miR-4428 and miR-185-5p as Key Modulators of Insulin Sensitivity and Glucose Homeostasis: Insights into Pathways and Therapeutic Potential in Type 2 Diabetes Mellitus
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. miRNA Extraction
2.3. Microarray Analysis
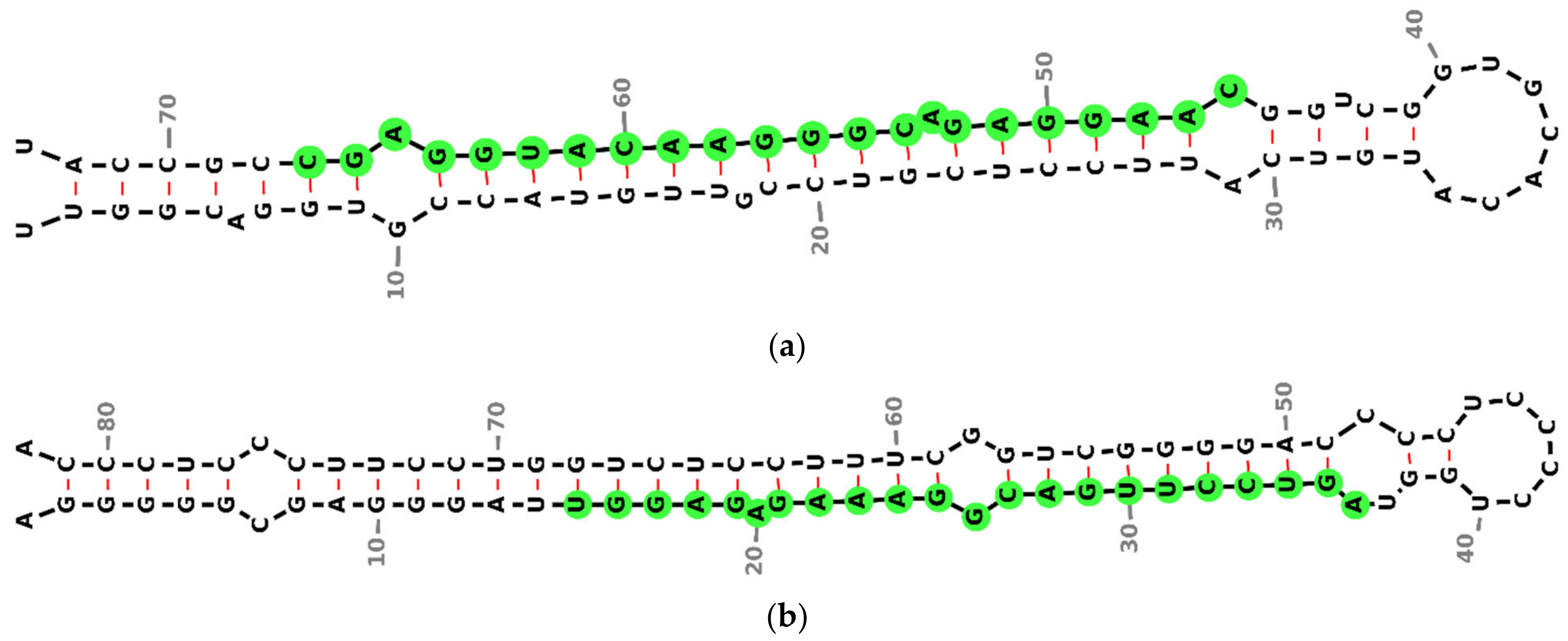
2.4. miRNA Secondary Structure Prediction
2.5. Target Prediction of miR-4428 and miR-185-5p
2.6. Network Analysis of miR-4428 and miR-185-5p Target Genes
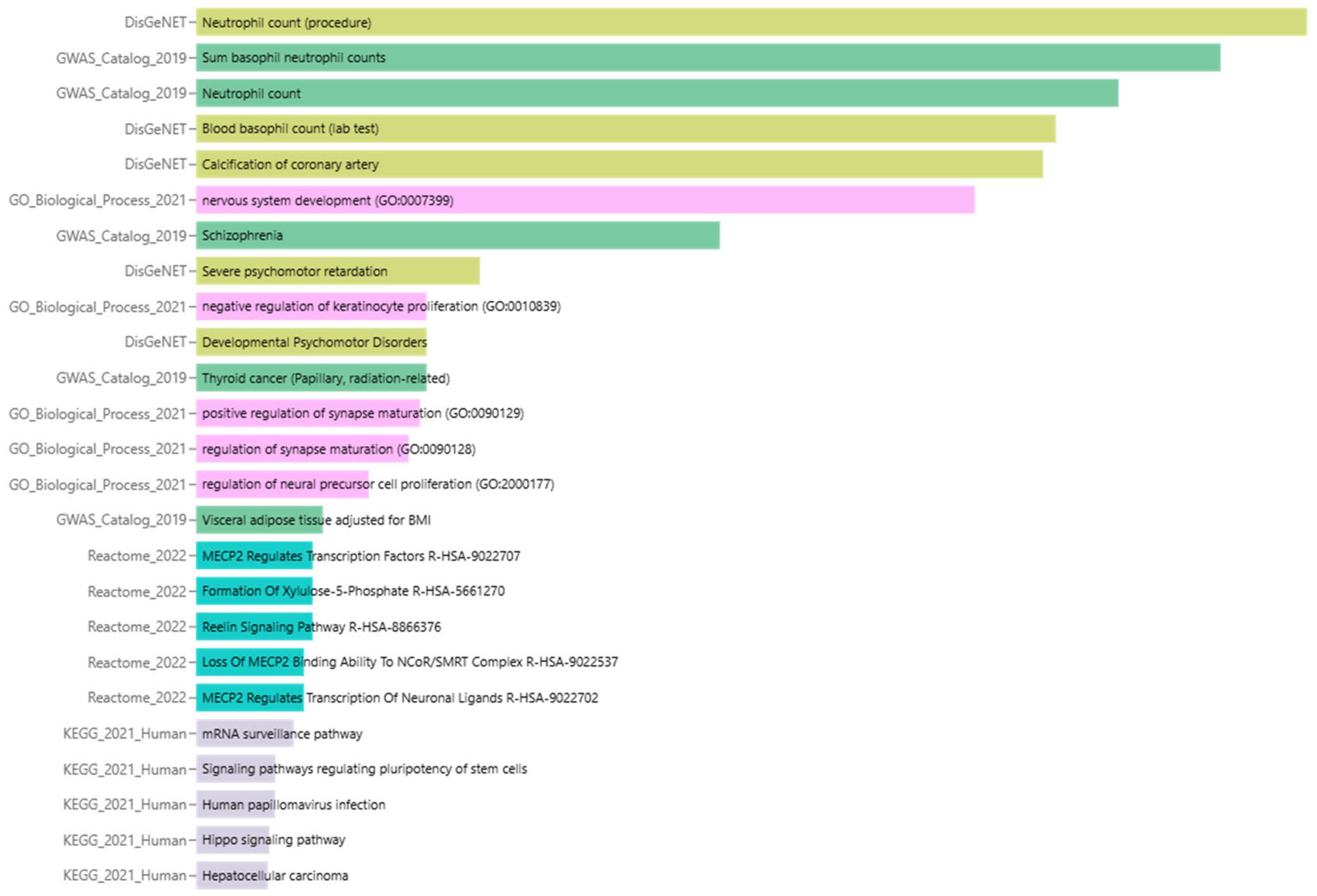
2.7. Enrichment Analysis of Biological Processes and Pathways
2.8. Summary of Enrichment Analysis Results
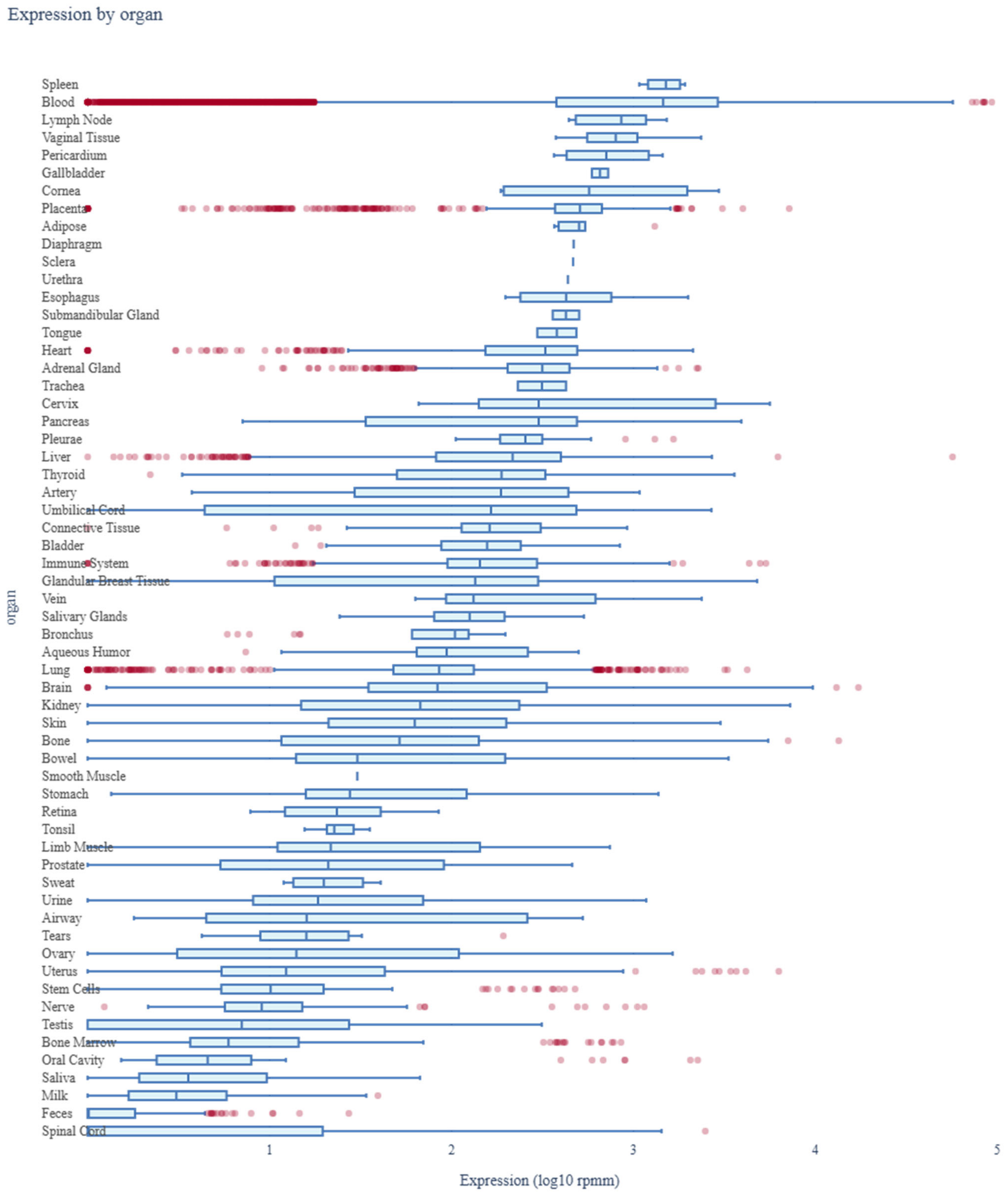
2.9. Assessment of miR-4428 and miR-185-5p Expression Profiles Across Human Organs Using miRNA Tissue Atlas 2025
2.10. Disease Association Analysis of miR-4428 and miR-185
2.11. Statistical Analysis
3. Results
3.1. Microarray Analysis
3.2. miRNA Secondary Structure Prediction
3.3. Target Prediction of miR-4428 and miR-185-5p
3.4. Network Analysis of miR-4428 and miR-185-5p Target Genes
3.5. Enrichment Analysis of Biological Processes and Pathways
3.6. Summary of Enrichment Analysis Results
3.7. Assessment of miR-4428 and miR-185-5p Expression Profiles Across Human Organs Using miRNA Tissue Atlas 2025
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| miRNA | MicroRNA |
| T1DM | Type 1 Diabetes Mellitus |
| T2DM | Type 2 Diabetes Mellitus |
| FBS | Fasting Blood Sugar |
| HbA1c | Hemoglobin A1c |
| lncRNA | Long Non-Coding RNA |
| FDR | False Discovery Rate |
| GO | Gene Ontology |
| β-cells | Beta cells |
References
- Fernandez-Valverde, S.L.; Taft, R.J.; Mattick, J.S. MicroRNAs in β-cell biology, insulin resistance, diabetes, and its complications. Diabetes 2011, 60, 1825–1831. [Google Scholar] [CrossRef] [PubMed]
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef] [PubMed]
