Unlocking the Potential of Animal Hair Shafts for Genomic Studies: A Comprehensive Evaluation of DNA Quality
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
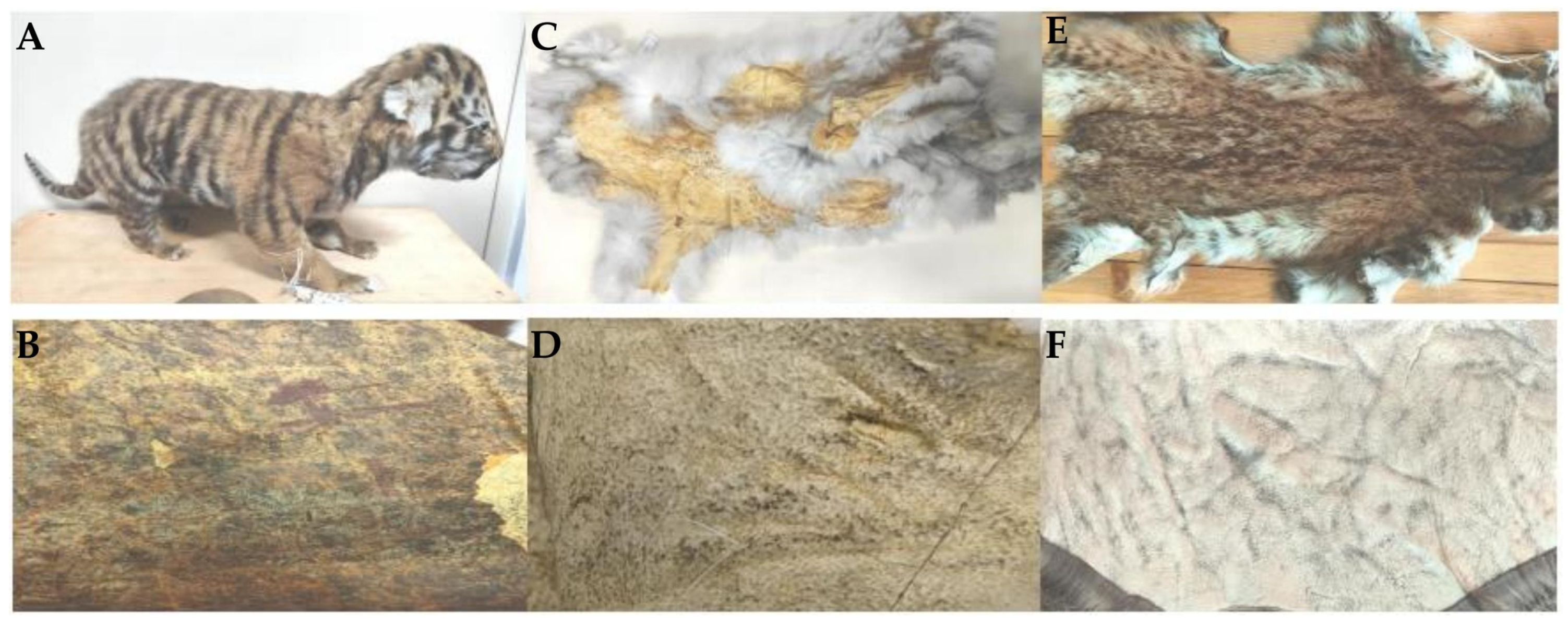
2.1. Sample Selection
2.2. Quality of Sequencing Data from Hair Shaft DNA
2.2.1. Different Types of Hair Shaft
2.2.2. Different Sections of Hair Shaft
2.2.3. Different Tanning Degree
2.2.4. Hair Shaft on Decaying Carcass
2.2.5. Hair Shaft in Faeces
2.3. Analysis of DNA Damage
2.4. Data Analysis and Statistics
3. Results
3.1. Different Types
3.2. Different Sections
3.3. Effect of Degree of Tanning
3.4. Hair Samples from Decaying Carcass and Digestive Tract
3.5. DNA Damage
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| mtDNA | Mitochondrial DNA |
| nuDNA | Nuclear DNA |
| aDNA | Ancient DNA |
| NCBI | National Center for Biotechnology Information |
| NGS | Next-generation sequencing |
References
- Junno, J.A.; Väre, T.; Tikkanen, J.; Heino, M.T.; Niskanen, M.; Kakko, I.; Aspi, J. Stable isotope analyses of carbon and nitrogen in hair keratin of suspected man-eating wolves from 1880s. Sci. Rep. 2024, 14, 4946. [Google Scholar]
- Saito, K.; Ito, R.; Sekiya, C.; Akiyama, H. Identification of animal species by nucleic acid chromatography of hair samples and investigation of its applicability to human biological samples. Forensic Sci. Int. 2023, 351, 111811. [Google Scholar] [CrossRef] [PubMed]
- González-Gómez, X.; Cambeiro-Pérez, N.; Martínez-Carballo, E.; Simal-Gándara, J. Screening of organic pollutants in pet hair samples and the significance of environmental factors. Sci. Total Environ. 2018, 625, 311–319. [Google Scholar] [CrossRef] [PubMed]
- McNevin, D.; Wilson-Wilde, L.; Robertson, J.; Kyd, J.; Lennard, C. Short tandem repeat (STR) genotyping of keratinised hair. Part 2. An optimised genomic DNA extraction procedure reveals donor dependence of STR profiles. Forensic Sci. Int. 2005, 153, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Parson, W.; Huber, G.; Moreno, L.; Madel, M.-B.; Brandhagen, M.D.; Nagl, S.; Xavier, C.; Eduardoff, M.; Callaghan, T.C.; Irwin, J.A. Massively parallel sequencing of complete mitochondrial genomes from hair shaft samples. Forensic Sci. Int. Genet. 2015, 15, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Colombo, F.M.; Cattaneo, P.; Bernardi, C. Selected results of DNA-based species identification on animal foods. J. Sci. Food Agric. 2018, 98, 2437–2439. [Google Scholar] [CrossRef] [PubMed]
- Szabo, S.; Jaeger, K.; Fischer, H.; Tschachler, E.; Parson, W.; Eckhart, L. In situ labeling of DNA reveals interindividual variation in nuclear DNA breakdown in hair and may be useful to predict success of forensic genotyping of hair. Int. J. Leg. Med. 2012, 126, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.; Engström, A.S.; Meyers, S.; Handt, O.; Saldeen, T.; von Haeseler, A.; Pääbo, S.; Gyllensten, U. Mitochondrial DNA sequencing of shed hairs and saliva on robbery caps: Sensitivity and matching probabilities. J. Forensic Sci. 1998, 43, 453–464. [Google Scholar] [PubMed]
