Transcriptomic Analysis Identifies Molecular Response of the Tolerant Alfalfa (Medicago sativa) Cultivar Nongjing 1 to Saline-Alkali Stress
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Quantitative Real-Time PCR Validation
2.3. Metabolite Extraction
2.4. Metabolite Detection
2.5. Physiological Measurements
2.6. Total RNA Isolation, Library Construction, and RNA Sequencing
2.7. Enrichment Analysis of Differentially Expressed Genes
3. Results
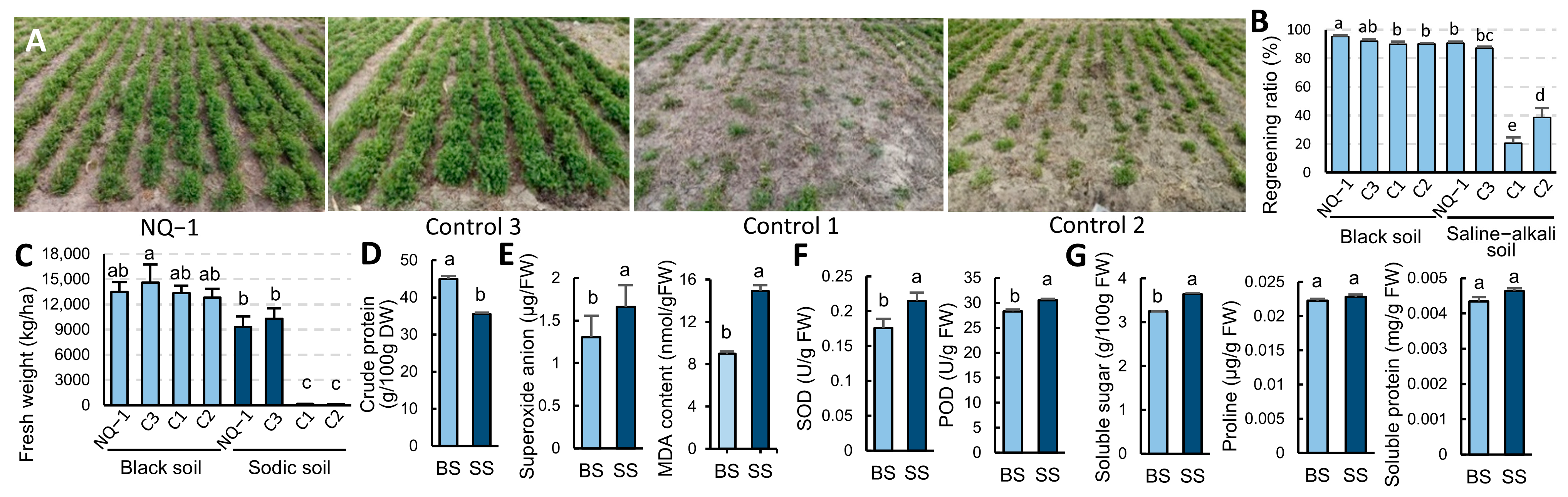
3.1. Nongjing 1 Is a High-Yielding, High-Quality, and Saline-Alkali-Tolerant Cultivar Suitable for Planting in Northeast China
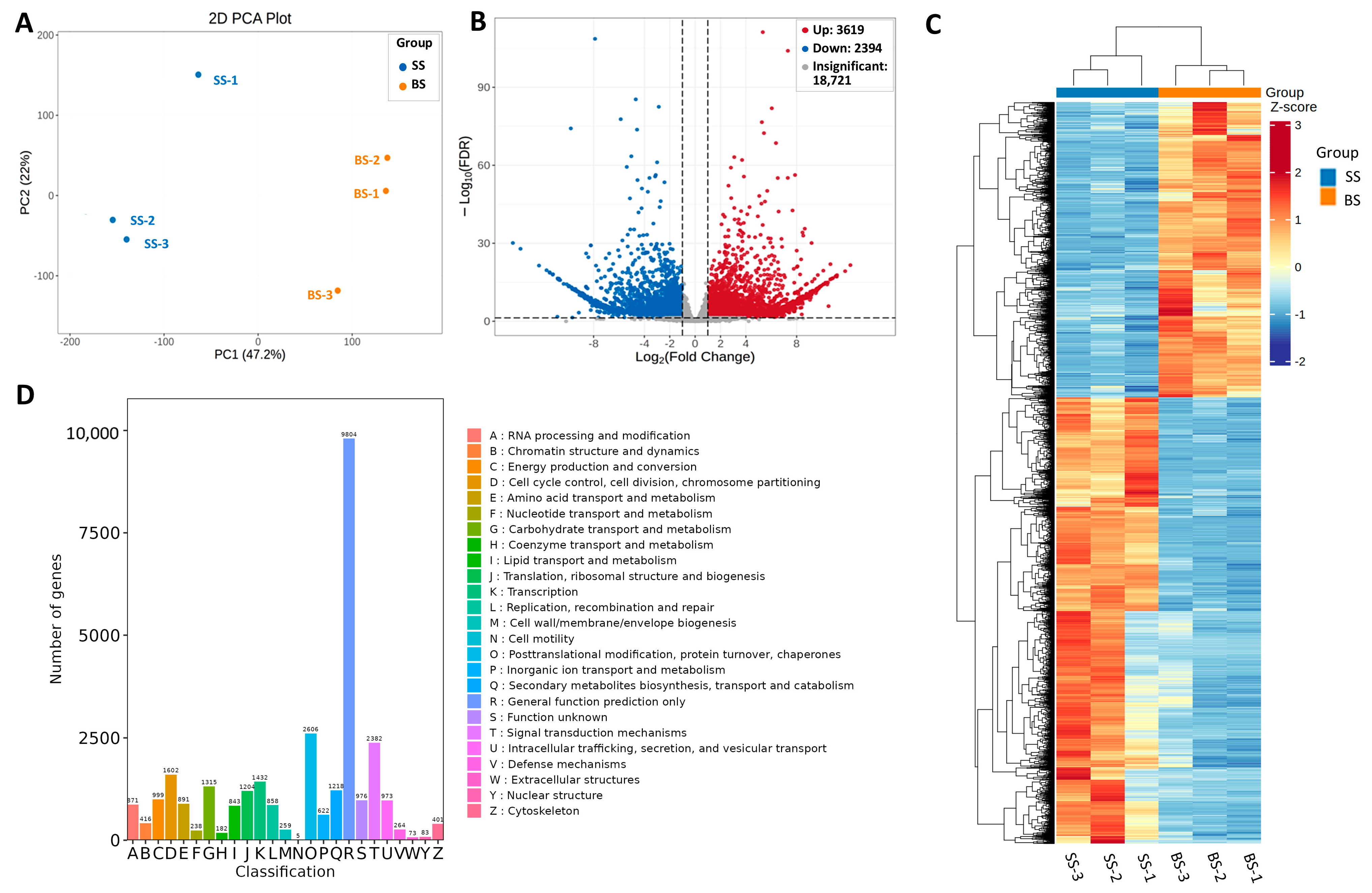
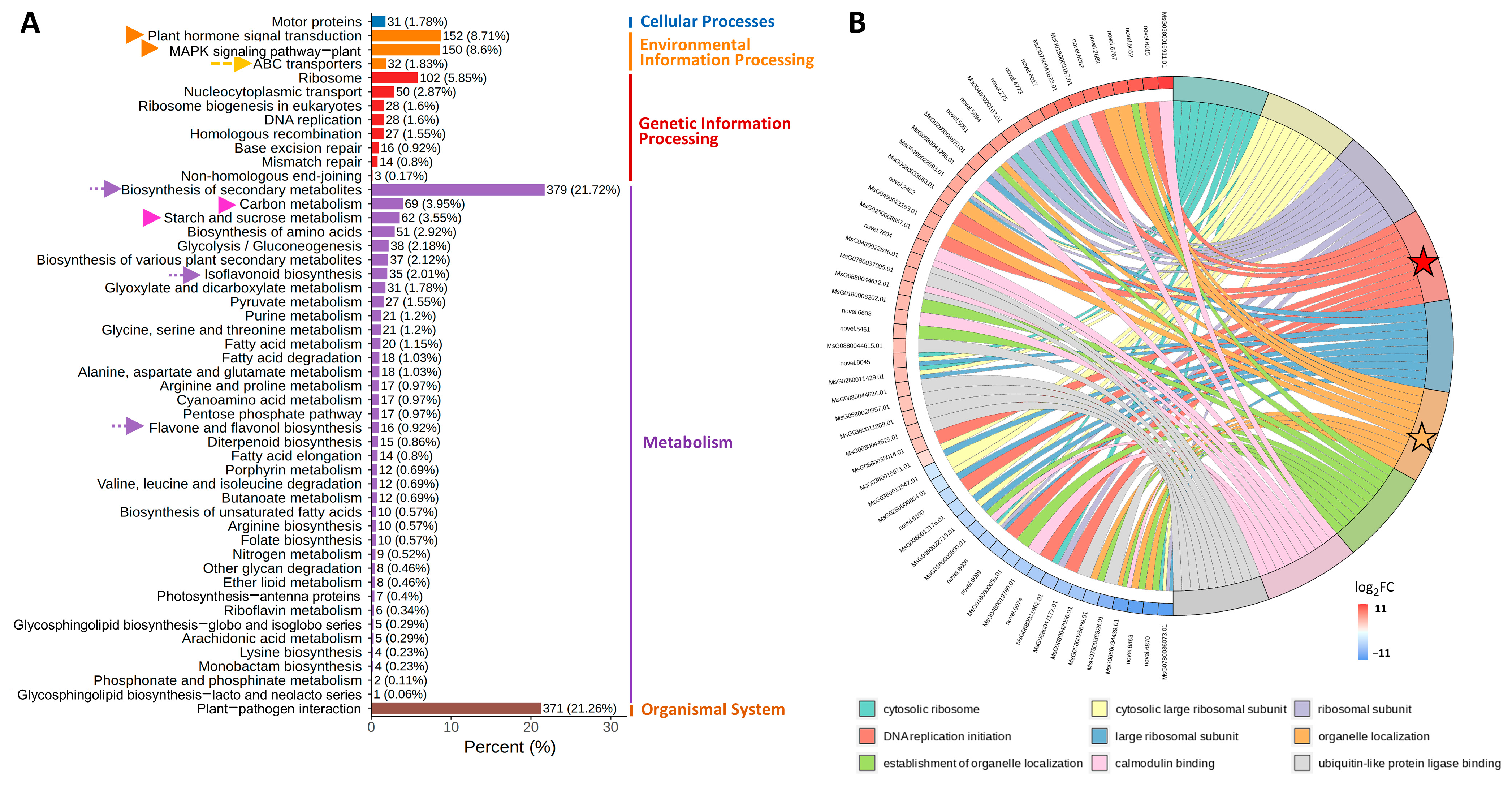
3.2. Comparative Transcriptome Analysis of NQ-1 Leaves
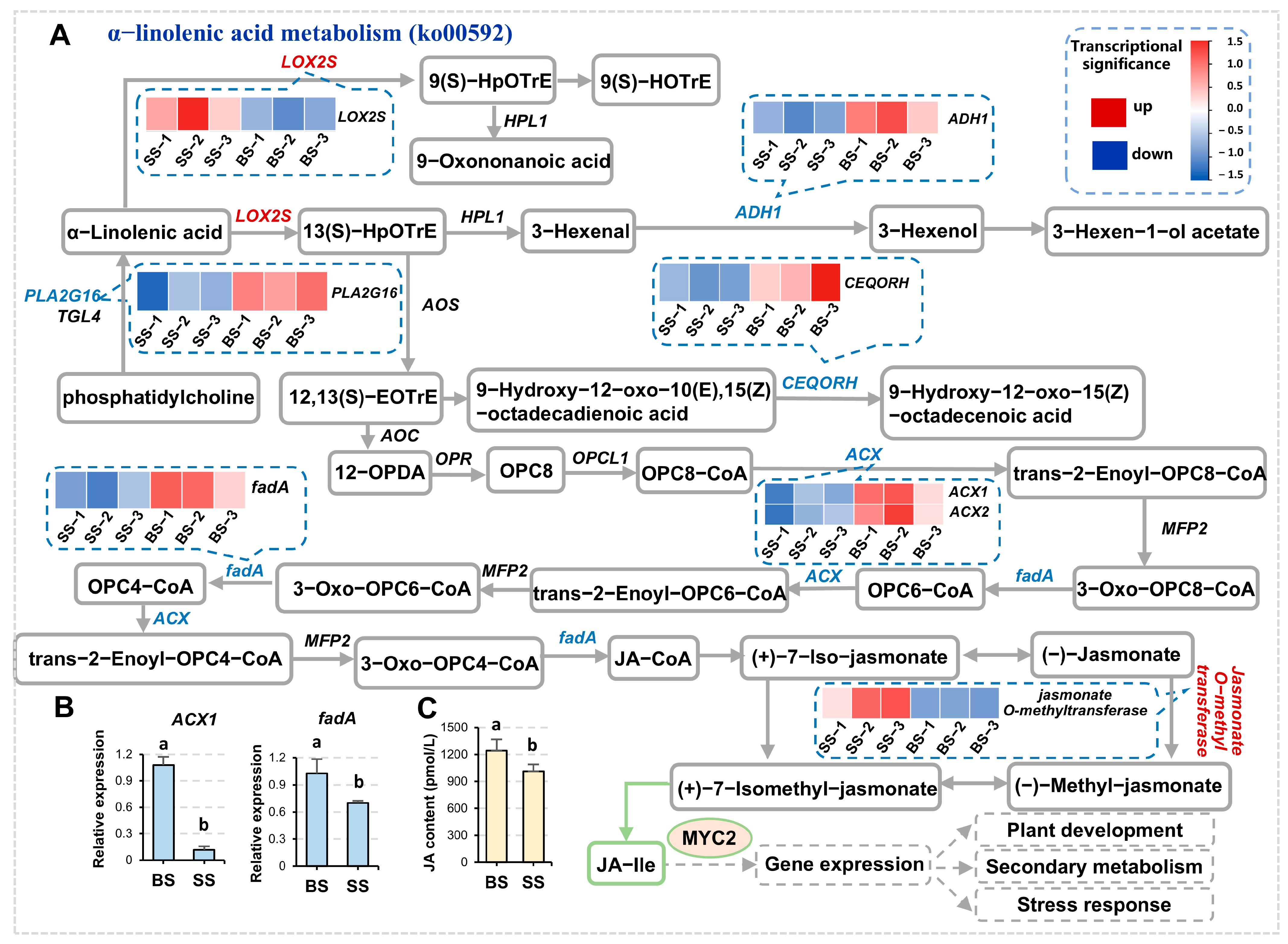
3.3. α-Linolenic Acid Metabolism Was Responsive to Saline-Alkali Stress
3.4. Saline-Alkali Stress Affected the Photosynthesis of NQ-1
3.5. Saline-Alkali Stress Did Not Inhibit Lignin Biosynthesis in NQ-1 Under Saline-Alkali Stress
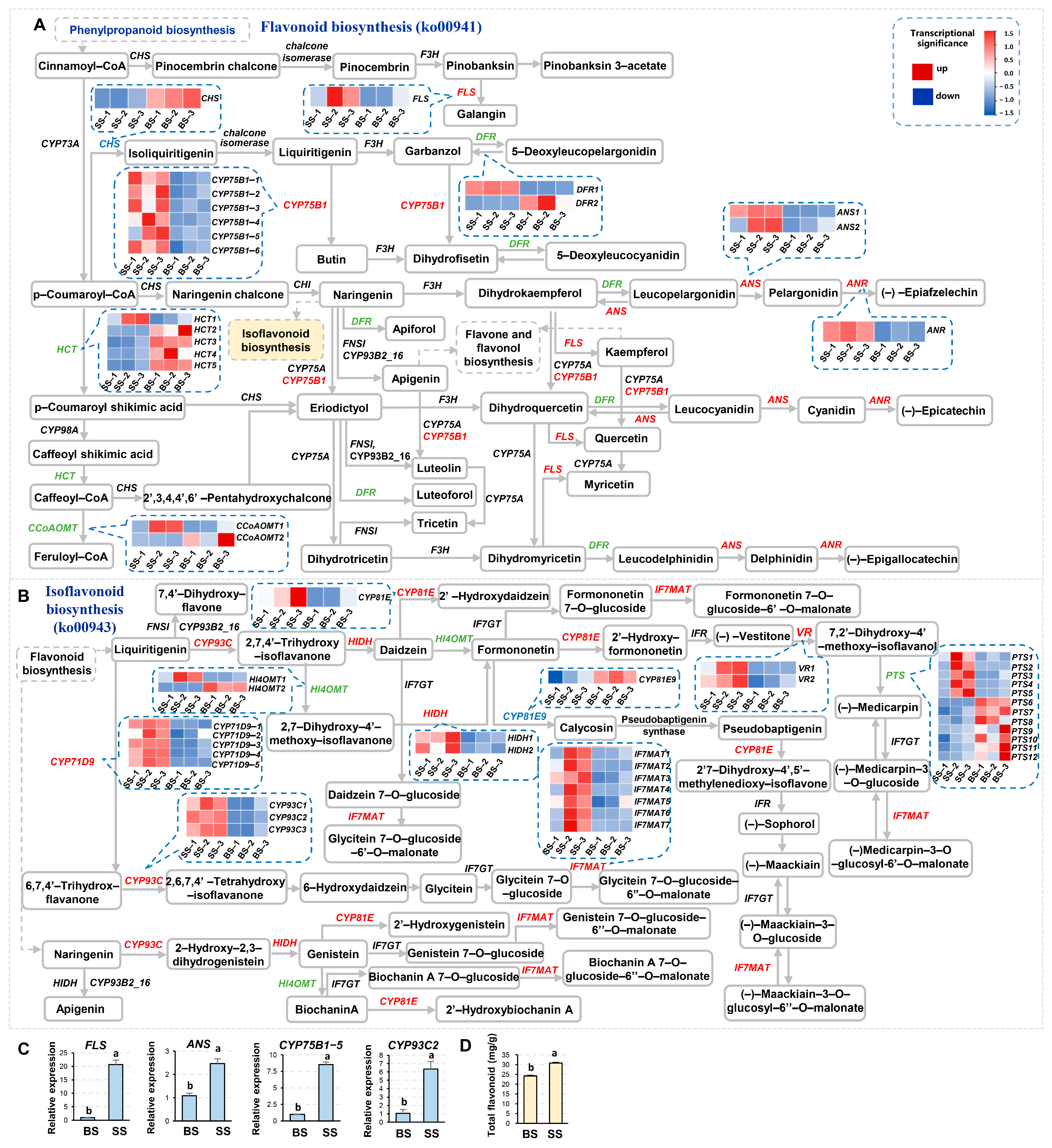
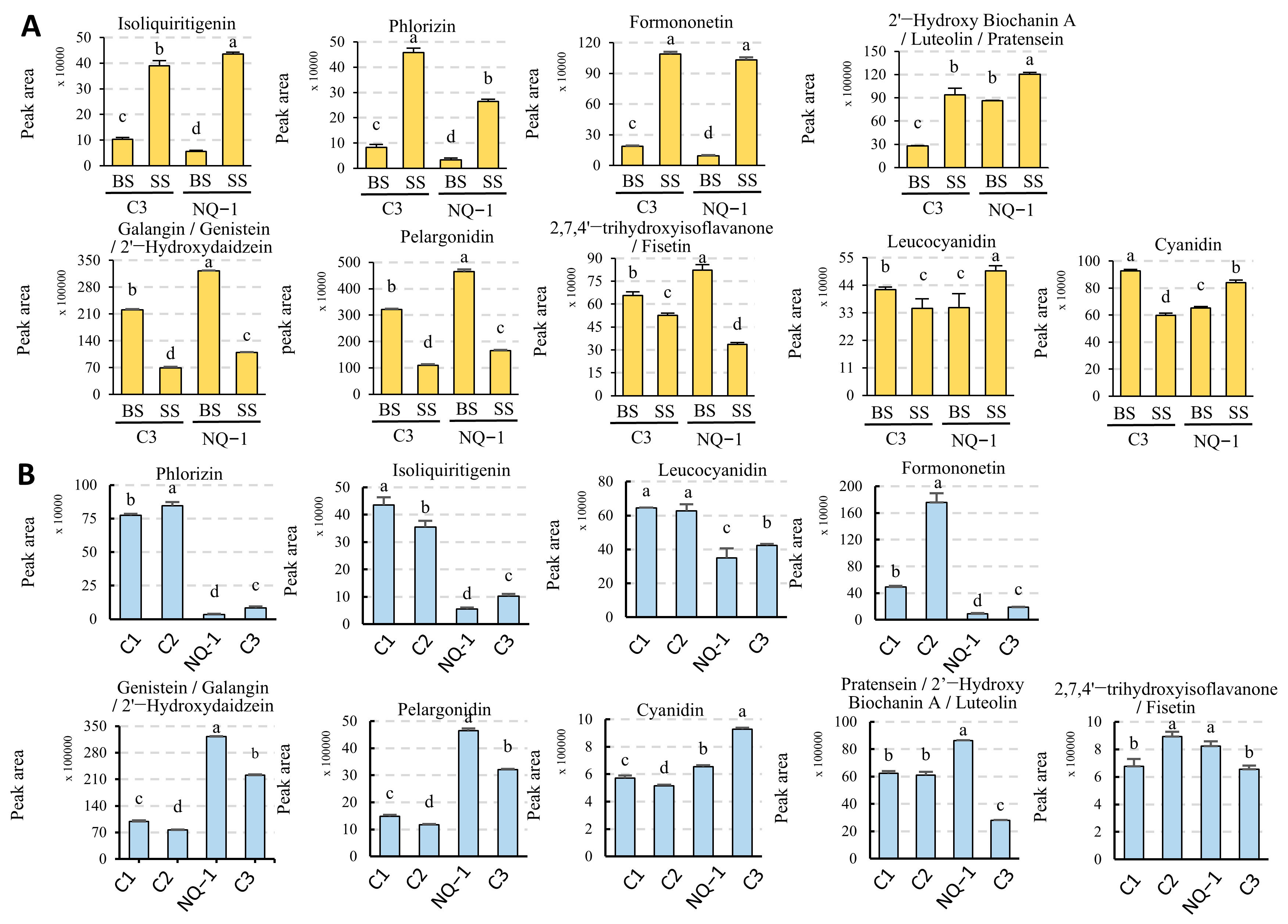
3.6. Flavonoid and Isoflavonoid Biosynthesis Pathways in NQ-1 Were Activated Under Saline-Alkali Stress
3.7. Identification and Analysis of Upstream Transcription Factors Potentially Functioning in Flavonoid and Isoflavonoid Biosynthesis in NQ-1 Under Saline-Alkali Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Munns, R.; Gilliham, M. Salinity tolerance of crops—What is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Dou, J.; Tang, Z.; Yu, J.; Wang, G.; An, W.; Zhang, Y.; Yang, Q. Effects of exogenous melatonin on the growth and photosynthetic characteristics of tomato seedlings under saline-alkali stress. Sci. Rep. 2025, 15, 5172. [Google Scholar] [CrossRef]
- Xu, S.; Na, M.; Huang, Y.; Zhang, J.; Zhou, J.; Li, L.-J. Changes in microbial carbon cycling functions along rice cultivation chronosequences in saline-alkali soils. Soil Biol. Biochem. 2025, 202, 109699. [Google Scholar] [CrossRef]
- Kang, P.; Bao, A.K.; Kumar, T.; Pan, Y.Q.; Bao, Z.; Wang, F.; Wang, S.M. Assessment of Stress Tolerance, Productivity, and Forage Quality in T(1) Transgenic Alfalfa Co-overexpressing ZxNHX and ZxVP1-1 from Zygophyllum xanthoxylum. Front. Plant Sci. 2016, 7, 1598. [Google Scholar] [CrossRef] [PubMed]