- Keegan, L.P.; Hajji, K.; O’Connell, M.A. Adenosine Deaminase Acting on RNA (ADAR) Enzymes: A Journey from Weird to Wondrous. Acc. Chem. Res. 2023, 56, 3165–3174. [Google Scholar] [CrossRef] [PubMed]
- Pollak, N.M.; Hoffman, M.; Goldberg, I.J.; Drosatos, K. Krüppel-like factors: Crippling and un-crippling metabolic pathways. JACC Basic Transl. Sci. 2018, 3, 132–156. [Google Scholar] [CrossRef]
- Cui, A.; Fan, H.; Zhang, Y.; Zhang, Y.; Niu, D.; Liu, S.; Liu, Q.; Ma, W.; Shen, Z.; Shen, L.; et al. Dexamethasone-induced Krüppel-like factor 9 expression promotes hepatic gluconeogenesis and hyperglycemia. J. Clin. Investig. 2019, 129, 2266–2278. [Google Scholar] [CrossRef]
- Vallin, J.; Grantham, J. The role of the molecular chaperone CCT in protein folding and mediation of cytoskeleton-associated processes: Implications for cancer cell biology. Cell Stress Chaperones 2019, 24, 17–27. [Google Scholar] [CrossRef]
- Tian, Y.; Zhan, Y.; Jiang, Q.; Lu, W.; Li, X. Expression and function of PDGF-C in development and stem cells. Open Biol. 2021, 11, 210268. [Google Scholar] [CrossRef]
- Rodríguez, A.G.; Rodríguez, J.Z.; Barreto, A.; Sanabria-Barrera, S.; Iglesias, J.; Morales, L. Impact of Acute High Glucose on Mitochondrial Function in a Model of Endothelial Cells: Role of PDGF-C. Int. J. Mol. Sci. 2023, 24, 4394. [Google Scholar] [CrossRef]
- Ortiz, G.G.; Huerta, M.; González-Usigli, H.A.; Torres-Sánchez, E.D.; Delgado-Lara, D.L.; Pacheco-Moisés, F.P.; Mireles-Ramírez, M.A.; Torres-Mendoza, B.M.; Moreno-Cih, R.I.; Velázquez-Brizuela, I.E. Cognitive disorder and dementia in type 2 diabetes mellitus. World J. Diabetes 2022, 13, 319–337. [Google Scholar] [CrossRef]
- Dudek, S.M.; Alexander, G.M.; Farris, S. Rediscovering area CA2: Unique properties and functions. Nat. Rev. Neurosci. 2016, 17, 89–102. [Google Scholar] [CrossRef]
