [Google Scholar] [PubMed]
- Irwin, D.E.; Irwin, J.H.; Price, T.D. Ring species as bridges between microevolution and speciation. Genetica 2001, 112, 223–243. [Google Scholar] [CrossRef]
- Irwin, D.E.; Bensch, S.; Irwin, J.H.; Price, T.D. Speciation by distance in a ring species. Science 2005, 307, 414–416. [Google Scholar] [CrossRef]
- Kovylov, N.S.; Marova, I.M.; Ivanitskii, V.V. Variation of song and plumage in the western (Phylloscopus trochiloides viridanus) and eastern (Phylloscopus trochiloides plumbeitarsus) forms of the greenish warbler in a sympatry zone: Is the hypothesis of ring speciation true? Biol. Bull. 2012, 39, 729–740. [Google Scholar] [CrossRef]
- Peterson, A.T.; Anamza, T. Reexamining Phylloscopus trochiloides complex as a ring species: A refugial counter-hypothesis. J. Avian Biol. 2017, 48, 1608–1613. [Google Scholar] [CrossRef]
- Alcaide, M.; Scordato, E.S.C.; Price, T.D.; Irwin, D.E. Genomic divergence in a ring species complex. Nature 2014, 511, 83–85. [Google Scholar] [CrossRef]
- Mason, N.A.; Bowie, R.C.K. Plumage patterns: Ecological functions, evolutionary origins, and advances in quantification. Auk 2020, 137, ukaa060. [Google Scholar] [CrossRef]
- Tanaka, Y. Social selection and the evolution of animal signals. Evolution 1996, 50, 512–523. [Google Scholar] [CrossRef]
- Hoi, H.; Griggio, M. Dual Utility of a Melanin-Based Ornament in Bearded Tits. Ethology 2008, 114, 1094–1100. [Google Scholar] [CrossRef]
- Patten, M.A.; Unitt, P. Diagnosability versus mean differences of sage sparrow subspecies. Auk 2002, 119, 26–35. [Google Scholar] [CrossRef]
- Paxton, E.H. The utility of plumage coloration for taxonomic and ecological studies. Open Ornithol. J. 2009, 2, 17–23. [Google Scholar] [CrossRef]
- Griggio, M.; Zanollo, V.; Hoi, H. UV plumage color is an honest signal of quality in male budgerigars. Ecol. Res. 2010, 25, 77–82. [Google Scholar] [CrossRef]
- Kabasakal, B.; Polacek, M.; Aslan, A.; Hoi, H.; Erdogan, A.; Griggio, M. Sexual and non-sexual social preferences in male and female white-eyed bulbuls. Sci. Rep. 2017, 7, 5847. [Google Scholar] [CrossRef] [PubMed]
- Servedio, M.R.; Van Doorn, G.S.; Kopp, M.; Frame, A.M.; Nosil, P. Magic traits in speciation: ‘Magic’ but not rare? Trends Ecol. Evol. 2011, 26, 389–397. [Google Scholar] [CrossRef]
- Pizarro, A.K.; DeRaad, D.A.; McCormack, J.E. Temporal stability of the hybrid zone between Calocitta magpie-jays revealed through comparison of museum specimens and iNaturalist photos. Ecol. Evol. 2023, 13, e9863. [Google Scholar] [CrossRef]
- Winger, B.M.; Bates, J.M. The tempo of trait divergence in geographic isolation: Avian speciation across the Maranon Valley of Peru. Evolution 2015, 69, 772–787. [Google Scholar] [CrossRef] [PubMed]
- Fiser, C.; Robinson, C.T.; Malard, F. Cryptic species as a window into the paradigm shift of the species concept. Mol. Ecol. 2018, 27, 613–635. [Google Scholar] [CrossRef]
- Del-Rio, G.; Rego, M.A.; Whitney, B.M.; Schunck, F.; Silveira, L.F.; Faircloth, B.C.; Brumfield, R.T. Displaced clines in an avian hybrid zone (Thamnophilidae: Rhegmatorhina) within an Amazonian interfluve. Evolution 2022, 76, 455–475. [Google Scholar] [CrossRef]