- Stritzler, M.; Elba, P.; Berini, C.; Gomez, C.; Ayub, N.; Soto, G. High-quality forage production under salinity by using a salt-tolerant AtNXH1-expressing transgenic alfalfa combined with a natural stress-resistant nitrogen-fixing bacterium. J. Biotechnol. 2018, 276–277, 42–45. [Google Scholar] [CrossRef]
- Hrbackova, M.; Dvorak, P.; Takac, T.; Ticha, M.; Luptovciak, I.; Samajova, O.; Ovecka, M.; Samaj, J. Biotechnological Perspectives of Omics and Genetic Engineering Methods in Alfalfa. Front. Plant Sci. 2020, 11, 592. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, L.; Zhang, T.; Sod, B.; Xu, Y.; Li, M.; Kang, J.; Yang, Q.; Li, X.; Long, R. Identification and Functional Prediction of Salt/Alkali-Responsive lncRNAs during Alfalfa Germination. Agriculture 2024, 14, 930. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Fang, S.; Hou, X.; Liang, X. Response Mechanisms of Plants Under Saline-Alkali Stress. Front. Plant Sci. 2021, 12, 667458. [Google Scholar] [CrossRef]
- Rao, Y.; Peng, T.; Xue, S. Mechanisms of plant saline-alkaline tolerance. J. Plant Physiol. 2023, 281, 153916. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Sánchez, M.C.; Domingo, R.; Torrecillas, A.; Pérez-Pastor, A. Water stress preconditioning to improve drought resistance in young apricot plants. Plant Sci. 2000, 156, 245–251. [Google Scholar] [CrossRef]
- Álvarez, S.; Sánchez-Blanco, M.J. Changes in growth rate, root morphology and water use efficiency of potted Callistemon citrinus plants in response to different levels of water deficit. Sci. Hortic. 2013, 156, 54–62. [Google Scholar] [CrossRef]
- Franco-Navarro, J.D.; Brumos, J.; Rosales, M.A.; Cubero-Font, P.; Talon, M.; Colmenero-Flores, J.M. Chloride regulates leaf cell size and water relations in tobacco plants. J. Exp. Bot. 2016, 67, 873–891. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Bellot, M.J.; Nortes, P.A.; Ortuño, M.F.; Romero, C.; Fernandez-Garcia, N.; Sánchez-Blanco, M.J. Influence of arbuscular mycorrhizal fungi and treated wastewater on water relations and leaf structure alterations of Viburnum tinus L. plants during both saline and recovery periods. J. Plant Physiol. 2015, 188, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Longstreth, D.J.; Nobel, P.S. Salinity effects on leaf anatomy: Consequences for photosynthesis. Plant Physiol. 1979, 63, 700–703. [Google Scholar] [CrossRef]