- Athanasaki, A.; Melanis, K.; Tsantzali, I.; Stefanou, M.I.; Ntymenou, S.; Paraskevas, S.G.; Kalamatianos, T.; Boutati, E.; Lambadiari, V.; Voumvourakis, K.I.; et al. Type 2 Diabetes Mellitus as a Risk Factor for Alzheimer’s Disease: Review and Meta-Analysis. Biomedicines 2022, 10, 778. [Google Scholar] [CrossRef] [PubMed]
- Khoo, S.; Griffen, S.C.; Xia, Y.; Baer, R.J.; German, M.S.; Cobb, M.H. Regulation of insulin gene transcription by ERK1 and ERK2 in pancreatic beta cells. J. Biol. Chem. 2003, 278, 32969–32977. [Google Scholar] [CrossRef] [PubMed]
- Cruz, C.D.; Cruz, F. The ERK 1 and 2 pathway in the nervous system: From basic aspects to possible clinical applications in pain and visceral dysfunction. Curr. Neuropharmacol. 2007, 5, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Šterk, M.; Križančić Bombek, L.; Skelin Klemen, M.; Slak Rupnik, M.; Marhl, M.; Stožer, A.; Gosak, M. NMDA receptor inhibition increases, synchronizes, and stabilizes the collective pancreatic beta cell activity: Insights through multilayer network analysis. PLoS Comput. Biol. 2021, 17, e1009002. [Google Scholar] [CrossRef]
- Hou, G.; Zhang, Z.W. NMDA Receptors Regulate the Development of Neuronal Intrinsic Excitability through Cell-Autonomous Mechanisms. Front. Cell. Neurosci. 2017, 11, 353. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Y.; Zhou, G.; Zhou, J.; Tian, Z.; Xu, J. circGRAMD1B contributes to migration, invasion and epithelial-mesenchymal transition of lung adenocarcinoma cells via modulating the expression of SOX4. Funct. Integr. Genom. 2023, 23, 75. [Google Scholar] [CrossRef]
- Huang, S.; Liang, S.; Huang, J.; Luo, P.; Mo, D.; Wang, H. LINC01806 mediated by STAT1 promotes cell proliferation, migration, invasion, and stemness in non-small cell lung cancer through Notch signaling by miR-4428/NOTCH2 axis. Cancer Cell Int. 2022, 22, 198. [Google Scholar] [CrossRef]