- Peñalba, J.V.; Peters, J.L.; Joseph, L. Sustained plumage divergence despite weak genomic differentiation and broad sympatry in sister species of Australian woodswallows (Artamus spp.). Mol. Ecol. 2022, 31, 5060–5073. [Google Scholar] [CrossRef]
- Wang, S.L.; Rohwer, S.; de Zwaan, D.R.; Toews, D.P.L.; Lovette, I.J.; Mackenzie, J.; Irwin, D. Selection on a small genomic region underpins differentiation in multiple color traits between two warbler species. Evol. Lett. 2020, 4, 502–515. [Google Scholar] [CrossRef] [PubMed]
- Kirschel, A.N.G.; Nwankwo, E.C.; Pierce, D.K.; Lukhele, S.M.; Moysi, M.; Ogolowa, B.O.; Hayes, S.C.; Monadjem, A.; Brelsford, A. CYP2J19 mediates carotenoid colour introgression across a natural avian hybrid zone. Mol. Ecol. 2020, 29, 4970–4984. [Google Scholar] [CrossRef]
- Uy, J.A.C.; Stein, A.C. Variable visual habitats may influence the spread of colourful plumage across an avian hybrid zone. J. Evol. Biol. 2007, 20, 1847–1858. [Google Scholar] [CrossRef] [PubMed]
- Cowles, S.A.; Uy, J.A.C. Rapid, complete reproductive isolation in two closely related Zosterops White-eye bird species despite broadly overlapping ranges. Evolution 2019, 73, 1647–1662. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.D.; Burri, R.; Liu, Y.; Suh, A.; Sundev, G.; Heckel, G.; Schweizer, M. Seasonal migration patterns and the maintenance of evolutionary diversity in a cryptic bird radiation. Mol. Ecol. 2022, 31, 632–645. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, R.; Cadena, V.; Danner, R.M.; Tattersall, G. Heat Loss May Explain Bill Size Differences between Birds Occupying Different Habitats. PLoS ONE 2012, 7, e40933. [Google Scholar] [CrossRef]
- Navalón, G.; Marugán-Lobón, J.; Bright, J.A.; Cooney, C.R.; Rayfield, E.J. The consequences of craniofacial integration for the adaptive radiations of Darwin’s finches and Hawaiian honeycreepers. Nat. Ecol. Evol. 2020, 4, 270–278. [Google Scholar] [CrossRef]
- Shao, S.; Quan, Q.; Cai, T.; Song, G.; Qu, Y.; Lei, F. Evolution of body morphology and beak shape revealed by a morphometric analysis of 14 Paridae species. Front. Zool. 2016, 13, 30. [Google Scholar] [CrossRef]
- Cheng, Y.; Gao, B.; Wang, H.; Han, N.; Shao, S.; Wu, S.; Song, G.; Zhang, Y.E.; Zhu, X.; Lu, X.; et al. Evolution of beak morphology in the Ground Tit revealed by comparative transcriptomics. Front. Zool. 2017, 14, 58. [Google Scholar] [CrossRef]
- Krishnan, A. Biomechanics illuminates form-function relationships in bird bills. J. Exp. Biol. 2023, 226, jeb245171. [Google Scholar] [CrossRef]
- Iqbal, A.; Moss, A.F. Review: Key tweaks to the chicken's beak: The versatile use of the beak by avian species and potential approaches for improvements in poultry production. Animal 2021, 15, 100119. [Google Scholar] [CrossRef]
- Eroukhmanoff, F.; Elgvin, T.O.; Gonzàlez Rojas, M.F.; Haas, F.; Hermansen, J.S.; Sætre, G.P. Effect of Species Interaction on Beak Integration in an Avian Hybrid Species Complex. Evol. Biol. 2014, 41, 452–458. [Google Scholar] [CrossRef]