- Zhou, X.; Li, J.; Wang, Y.; Liang, X.; Zhang, M.; Lu, M.; Guo, Y.; Qin, F.; Jiang, C. The classical SOS pathway confers natural variation of salt tolerance in maize. New Phytol. 2022, 236, 479–494. [Google Scholar] [CrossRef]
- Hauser, F.; Horie, T. A conserved primary salt tolerance mechanism mediated by HKT transporters: A mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environ. 2010, 33, 552–565. [Google Scholar] [CrossRef]
- Gholizadeh, F.; Mirmazloum, I.; Janda, T. Genome-wide identification of HKT gene family in wheat (Triticum aestivum L.): Insights from the expression of multiple genes (HKT, SOS, TVP and NHX) under salt stress. Plant Stress 2024, 13, 100539. [Google Scholar] [CrossRef]
- Dikilitas, M.; Simsek, E.; Roychoudhury, A. Role of proline and glycine betaine in overcoming abiotic stresses. In Protective Chemical Agents in the Amelioration of Plant Abiotic Stress: Biochemical and Molecular Perspectives; John Wiley & Sons: Hoboken, NJ, USA, 2020; pp. 1–23. [Google Scholar]
- Rhaman, M.S.; Rauf, F.; Tania, S.S.; Bayazid, N.; Tahjib-ul-Arif, M.; Robin, A.H.K.; Hoque, M.A.; Yang, X.; Murata, Y.; Brestic, M. Proline and glycine betaine: A dynamic duo for enhancing salt stress resilience in maize by regulating growth, Stomatal size, and Oxidative stress responses. Plant Stress 2024, 14, 100563. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Raihan, M.R.H.; Masud, A.A.C.; Rahman, K.; Nowroz, F.; Rahman, M.; Nahar, K.; Fujita, M. Regulation of Reactive Oxygen Species and Antioxidant Defense in Plants under Salinity. Int. J. Mol. Sci. 2021, 22, 9326. [Google Scholar] [CrossRef] [PubMed]
- Kesawat, M.S.; Satheesh, N.; Kherawat, B.S.; Kumar, A.; Kim, H.U.; Chung, S.M.; Kumar, M. Regulation of Reactive Oxygen Species during Salt Stress in Plants and Their Crosstalk with Other Signaling Molecules-Current Perspectives and Future Directions. Plants 2023, 12, 864. [Google Scholar] [CrossRef] [PubMed]