- Ma, X.; Liu, H.; Zhu, J.; Zhang, C.; Peng, Y.; Mao, Z.; Jing, Y.; Chen, F. miR-185-5p Regulates Inflammation and Phagocytosis through CDC42/JNK Pathway in Macrophages. Genes 2022, 13, 468. [Google Scholar] [CrossRef]
- Wang, T.; Li, N.; Yuan, L.; Zhao, M.; Li, G.; Chen, Y.; Zhou, H. MALAT1/miR-185-5p mediated high glucose-induced oxidative stress, mitochondrial injury and cardiomyocyte apoptosis via the RhoA/ROCK pathway. J. Cell. Mol. Med. 2023, 27, 2495–2506. [Google Scholar] [CrossRef]
- Yuan, Q.; Xu, T.; Chen, Y.; Qu, W.; Sun, D.; Liu, X.; Sun, L. MiR-185-5p ameliorates endoplasmic reticulum stress and renal fibrosis by downregulation of ATF6. Lab. Investig. 2020, 100, 1436–1446. [Google Scholar] [CrossRef]
- Ying, K.; Wang, L.; Long, G.; Lian, C.; Chen, Z.; Lin, W. ACTA2-AS1 suppresses lung adenocarcinoma progression via sequestering miR-378a-3p and miR-4428 to elevate SOX7 expression. Cell Biol. Int. 2020, 44, 2438–2449. [Google Scholar] [CrossRef] [PubMed]
- Jordan-Alejandre, E.; Campos-Parra, A.D.; Castro-López, D.L.; Silva-Cázares, M.B. Potential miRNA use as a biomarker: From breast cancer diagnosis to metastasis. Cells 2023, 12, 525. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Huang, Y.; Wang, Y.; Li, D.; Lei, C. LncRNA ACTA2-AS1 suppresses colon adenocarcinoma progression by sponging miR-4428 upregulation BCL2L11. Cancer Cell Int. 2021, 21, 4. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Wang, B.; Zhang, K.; Wang, C.; Wang, J.; An, Z.; Shu, L. RGMB-AS1/miR-4428/PBX1 axis drives the progression of cervical cancer. Transl. Cancer Res. 2020, 9, 3180. [Google Scholar] [CrossRef]
- Zhao, Y.; Guo, L.; Tian, J.; Wang, H. Downregulation of microRNA-185 expression in diabetic patients increases the expression of NOS2 and results in vascular injury. Exp. Ther. Med. 2021, 22, 1458. [Google Scholar] [CrossRef]