- Grant, B.R.; Grant, P.R. Simulating secondary contact in allopatric speciation: An empirical test of premating isolation. Biol. J. Linn. Soc. 2002, 76, 545–556. [Google Scholar] [CrossRef]
- Lamichhaney, S.; Berglund, J.; Almén, M.S.; Maqbool, K.; Grabherr, M.; Martinez-Barrio, A.; Promerova, M.; Rubin, C.J.; Wang, C.; Zamani, N.; et al. Evolution of Darwin's finches and their beaks revealed by genome sequencing. Nature 2015, 518, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Grant, P.R.; Grant, B.R. The secondary contact phase of allopatric speciation in Darwin's finches. Proc. Natl. Acad. Sci. USA 2009, 106, 20141–20148. [Google Scholar] [CrossRef]
- Masello, J.F.; Ryan, P.G.; Shepherd, L.D.; Quillfeldt, P.; Cherel, Y.; Tennyson, A.J.D.; Alderman, R.; Calderon, L.; Cole, T.L.; Cuthbert, R.J.; et al. Independent evolution of intermediate bill widths in a seabird clade. Mol. Genet. Genom. 2022, 297, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Lamichhaney, S.; Han, F.; Webster, M.T.; Andersson, L.; Grant, B.R.; Grant, P.R. Rapid hybrid speciation in Darwin’s finches. Science 2018, 359, 224–227. [Google Scholar] [CrossRef]
- Turbek, S.P.; Scordato, E.S.C.; Safran, R.J. The Role of Seasonal Migration in Population Divergence and Reproductive Isolation. Trends Ecol. Evol. 2018, 33, 164–175. [Google Scholar] [CrossRef]
- Irwin, D.E.; Irwin, J.H.; Smith, T.B. Genetic variation and seasonal migratory connectivity in Wilson's warblers (Wilsonia pusilla): Species-level differences in nuclear DNA between western and eastern populations. Mol. Ecol. 2011, 20, 3102–3115. [Google Scholar] [CrossRef]
- Turbek, S.P.; Schield, D.R.; Scordato, E.S.C.; Contina, A.; Da, X.-W.; Liu, Y.; Liu, Y.; Pagani-Nunez, E.; Ren, Q.-M.; Smith, C.C.R.; et al. A migratory divide spanning two continents is associated with genomic and ecological divergence. Evolution 2022, 76, 722–736. [Google Scholar] [CrossRef]
- Sokolovskis, K.; Lundberg, M.; Akesson, S.; Willemoes, M.; Zhao, T.; Caballero-Lopez, V.; Bensch, S. Migration direction in a songbird explained by two loci. Nat. Commun. 2023, 14, 165. [Google Scholar] [CrossRef]
- Uy J A, C.; Irwin D, E.; Webster M, S. Behavioral isolation and incipient speciation in birds. Annu. Rev. Ecol. Evol. Syst. 2018, 49, 1–24. [Google Scholar]
- Toews, D.P.L.; Delmore, K.E.; Osmond, M.M.; Taylor, P.D.; Irwin, D.E. Migratory orientation in a narrow avian hybrid zone. PeerJ 2017, 5, e3201. [Google Scholar] [CrossRef] [PubMed]
- Buggs, R.J.A.; Pannell, J.R. Ecological differentiation and diploid superiority across a moving ploidy contact zone. Evolution 2007, 61, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.L.; Hale, J.M.; Gay, L.; Kearney, M.; Austin, J.J.; Parris, K.M.; Melville, J. Spatio-temporal changes in the structure of an australian frog hybrid zone: A 40-year perspective. Evolution 2013, 67, 3442–3454. [Google Scholar] [CrossRef]