- Hameed, A.; Gulzar, S.; Aziz, I.; Hussain, T.; Gul, B.; Khan, M.A. Effects of salinity and ascorbic acid on growth, water status and antioxidant system in a perennial halophyte. AoB Plants 2015, 7, plv004. [Google Scholar] [CrossRef]
- Seifikalhor, M.; Aliniaeifard, S.; Shomali, A.; Azad, N.; Hassani, B.; Lastochkina, O.; Li, T. Calcium signaling and salt tolerance are diversely entwined in plants. Plant Signal. Behav. 2019, 14, 1665455. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Bheri, M.; Bisht, D.; Pandey, G.K. Calcium signaling and transport machinery: Potential for development of stress tolerance in plants. Curr. Plant Biol. 2022, 29, 100235. [Google Scholar] [CrossRef]
- Kiegerl, S.; Cardinale, F.; Siligan, C.; Gross, A.; Baudouin, E.; Liwosz, A.; Eklöf, S.; Till, S.; Bögre, L.; Hirt, H. SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. Plant Cell 2000, 12, 2247–2258. [Google Scholar]
- Ding, R.; Li, J.; Wang, J.; Li, Y.; Ye, W.; Yan, G.; Yin, Z. Molecular traits of MAPK kinases and the regulatory mechanism of GhMAPKK5 alleviating drought/salt stress in cotton. Plant Physiol. 2024, 196, 2030–2047. [Google Scholar] [CrossRef]
- Zhang, J.; Jia, W.; Yang, J.; Ismail, A.M. Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Res. 2006, 97, 111–119. [Google Scholar] [CrossRef]
- Hussain, Q.; Asim, M.; Zhang, R.; Khan, R.; Farooq, S.; Wu, J. Transcription Factors Interact with ABA through Gene Expression and Signaling Pathways to Mitigate Drought and Salinity Stress. Biomolecules 2021, 11, 1159. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, J.; Liu, B.; Xu, Z.Y. Dynamic regulation of DNA methylation and histone modifications in response to abiotic stresses in plants. J. Integr. Plant Biol. 2022, 64, 2252–2274. [Google Scholar] [CrossRef]
- Czajka, K.; Mehes-Smith, M.; Nkongolo, K. DNA methylation and histone modifications induced by abiotic stressors in plants. Genes Genom. 2022, 44, 279–297. [Google Scholar] [CrossRef]