- Zheng, H.; Wan, J.; Shan, Y.; Song, X.; Jin, J.; Su, Q.; Chen, S.; Lu, X.; Yang, J.; Li, Q.; et al. MicroRNA-185-5p inhibits hepatic gluconeogenesis and reduces fasting blood glucose levels by suppressing G6Pase. Theranostics 2021, 11, 7829. [Google Scholar] [CrossRef]
- Galicia-Garcia, U.; Benito-Vicente, A.; Jebari, S.; Larrea-Sebal, A.; Siddiqi, H.; Uribe, K.B.; Ostolaza, H.; Martín, C. Pathophysiology of type 2 diabetes mellitus. Int. J. Mol. Sci. 2020, 21, 6275. [Google Scholar] [CrossRef]
- Chen, D.L.; Shen, D.Y.; Han, C.K.; Tian, Y. LncRNA MEG3 aggravates palmitate-induced insulin resistance by regulating miR-185-5p/Egr2 axis in hepatic cells. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5456–5467. [Google Scholar] [CrossRef]
- Qi, S.; Wang, X. Decreased Expression of miR-185 in serum and placenta of patients with gestational diabetes mellitus. Clin. Lab. 2019, 65, 2367–2372. [Google Scholar] [CrossRef]

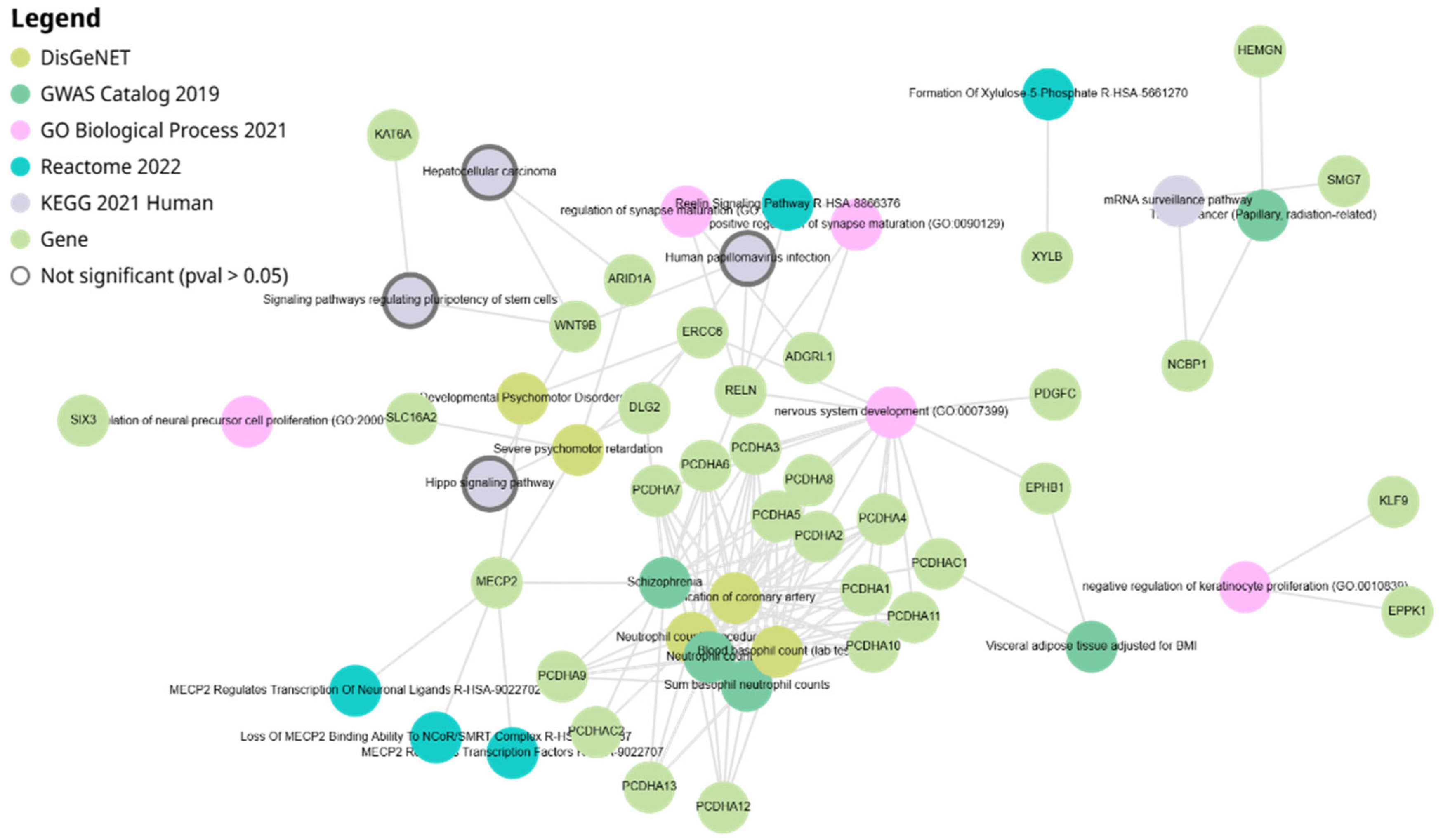

| Target Rank | Target Score | Gene Symbol | Gene Description |
|---|---|---|---|
| Top Predicted Targets of miR-4428 | |||
| 1 | 100 | EPHB1 | EPH receptor B1 |
| 2 | 99 | MECP2 | methyl-CpG binding protein 2 |
| 3 | 98 | KAT6A | lysine acetyltransferase 6A |
| 4 | 98 | ADAR | adenosine deaminase, RNA specific |
| 5 | 98 | RELN | reelin |
| 6 | 97 | CCT8 | chaperonin containing TCP1 subunit 8 |
| 7 | 97 | KLF9 | Kruppel like factor 9 |
| 8 | 96 | NCBP1 | nuclear cap binding protein subunit 1 |
| 9 | 95 | ST8SIA6 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| 10 | 95 | ZFP91 | ZFP91 zinc finger protein |
| 11 | 95 | PDGFC | platelet-derived growth factor C |
| 12 | 95 | EGR3 | early growth response 3 |
| 13 | 95 | HEMGN | hemogen |
| Top Predicted Targets of miR-185-5p | |||
| 1 | 99 | SMG7 | SMG7, nonsense-mediated mRNA decay factor |
| 2 | 99 | SLC16A2 | solute carrier family 16-member 2 |
| 3 | 98 | SOGA1 | suppressor of glucose, autophagy associated 1 |
| 4 | 98 | EPPK1 | epiplakin 1 |