- Prieto, L.; Montesino, M.; Salas, A.; Alonso, A.; Albarrán, C.; Álvarez, S.; Crespillo, M.; Di Lonardo, A.M.; Doutremepuich, C.; Fernández-Fernández, I.; et al. The 2000–2001 GEP-ISFG Collaborative Exercise on mtDNA: Assessing the cause of unsuccessful mtDNA PCR amplification of hair shaft samples. Forensic Sci. Int. 2003, 134, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Andréasson, H.; Asp, A.; Alderborn, A.; Gyllensten, U.; Allen, M. Mitochondrial sequence analysis for forensic identification using pyrosequencing technology. Biotechniques 2002, 32, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, R.; von Beroldingen, C.H.; Sensabaugh, G.F.; Erlich, H.A. DNA typing from single hairs. Nature 1988, 332, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Amory, S.; Keyser, C.; Crubézy, E.; Ludes, B. STR typing of ancient DNA extracted from hair shafts of Siberian mummies. Forensic Sci. Int. 2007, 166, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Heywood, D.M.; Skinner, R.; Cornwell, P.A. Analysis of DNA in hair fibers. J. Cosmet. Sci. 2003, 54, 21–27. [Google Scholar] [PubMed]
- Linch, C.A.; Whiting, D.A.; Holland, M.M. Human hair histogenesis for the mitochondrial DNA forensic scientist. J. Forensic Sci. 2001, 46, 844–853. [Google Scholar] [CrossRef] [PubMed]
- Brandhagen, M.D.; Loreille, O.; Irwin, J.A. Fragmented Nuclear DNA is the Predominant Genetic Material in Human Hair Shafts. Genes 2018, 9, 640. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Waits, L.P.; Luikart, G. Noninvasive genetic sampling: Look before you leap. Trends Ecol. Evol. 1999, 14, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Gaag, K.J.V.; Desmyter, S.; Smit, S.; Prieto, L.; Sijen, T. Reducing the Number of Mismatches between Hairs and Buccal References When Analysing mtDNA Heteroplasmic Variation by Massively Parallel Sequencing. Genes 2020, 11, 1355. [Google Scholar] [CrossRef] [PubMed]
- Lewis, E.J.; Benefiel, O.; Fronda, M.V.; Podini, D. Nuclear DNA SNP profiles derived from human hair shaft. Forensic Sci. Int. Genet. Suppl. Ser. 2022, 8, 333–335. [Google Scholar] [CrossRef]
- Gilbert, M.T.; Menez, L.; Janaway, R.C.; Tobin, D.J.; Cooper, A.; Wilson, A.S. Resistance of degraded hair shafts to contaminant DNA. Forensic Sci. Int. 2006, 156, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson, C.F.; Olsen, M.E.; Brandt, L.Ø.; Bertelsen, M.F.; Willerslev, E.; Tobin, D.J.; Wilson, A.S.; Gilbert, M.T. DNA from keratinous tissue. Part I: Hair and nail. Ann. Anat. 2012, 194, 17–25. [Google Scholar] [CrossRef]
- An, G.C. Identification of 15 kinds of animal hair fibers by combining scanning electron microscopy and infrared spectroscopy. Wool. Text. J. 2024, 9, 111–116. [Google Scholar]
- Neukamm, J.; Peltzer, A.; Nieselt, K. DamageProfiler: Fast damage pattern calculation for ancient DNA. Bioinformatics 2021, 37, 3652–3653. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yang, X.; Sun, Y.; Wang, Y.; Song, L.; Qiao, Z.; Fang, Z.; Wang, Z.; Liu, L.; Chen, Y.; et al. Test development, optimization and validation of a WGS pipeline for genetic disorders. BMC Med. Genom. 2023, 16, 74. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transforms. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- De Summa, S.; Malerba, G.; Pinto, R.; Mori, A.; Mijatovic, V.; Tommasi, S. GATK hard filtering: Tunable parameters to improve variant calling for next generation sequencing targeted gene panel data. BMC Bioinform. 2017, 18 (Suppl. S5), 119. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- McKittrick, J.; Chen, P.Y.; Bodde, S.G.; Yang, W.; Novitskaya, E.E.; Meyers, M.A. The structure, functions, and mechanical properties of keratin. JOM 2012, 64, 449–468. [Google Scholar] [CrossRef]