- Chen, S.; Wang, X.; Cheng, Y.; Gao, H.; Chen, X. A Review of Classification, Biosynthesis, Biological Activities and Potential Applications of Flavonoids. Molecules 2023, 28, 4982. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Zhu, L.; Ma, D.; Zhang, F.; Cai, Z.; Bai, H.; Hui, J.; Li, S.; Xu, X.; Li, M. Integrated Metabolomics and Transcriptomics Analyses Highlight the Flavonoid Compounds Response to Alkaline Salt Stress in Glycyrrhiza uralensis Leaves. J. Agric. Food Chem. 2024, 72, 5477–5490. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Lv, L.; Meng, C.; Zhang, C.; Li, Y. Integrative Analysis of the Metabolome and Transcriptome of Sorghum bicolor Reveals Dynamic Changes in Flavonoids Accumulation under Saline-Alkali Stress. J. Agric. Food Chem. 2020, 68, 14781–14789. [Google Scholar] [CrossRef]
- Gao, Y.; Dong, X.; Wang, R.; Hao, F.; Zhang, H.; Zhang, Y.; Lin, G. Exogenous Calcium Alleviates Oxidative Stress Caused by Salt Stress in Peanut Seedling Roots by Regulating the Antioxidant Enzyme System and Flavonoid Biosynthesis. Antioxidants 2024, 13, 233. [Google Scholar] [CrossRef]
- Peng, W.; Cai, W.; Pan, J.; Su, X.; Dou, L. Molecular Mechanisms of Alfalfa Response to Abiotic Stresses. Plants 2025, 14, 487. [Google Scholar] [CrossRef] [PubMed]
- Qian, G.; Li, X.; Zhang, H.; Zhang, H.; Zhou, J.; Ma, X.; Sun, W.; Yang, W.; He, R.; Wahab, A.-t.; et al. Metabolomics analysis reveals the accumulation patterns of flavonoids and phenolic acids in quinoa (Chenopodium quinoa Willd.) grains of different colors. Food Chem. X 2023, 17, 100594. [Google Scholar] [CrossRef]
- Qian, G.; Wang, M.; Zhou, J.; Wang, X.; Zhang, Y.; Liu, Y.; Zhu, P.; Han, L.; Li, X.; Liu, C.; et al. Analysis of widely targeted metabolites of quinoa sprouts (Chenopodium quinoa Willd.) under saline-alkali stress provides new insights into nutritional value. Food Chem. 2024, 448, 138575. [Google Scholar] [CrossRef]
- Qian, G.; Wang, M.; Wang, X.; Liu, K.; Li, Y.; Bu, Y.; Li, L. Integrated Transcriptome and Metabolome Analysis of Rice Leaves Response to High Saline-Alkali Stress. Int. J. Mol. Sci. 2023, 24, 4062. [Google Scholar] [CrossRef]
- Ahmadi, A.; Shahidi, S.-A.; Safari, R.; Motamedzadegan, A.; Ghorbani-HasanSaraei, A. Evaluation of stability and antibacterial properties of extracted chlorophyll from alfalfa (Medicago sativa L.). Food Chem. Toxicol. 2022, 163, 112980. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, H.; Zhao, X.; Zhou, J.; Qin, G.; Liu, Y.; Kou, X.; Zhao, Z.; Wu, T.; Zhu, J.