- Mettler, R.D.; Spellman, G.M. A hybrid zone revisited: Molecular and morphological analysis of the maintenance, movement, and evolution of a Great Plains avian (Cardinalidae: Pheucticus) hybrid zone. Mol. Ecol. 2009, 18, 3256–3267. [Google Scholar] [CrossRef]
- Taylor, S.A.; White, T.A.; Hochachka, W.M.; Ferretti, V.; Curry, R.L.; Lovette, I. Climate-Mediated Movement of an Avian Hybrid Zone. Curr. Biol. 2014, 24, 671–676. [Google Scholar] [CrossRef]
- Dasmahapatra, K.K.; Blum, M.J.; Aiello, A.; Hackwell, S.; Davies, N.; Bermingham, E.P.; Mallett, T. Inferences from a rapidly moving hybrid zone. Evolution 2002, 56, 741–753. [Google Scholar]
- Goodisman, M.A.D.; Shoemaker, D.D.; Asmussen, M.A. Cytonuclear theory for haplodiploid species and X-linked genes. II. Stepping-stone models of gene flow and application to a fire ant hybrid zone. Evolution 1998, 52, 1423–1440. [Google Scholar] [CrossRef]
- Bronson, C.L.; Grubb, T.C.; Braun, M.J. A test of the endogenous and exogenous selection hypotheses for the maintenance of a narrow avian hybrid zone. Evolution 2003, 57, 630–637. [Google Scholar]
- Mallet, J. Hybrid zones of heliconius butterflies in panama and the stability and movement of warning color clines. Heredity 1986, 56, 191–202. [Google Scholar] [CrossRef]
- Britch, S.C.; Cain, M.L.; Howard, D.J. Spatio-temporal dynamics of the Allonemobius fasciatus A. socius mosaic hybrid zone: A 14-year perspective. Mol. Ecol. 2001, 10, 627–638. [Google Scholar]
- Key, K.H.L. Concept of stasipatric speciation. Syst. Zool. 1968, 17, 14–22. [Google Scholar] [CrossRef]
- May, R.M.; Endler, J.A.; McMurtrie, R.E. Gene frequency clines in presence of selection opposed by gene flow. Am. Nat. 1975, 109, 659–676. [Google Scholar] [CrossRef] [PubMed]
- Carling, M.D.; Zuckerberg, B. Spatio-temporal changes in the genetic structure of the Passerina bunting hybrid zone. Mol. Ecol. 2011, 20, 1166–1175. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, E.S.; Henriques, R.; Beger, M.; Toonen, R.J.; von der Heyden, S. Multi-model seascape genomics identifies distinct environmental drivers of selection among sympatric marine species. BMC Evol. Biol. 2020, 20, 121. [Google Scholar] [CrossRef] [PubMed]
- Goodman, S.J.; Barton, N.H.; Swanson, G.; Abernethy, K.; Pemberton, J.M. Introgression through rare hybridization: A genetic study of a hybrid zone between red and sika deer (genus Cervus) in Argyll, Scotland. Genetics 1999, 152, 355–371. [Google Scholar] [CrossRef]
- Moore, W.S. Evaluation of narrow hybrid zones in vertebrates. Q. Rev. Biol. 1977, 52, 263–277. [Google Scholar] [CrossRef]
- Moore, W.S.; Price, J.T. Nature of selection in the northern flicker hybrid zone and its implications for speciation theory. Hybrid. Zones Evol. Process 1993, 196, 225. [Google Scholar]
- Sequeira, F.; Arntzen, J.W.; van Gulik, D.; Hajema, S.; Diaz, R.L.; Wagt, M.; van Riemsdijk, I. Genetic traces of hybrid zone movement across a fragmented habitat. J. Evol. Biol. 2022, 35, 400–412. [Google Scholar] [CrossRef]
- Leaché, A.D.; Grummer, J.A.; Harris, R.B.; Breckheimer, I.K. Evidence for concerted movement of nuclear and mitochondrial clines in a lizard hybrid zone. Mol. Ecol. 2017, 26, 2306–2316. [Google Scholar] [CrossRef] [PubMed]