| 5 | 97 | WNT9B | Wnt family member 9B |
| 6 | 97 | SOX13 | SRY-box 13 |
| 7 | 97 | XYLB | xylulokinase |
| 8 | 97 | HP1BP3 | heterochromatin protein 1 binding protein 3 |
| 9 | 96 | PCDHA8 | protocadherin alpha 8 |
| 10 | 96 | PCDHAC1 | protocadherin alpha subfamily C, 1 |
| 11 | 96 | DLG2 | discs large MAGUK scaffold protein 2 |
| 12 | 96 | PCDHAC2 | protocadherin alpha subfamily C, 2 |
| 13 | 96 | PCDHA13 | protocadherin alpha 13 |
| 14 | 96 | ERCC6 | ERCC excision repair 6, chromatin remodeling factor |
| 15 | 96 | PCDHA10 | protocadherin alpha 10 |
| 16 | 96 | PCDHA5 | protocadherin alpha 5 |
| 17 | 96 | ADGRL1 | adhesion G protein-coupled receptor L1 |
| 18 | 96 | PCDHA6 | protocadherin alpha 6 |
| 19 | 96 | PCDHA11 | protocadherin alpha 11 |
| 20 | 96 | PCDHA3 | protocadherin alpha 3 |
| 21 | 96 | PCDHA9 | protocadherin alpha 9 |
| 22 | 96 | ZNF704 | zinc finger protein 704 |
| 23 | 96 | RAE1 | ribonucleic acid export 1 |
| 24 | 96 | PCDHA4 | protocadherin alpha 4 |
| 25 | 96 | ABCG4 | ATP binding cassette subfamily G member 4 |
| 26 | 96 | PCDHA1 | protocadherin alpha 1 |
| 27 | 96 | PCDHA7 | protocadherin alpha 7 |
| 28 | 96 | PCDHA2 | protocadherin alpha 2 |
| 29 | 96 | CDH4 | cadherin 4 |
| 30 | 96 | CA10 | carbonic anhydrase 10 |
| 31 | 96 | PCDHA12 | protocadherin alpha 12 |
| 32 | 95 | TUBGCP3 | tubulin gamma complex associated protein 3 |
| 33 | 95 | FAM234B | family with sequence similarity 234 member B |
| 34 | 95 | ARID1A | AT-rich interaction domain 1A |
| 35 | 95 | C17orf77 | chromosome 17 open reading frame 77 |
| 36 | 95 | SIX3 | SIX homeobox 3 |
| 37 | 95 | RAB35 | RAB35, member RAS oncogene family |
| 38 | 95 | ZNF236 | zinc finger protein 236 |
| Term | Library | p-Value | q-Value | z-Score | Combined Score |
|---|---|---|---|---|---|
| Neutrophil count (procedure) | DisGeNET | 8.11 × 10−19 | 8.71 × 10−16 | 70.02 | 2917 |