-K.; et al. SYNTAXIN OF PLANTS81 regulates root meristem activity and stem cell niche maintenance via ROS signaling. Plant Physiol. 2023, 191, 1365–1382. [Google Scholar] [CrossRef] [PubMed]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.; Harris, P.J.C. Photosynthesis under stressful environments: An overview. Photosynthetica 2013, 51, 163–190. [Google Scholar] [CrossRef]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.-K.; Bohnert, H.J. Plantcellular Andmolecularresponses Tohighsalinity. Annu. Rev. Plant Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef]
- Tester, R.F.; Karkalas, J.; Qi, X. Starch—Composition, fine structure and architecture. J. Cereal Sci. 2004, 39, 151–165. [Google Scholar] [CrossRef]
- Slewinski, T.L.; Braun, D.M. Current perspectives on the regulation of whole-plant carbohydrate partitioning. Plant Sci. 2010, 178, 341–349. [Google Scholar] [CrossRef]
- Alcázar-Alay, S.C.; Meireles, M.A.A. Physicochemical properties, modifications and applications of starches from different botanical sources. Food Sci. Technol. 2015, 35, 215–236. [Google Scholar] [CrossRef]
- Aluko, O.O.; Li, C.; Wang, Q.; Liu, H. Sucrose Utilization for Improved Crop Yields: A Review Article. Int. J. Mol. Sci. 2021, 22, 4704. [Google Scholar] [CrossRef]
- Wang, X.; Wang, M.; Huang, Y.; Zhu, P.; Qian, G.; Zhang, Y.; Liu, Y.; Zhou, J.; Li, L. Genome-Wide Identification and Analysis of Stress Response of Trehalose-6-Phosphate Synthase and Trehalose-6-Phosphate Phosphatase Genes in Quinoa. Int. J. Mol. Sci. 2023, 24, 6950. [Google Scholar] [CrossRef]
- O’Hara, L.E.; Paul, M.J.; Wingler, A. How do sugars regulate plant growth and development? New insight into the role of trehalose-6-phosphate. Mol. Plant 2013, 6, 261–274. [Google Scholar] [CrossRef]
- Sun, T.; Wang, P.; Lu, S.; Yuan, H.; Yang, Y.; Fish, T.; Thannhauser, T.; Liu, J.; Mazourek, M.; Grimm, B.; et al. Orchestration of Chlorophyll and Carotenoid Biosynthesis by ORANGE Family Proteins in Plant. bioRxiv 2022. bioRxiv:2022.2002.2008.479616. [Google Scholar] [CrossRef]