- Ryan, S.F.; Deines, J.M.; Scriber, J.M.; Pfrender, M.E.; Jones, S.E.; Emrich, S.J.; Hellmann, J.J. Climate-mediated hybrid zone movement revealed with genomics, museum collection, and simulation modeling. Proc. Natl. Acad. Sci. USA 2018, 115, E2284–E2291. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.A.; Larson, E.L.; Harrison, R.G. Hybrid zones: Windows on climate change. Trends Ecol. Evol. 2015, 30, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Morales-Rozo, A.; Tenorio, E.A.; Carling, M.D.; Cadena, C.D. Origin and cross-century dynamics of an avian hybrid zone. BMC Evol. Biol. 2017, 17, 257. [Google Scholar] [CrossRef]
- Thurman, T.J.; Szejner-Sigal, A.; McMillan, W.O. Movement of a Heliconius hybrid zone over 30 years: A Bayesian approach. J. Evol. Biol. 2019, 32, 974–983. [Google Scholar] [CrossRef]
- Lopez-Delgado, J.; van Riemsdijk, I.; Arntzen, J.W. Tracing species replacement in Iberian marbled newts. Ecol. Evol. 2021, 11, 402–414. [Google Scholar] [CrossRef]
- Barton, N.H.; Hewitt, G.M. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 1985, 16, 113–148. [Google Scholar] [CrossRef]
- Pinto, B.J.; Titus-McQuillan, J.; Daza, J.D.; Gamble, T. Persistence of a Geographically-Stable Hybrid Zone in Puerto Rican Dwarf Geckos. J. Hered. 2019, 110, 523–534. [Google Scholar] [CrossRef]
- Alexander, A.; Robbins, M.B.; Holmes, J.; Moyle, R.G.; Peterson, A.T. Limited movement of an avian hybrid zone in relation to regional variation in magnitude of climate change. Mol. Ecol. 2022, 31, 6634–6648. [Google Scholar] [CrossRef]
- Aguillon, S.M.; Rohwer, V.G. Revisiting a classic hybrid zone: Movement of the northern flicker hybrid zone in contemporary times. Evolution 2022, 76, 1082–1090. [Google Scholar] [CrossRef]
- Krosby, M.; Rohwer, S. Ongoing movement of the hermit warbler X Townsend’s warbler hybrid zone. PLoS ONE 2010, 5, e14164. [Google Scholar] [CrossRef]
- Vali, U.; Treinys, R.; Bergmanis, U.; Daroczi, S.; Demerdzhiev, D.; Dombrovski, V.; Dravecky, M.; Ivanovski, V.; Kicko, J.; Langgemach, T.; et al. Contrasting patterns of genetic diversity and lack of population structure in the lesser spotted eagle Clanga pomarina (Aves: Accipitriformes) across its breeding range. Biol. J. Linn. Soc. 2022, 136, 506–519. [Google Scholar] [CrossRef]
- Vali, U.; Dombrovski, V.; Dzmitranok, M.; Maciorowski, G.; Meyburg, B.-U. High genetic diversity and low differentiation retained in the European fragmented and declining Greater Spotted Eagle (Clanga clanga) population. Sci. Rep. 2019, 9, 3064. [Google Scholar] [CrossRef] [PubMed]
- Maciorowski, G.; Mirski, P.; Vaeli, U. Hybridisation dynamics between the Greater Spotted Eagles Aquila clanga and Lesser Spotted Eagles Aquila pomarina in the Biebrza River Valley (NE Poland). Acta Ornithol. 2015, 50, 33–41. [Google Scholar] [CrossRef]
- Wilkins, A.S. Speciation patterns and mechanisms: A symposium to honor Ernst Mayr. Bioessays 2005, 27, 661–663. [Google Scholar] [CrossRef] [PubMed]