| Sum basophil neutrophil counts | GWAS_Catalog_2019 | 2.08 × 10−17 | 2.52 × 10−15 | 53.4 | 2051 |
| Neutrophil count | GWAS_Catalog_2019 | 9.56 × 10−16 | 5.79 × 10−14 | 38.88 | 1345 |
| Blood basophil count (lab test) | DisGeNET | 1.01 × 10−14 | 5.40 × 10−12 | 32 | 1031 |
| Calcification of coronary artery | DisGeNET | 1.63 × 10−14 | 5.82 × 10−12 | 22.67 | 719.8 |
| nervous system development (GO:0007399) | GO_Biological_Process_2021 | 2.10 × 10−13 | 1.06 × 10−10 | 18.82 | 549.5 |
| Schizophrenia | GWAS_Catalog_2019 | 2.97 × 10−9 | 1.20 × 10−7 | 9.171 | 180.1 |
| Severe psychomotor retardation | DisGeNET | 0.00002405 | 0.006457 | 27.3 | 290.3 |
| Negative regulation of keratinocyte proliferation (GO:0010839) | GO_Biological_Process_2021 | 0.0001768 | 0.03812 | 135.7 | 1172 |
| Developmental Psychomotor Disorders | DisGeNET | 0.0001768 | 0.03164 | 135.7 | 1172 |
| Thyroid cancer (Papillary, radiation-related) | GWAS_Catalog_2019 | 0.0001768 | 0.005347 | 135.7 | 1172 |
| positive regulation of synapse maturation (GO:0090129) | GO_Biological_Process_2021 | 0.0002269 | 0.03812 | 116.3 | 975.7 |
| regulation of synapse maturation (GO:0090128) | GO_Biological_Process_2021 | 0.0003455 | 0.04353 | 90.43 | 720.8 |
| regulation of neural precursor cell proliferation (GO:2000177) | GO_Biological_Process_2021 | 0.001559 | 0.1179 | 38.73 | 250.4 |
| Visceral adipose tissue adjusted for BMI | GWAS_Catalog_2019 | 0.008685 | 0.1631 | 15.32 | 72.72 |
| MECP2 Regulates Transcription Factors R-HSA-9022707 | Reactome_2022 | 0.01269 | 0.3185 | 99.72 | 435.5 |
| Formation Of Xylulose-5-Phosphate R-HSA-5661270 | Reactome_2022 | 0.01269 | 0.3185 | 99.72 | 435.5 |
| Reelin Signaling Pathway R-HSA-8866376 | Reactome_2022 | 0.01269 | 0.3185 | 99.72 | 435.5 |
| Loss Of MECP2 Binding Ability To NCoR/SMRT Complex R-HSA-9022537 | Reactome_2022 | 0.01772 | 0.3185 | 66.48 | 268.1 |