- Ullah, N.; Yüce, M.; Neslihan Öztürk Gökçe, Z.; Budak, H. Comparative metabolite profiling of drought stress in roots and leaves of seven Triticeae species. BMC Genom. 2017, 18, 969. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.-F.; Chao, D.-Y.; Shi, M.; Zhu, M.-Z.; Gao, J.-P.; Lin, H.-X. Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 2008, 228, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Satoh-Nagasawa, N.; Nagasawa, N.; Malcomber, S.; Sakai, H.; Jackson, D. A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 2006, 441, 227–230. [Google Scholar] [CrossRef]
- Wingler, A.; Delatte, T.L.; O’Hara, L.E.; Primavesi, L.F.; Jhurreea, D.; Paul, M.J.; Schluepmann, H. Trehalose 6-Phosphate Is Required for the Onset of Leaf Senescence Associated with High Carbon Availability. Plant Physiol. 2012, 158, 1241–1251. [Google Scholar] [CrossRef]
- Martínez-Barajas, E.; Delatte, T.; Schluepmann, H.; de Jong, G.J.; Somsen, G.W.; Nunes, C.; Primavesi, L.F.; Coello, P.; Mitchell, R.A.C.; Paul, M.J. Wheat Grain Development Is Characterized by Remarkable Trehalose 6-Phosphate Accumulation Pregrain Filling: Tissue Distribution and Relationship to SNF1-Related Protein Kinase1 Activity. Plant Physiol. 2011, 156, 373–381. [Google Scholar] [CrossRef]
- Nägele, T.; Weckwerth, W. Mathematical modeling reveals that metabolic feedback regulation of SnRK1 and hexokinase is sufficient to control sugar homeostasis from energy depletion to full recovery. Front. Plant Sci. 2014, 5, 365. [Google Scholar] [CrossRef]
- Kim, S.-J.; Jeong, D.-H.; An, G.; Kim, S.-R. Characterization of a drought-responsive gene, OsTPS1, identified by the T-DNA Gene-Trap system in rice. J. Plant Biol. 2005, 48, 371–379. [Google Scholar] [CrossRef]
- Kondrák, M.; Marincs, F.; Antal, F.; Juhász, Z.; Bánfalvi, Z. Effects of yeast trehalose-6-phosphate synthase 1 on gene expression and carbohydrate contents of potato leaves under drought stress conditions. BMC Plant Biol. 2012, 12, 74. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kawai, T.; Inukai, Y.; Aoki, D.; Feng, Z.; Xiao, Y.; Fukushima, K.; Lin, X.; Shi, W.; Busch, W.; et al. A lignin-derived material improves plant nutrient bioavailability and growth through its metal chelating capacity. Nat. Commun. 2023, 14, 4866. [Google Scholar] [CrossRef]