- Kulathinal, R.J.; Singh, R.S. The molecular basis of speciation: From patterns to processes, rules to mechanisms. J. Genet. 2008, 87, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Wollenberg Valero, K.C.; Marshall, J.C.; Bastiaans, E.; Caccone, A.; Camargo, A.; Morando, M.; Niemiller, M.L.; Pabijan, M.; Russello, M.A.; Sinervo, B.; et al. Patterns, Mechanisms and Genetics of Speciation in Reptiles and Amphibians. Genes 2019, 10, 646. [Google Scholar] [CrossRef]
- Lei, F.; Qu, Y.; Song, G.; Alström, P.; Fjeldså, J. The potential drivers in forming avian biodiversity hotspots in the East Himalaya Mountains of Southwest China. Integr. Zool. 2015, 10, 171–181. [Google Scholar] [CrossRef]
- Zhou, P.; Li, J.; Möller, M. Secondary contact, hybridization and polyploidization add to the biodiversity in the Hengduan Mountains, exemplified by the widespread Corallodiscus lanuginosus (Gesneriaceae). Plant Syst. Evol. 2017, 303, 587–602. [Google Scholar] [CrossRef]
- Rogers, A.D. Evolution and biodiversity of Antarctic organisms: A molecular perspective. Philos. Trans. R. Soc. B Biol. Sci. 2007, 362, 2191–2214. [Google Scholar] [CrossRef]
- Gaboriau, T.; Leprieur, F.; Mouillot, D.; Hubert, N. Influence of the geography of speciation on current patterns of coral reef fish biodiversity across the Indo-Pacific. Ecography 2018, 41, 1295–1306. [Google Scholar] [CrossRef]
- Meraner, A.; Cornetti, L.; Gandolfi, A. Defining conservation units in a stocking-induced genetic melting pot: Unraveling native and multiple exotic genetic imprints of recent and historical secondary contact in Adriatic grayling. Ecol. Evol. 2014, 4, 1313–1327. [Google Scholar] [CrossRef]
- Porto-Hannes, I.; Burlakova, L.E.; Zanatta, D.T.; Lasker, H.R. Boundaries and hybridization in a secondary contact zone between freshwater mussel species (Family:Unionidae). Heredity 2021, 126, 955–973. [Google Scholar] [CrossRef]
- Grünig, S.; Fischer, M.; Parisod, C. Recent hybrid speciation at the origin of the narrow endemic Pulmonaria helvetica. Ann. Bot. 2021, 127, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Hermansen, J.S.; Sæther, S.A.; Elgvin, T.O.; Borge, T.; Hjelle, E.; Sætre, G.P. Hybrid speciation in sparrows I: Phenotypic intermediacy, genetic admixture and barriers to gene flow. Mol. Ecol. 2011, 20, 3812–3822. [Google Scholar] [CrossRef]
- Elgvin, T.O.; Hermansen, J.S.; Fijarczyk, A.; Bonnet, T.; Borge, T.; Sæther, S.A.; Voje, K.L.; Sætre, G.P. Hybrid speciation in sparrows II: A role for sex chromosomes? Mol. Ecol. 2011, 20, 3823–3837. [Google Scholar] [CrossRef] [PubMed]
- Lavretsky, P.; Engilis, A.; Eadie, J.M.; Peters, J.L. Genetic admixture supports an ancient hybrid origin of the endangered Hawaiian duck. J. Evol. Biol. 2015, 28, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Töpfer, T. The taxonomic status of the Italian Sparrow Passer italiae (Vieillot 1817): Speciation by stabilised hybridisation? A critical analysis. Zootaxa 2006, 1325, 117. [Google Scholar] [CrossRef]
- Higgie, M.; Chenoweth, S.; Blows, M.W. Natural selection and the reinforcement of mate recognition. Science 2000, 290, 519–521. [Google Scholar] [CrossRef] [PubMed]