| MECP2 Regulates Transcription Of Neuronal Ligands R-HSA-9022702 | Reactome_2022 | 0.01772 | 0.3185 | 66.48 | 268.1 |
| mRNA surveillance pathway | KEGG_2021_Human | 0.02593 | 0.4923 | 8.441 | 30.83 |
| Signaling pathways regulating pluripotency of stem cells | KEGG_2021_Human | 0.05155 | 0.4923 | 5.734 | 17 |
| Human papillomavirus infection | KEGG_2021_Human | 0.05225 | 0.4923 | 3.739 | 11.04 |
| Hippo signaling pathway | KEGG_2021_Human | 0.06493 | 0.4923 | 5.017 | 13.72 |
| Hepatocellular carcinoma | KEGG_2021_Human | 0.06844 | 0.4923 | 4.864 | 13.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rattanapan, Y.; Duangchan, T.; Sai-ong, T.; Chareonsirisuthigul, T. miR-4428 and miR-185-5p as Key Modulators of Insulin Sensitivity and Glucose Homeostasis: Insights into Pathways and Therapeutic Potential in Type 2 Diabetes Mellitus. Biology 2025, 14, 424. https://doi.org/10.3390/biology14040424
Rattanapan Y, Duangchan T, Sai-ong T, Chareonsirisuthigul T. miR-4428 and miR-185-5p as Key Modulators of Insulin Sensitivity and Glucose Homeostasis: Insights into Pathways and Therapeutic Potential in Type 2 Diabetes Mellitus. Biology. 2025; 14(4):424. https://doi.org/10.3390/biology14040424
Chicago/Turabian StyleRattanapan, Yanisa, Thitinat Duangchan, Thaveesak Sai-ong, and Takol Chareonsirisuthigul. 2025. "miR-4428 and miR-185-5p as Key Modulators of Insulin Sensitivity and Glucose Homeostasis: Insights into Pathways and Therapeutic Potential in Type 2 Diabetes Mellitus" Biology 14, no. 4: 424. https://doi.org/10.3390/biology14040424
APA StyleRattanapan, Y., Duangchan, T., Sai-ong, T., & Chareonsirisuthigul, T. (2025). miR-4428 and miR-185-5p as Key Modulators of Insulin Sensitivity and Glucose Homeostasis: Insights into Pathways and Therapeutic Potential in Type 2 Diabetes Mellitus. Biology, 14(4), 424. https://doi.org/10.3390/biology14040424