- Wang, B.; Yang, W.; Sherman, V.R.; Meyers, M.A. Pangolin armor: Overlapping, structure, and mechanical properties of the keratinous scales. Acta Biomater. 2016, 41, 60–74. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, W.; Hayashi, C.; Gatesy, J.; McKittrick, J. Microstructure and mechanical properties of different keratinous horns. J. R. Soc. Interface 2018, 15, 20180093. [Google Scholar] [PubMed]
- Borowczyk, K.; Suliburska, J.; Jakubowski, H. Demethylation of methionine and keratin damage in human hair. Amino Acids 2018, 50, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekara, M.N.; Ranganathaiah, C. Chemical and photochemical degradation of human hair: A free-volume microprobe study. J. Photochem. Photobiol. B 2010, 101, 286–294. [Google Scholar] [PubMed]
- Tridico, S.R.; Koch, S.; Michaud, A.; Thomson, G.; Kirkbride, K.P.; Bunce, M. Interpreting biological degradative processes acting on mammalian hair in the living and the dead: Which ones are taphonomic? Proc. R. Soc. B 2014, 281, 20141755. [Google Scholar] [PubMed]
- Tie, J.; Uchigasaki, S.; Isobe, E.; Iwakami, E.; Okuda, T. Detection of deletion/insertion polymorphism profiles from single human hair shafts. Mol. Biol. Rep. 2022, 49, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Martins, C.; Ferreira, P.M.; Carvalho, R.; Costa, S.C.; Farinha, C.; Azevedo, L.; Oliveira, M. Evaluation of InnoQuant® HY and InnoTyper® 21 kits in the DNA analysis of rootless hair samples. Forensic Sci. Int. Genet. 2019, 39, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wang, J.; Zhang, Z.; Xiao, Q.; Li, M.; Pan, Y. Re-alignment of the unmapped reads with base quality score. BMC Bioinform. 2015, 16 (Suppl. S5), S8. [Google Scholar] [CrossRef]
- Cui, L.Y.; Liu, B.Y.; Li, H.M.; Zhu, Y.X.; Zhou, Y.H.; Su, C.; Tian, Y.P.; Xu, H.T.; Liu, D.; Li, X.P.; et al. A simple and effective method to enrich endogenous DNA from mammalian faeces. Mol. Ecol. Resour. 2024, 24, e13939. [Google Scholar] [CrossRef] [PubMed]
- Yonekura, S.I.; Nakamura, N.; Yonei, S.; Zhang-Akiyama, Q.M. Generation, biological consequences and repair mechanisms of cytosine deamination in DNA. J. Radiat. Res. 2009, 50, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Briggs, A.W.; Stenzel, U.; Johnson, P.L.; Green, R.E.; Kelso, J.; Prüfer, K.; Pääbo, S. Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl. Acad. Sci. USA 2007, 104, 14616–14621. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Zhang, Q.; Gao, P.; Yang, S.; Xu, Y. Unlocking the Potential of Animal Hair Shafts for Genomic Studies: A Comprehensive Evaluation of DNA Quality. Biology 2025, 14, 353. https://doi.org/10.3390/biology14040353
Zhou Y, Zhang Q, Gao P, Yang S, Xu Y. Unlocking the Potential of Animal Hair Shafts for Genomic Studies: A Comprehensive Evaluation of DNA Quality. Biology. 2025; 14(4):353. https://doi.org/10.3390/biology14040353
Chicago/Turabian StyleZhou, Yongheng, Qi Zhang, Peng Gao, Shuhui Yang, and Yanchun Xu. 2025. "Unlocking the Potential of Animal Hair Shafts for Genomic Studies: A Comprehensive Evaluation of DNA Quality" Biology 14, no. 4: 353. https://doi.org/10.3390/biology14040353
APA StyleZhou, Y., Zhang, Q., Gao, P., Yang, S., & Xu, Y. (2025). Unlocking the Potential of Animal Hair Shafts for Genomic Studies: A Comprehensive Evaluation of DNA Quality. Biology, 14(4), 353. https://doi.org/10.3390/biology14040353