- Nosil, P.; Crespi, B.J.; Gries, R.; Gries, G. Natural selection and divergence in mate preference during speciation. Genetica 2007, 129, 309–327. [Google Scholar] [CrossRef]
- Latour, Y.; Perriat-Sanguinet, M.; Caminade, P.; Boursot, P.; Smadja, C.M.; Ganem, G. Sexual selection against natural hybrids may contribute to reinforcement in a house mouse hybrid zone. Proc. R. Soc. B Biol. Sci. 2014, 281, 20132733. [Google Scholar] [CrossRef] [PubMed]
- Vonlanthen, P.; Bittner, D.; Hudson, A.G.; Young, K.A.; Müller, R.; Lundsgaard-Hansen, B.; Roy, D.; Di Piazza, S.; Largiader, C.R.; Seehausen, O. Eutrophication causes speciation reversal in whitefish adaptive radiations. Nature 2012, 482, 357-U1500. [Google Scholar] [CrossRef] [PubMed]
- Webb, W.C.; Marzluff, J.M.; Omland, K.E. Random interbreeding between cryptic lineages of the Common Raven: Evidence for speciation in reverse. Mol. Ecol. 2011, 20, 2390–2402. [Google Scholar] [CrossRef]
- Rudman, S.M.; Schluter, D. Ecological Impacts of Reverse Speciation in Threespine Stickleback. Curr. Biol. 2016, 26, 490–495. [Google Scholar] [CrossRef]
- Barmentlo, S.H.; Meirmans, P.G.; Luijten, S.H.; Triest, L.; Oostermeijer, J.G.B. Outbreeding depression and breeding system evolution in small, remnant populations of Primula vulgaris: Consequences for genetic rescue. Conserv. Genet. 2018, 19, 545–554. [Google Scholar] [CrossRef]
- Wolf, D.E.; Takebayashi, N.; Rieseberg, L.H. Predicting the risk of extinction through hybridization. Conserv. Biol. 2001, 15, 1039–1053. [Google Scholar] [CrossRef]
- Billerman, S.M.; Murphy, M.A.; Carling, M.D. Changing climate mediates sapsucker (Aves: Sphyrapicus) hybrid zone movement. Ecol. Evol. 2016, 6, 7976–7990. [Google Scholar] [CrossRef]
- Wegener, J.E.; Pita-Aquino, J.N.; Atutubo, J.; Moreno, A.; Kolbe, J.J. Hybridization and rapid differentiation after secondary contact between the native green anole (Anolis carolinensis) and the introduced green anole (Anolis porcatus). Ecol. Evol. 2019, 9, 4138–4148. [Google Scholar] [PubMed]
- Sætre, G.P.; Sæther, S.A. Ecology and genetics of speciation in Ficedula flycatchers. Mol. Ecol. 2010, 19, 1091–1106. [Google Scholar] [CrossRef]
- Pirani, R.M.; Tonini, J.F.R.; Thomaz, A.T.; Napoli, M.F.; Encarnacao, L.C.; Knowles, L.L.; Werneck, F.P. Deep Genomic Divergence and Phenotypic Admixture of the Treefrog Dendropsophus elegans (Hylidae: Amphibia) Coincide with Riverine Boundaries at the Brazilian Atlantic Forest. Front. Ecol. Evol. 2022, 10, 765977. [Google Scholar]
- Reding, D.M.; Castaneda-Rico, S.; Shirazi, S.; Hofman, C.A.; Cancellare, I.A.; Lance, S.L.; Beringer, J.; Clark, W.R.; Maldonado, J.E. Mitochondrial Genomes of the United States Distribution of Gray Fox (Urocyon cinereoargenteus) Reveal a Major Phylogeographic Break at the Great Plains Suture Zone. Front. Ecol. Evol. 2021, 9, 666800. [Google Scholar] [CrossRef]
- Knowles, L.L. Tests of Pleistocene speciation in montane grasshoppers (genus Melanoplus) from the sky islands of western North America. Evolution 2000, 54, 1337–1348. [Google Scholar]
- Oziolor, E.M.; Reid, N.M.; Yair, S.; Lee, K.M.; VerPloeg, S.G.; Bruns, P.C.; Shaw, J.R.; Whitehead, A.; Matson, C.W. Adaptive introgression enables evolutionary rescue from extreme environmental pollution. Science 2019, 364, 455–457. [Google Scholar] [CrossRef] [PubMed]
- Yeager, K.M.; Santschi, P.H.; Rifai, H.S.; Suarez, M.P.; Brinkmeyer, R.; Hung, C.-C.; Schindler, K.J.; Andres, M.J.; Weaver, E.A. Dioxin chronology and fluxes in sediments of the Houston Ship Channel, Texas: Influences of non-steady-state sediment transport and total organic carbon. Environ. Sci. Technol. 2007, 41, 5291–5298. [Google Scholar] [CrossRef]
- Lucek, K.; Bouaouina, S.; Jospin, A.; Grill, A.; de Vos, J.M. Prevalence and relationship of endosymbiotic Wolbachia in the butterfly genus Erebia. BMC Ecol. Evol. 2021, 21, 95. [Google Scholar] [CrossRef] [PubMed]
- Tusso, S.; Nieuwenhuis, B.P.S.; Sedlazeck, F.J.; Davey, J.W.; Jeffares, D.C.; Wolf, J.B.W. Ancestral Admixture Is the Main Determinant of Global Biodiversity in Fission Yeast. Mol. Biol. Evol. 2019, 36, 1975–1989. [Google Scholar] [CrossRef]
- Longo, G.C.; Lam, L.; Basnett, B.; Samhouri, J.; Hamilton, S.; Andrews, K.; Williams, G.; Goetz, G.; McClure, M.; Nichols, K.M. Strong population differentiation in lingcod (Ophiodon elongatus) is driven by a small portion of the genome. Evol. Appl. 2020, 13, 2536–2554. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, I.R.; Hamilton, L.C.; Dempson, B.; Robertson, M.J.; Bourret, V.; Bernatchez, L.; Verspoor, E. Transatlantic secondary contact in Atlantic Salmon, comparing microsatellites, a single nucleotide polymorphism array and restriction-site associated DNA sequencing for the resolution of complex spatial structure. Mol. Ecol. 2015, 24, 5130–5144. [Google Scholar] [CrossRef]
- Lundberg, M.; Mackintosh, A.; Petri, A.; Bensch, S. Inversions maintain differences between migratory phenotypes of a songbird. Nat. Commun. 2023, 14, 452. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Wu, L.; Zhu, Q.; Wu, J.; Tang, S.; Zhao, Y.; Cheng, Y.; Zhang, D.; Qiao, G.; Zhang, R.; et al. Trait Variation and Spatiotemporal Dynamics across Avian Secondary Contact Zones. Biology 2024, 13, 643. https://doi.org/10.3390/biology13080643
Wang S, Wu L, Zhu Q, Wu J, Tang S, Zhao Y, Cheng Y, Zhang D, Qiao G, Zhang R, et al. Trait Variation and Spatiotemporal Dynamics across Avian Secondary Contact Zones. Biology. 2024; 13(8):643. https://doi.org/10.3390/biology13080643
Chicago/Turabian StyleWang, Shangyu, Lei Wu, Qianghui Zhu, Jiahao Wu, Shiyu Tang, Yifang Zhao, Yalin Cheng, Dezhi Zhang, Gexia Qiao, Runzhi Zhang, and et al. 2024. "Trait Variation and Spatiotemporal Dynamics across Avian Secondary Contact Zones" Biology 13, no. 8: 643. https://doi.org/10.3390/biology13080643
APA StyleWang, S., Wu, L., Zhu, Q., Wu, J., Tang, S., Zhao, Y., Cheng, Y., Zhang, D., Qiao, G., Zhang, R., & Lei, F. (2024). Trait Variation and Spatiotemporal Dynamics across Avian Secondary Contact Zones. Biology, 13(8), 643. https://doi.org/10.3390/biology13080643