- Yuan, L.; Dang, J.; Zhang, J.; Wang, L.; Zheng, H.; Li, G.; Li, J.; Zhou, F.; Khan, A.; Zhang, Z.; et al. A glutathione S-transferase regulates lignin biosynthesis and enhances salt tolerance in tomato. Plant Physiol. 2024, 196, 2989–3006. [Google Scholar] [CrossRef]
- Song, H.; Cao, Y.; Zhao, X.; Zhang, L. Na+-preferential ion transporter HKT1;1 mediates salt tolerance in blueberry. Plant Physiol. 2024, 194, 511–529. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Yang, W.; Jing, W.; Shahid, M.O.; Liu, Y.; Qiu, X.; Choisy, P.; Xu, T.; Ma, N.; Gao, J.; et al. Multi-omics analysis reveals key regulatory defense pathways and genes involved in salt tolerance of rose plants. Hortic. Res. 2024, 11, uhae068. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Yang, J.; Li, Q.; Jiang, W.; Pang, Y. Medicago truncatula β-glucosidase 17 contributes to drought and salt tolerance through antioxidant flavonoid accumulation. Plant Cell Environ. 2024, 47, 3076–3089. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ma, L.; Liu, P.; Luo, Z.; Li, Z.; Wu, M.; Xu, X.; Pu, W.; Huang, P.; Yang, J. Transcription factor NtWRKY33a modulates the biosynthesis of polyphenols by targeting NtMYB4 and NtHCT genes in tobacco. Plant Sci. 2023, 326, 111522. [Google Scholar] [CrossRef]



| Sample | Raw Reads (Mb) | Clean Reads (Mb) | Clean Base (G) | Reads Mapped | Unique Mapped |
|---|---|---|---|---|---|
| BS | 54.93 | 53.56 | 8.04 | 42.90 (80.07%) | 40.49 (75.58%) |
| SS | 52.29 | 50.97 | 7.32 | 39.83 (78.14%) | 37.66 (73.89%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, D.; Li, J.; Zhang, Y.; Zhang, Y.; Wang, W.; Li, Z.; Zhu, P.; Huang, Y.; Han, L.; Wang, M.; et al. Transcriptomic Analysis Identifies Molecular Response of the Tolerant Alfalfa (Medicago sativa) Cultivar Nongjing 1 to Saline-Alkali Stress. Biology 2025, 14, 439. https://doi.org/10.3390/biology14040439
Zhang D, Li J, Zhang Y, Zhang Y, Wang W, Li Z, Zhu P, Huang Y, Han L, Wang M, et al. Transcriptomic Analysis Identifies Molecular Response of the Tolerant Alfalfa (Medicago sativa) Cultivar Nongjing 1 to Saline-Alkali Stress. Biology. 2025; 14(4):439. https://doi.org/10.3390/biology14040439
Chicago/Turabian StyleZhang, Dongmei, Jinxia Li, Yiming Zhang, Yuanhao Zhang, Wenhui Wang, Zhaohui Li, Peng Zhu, Yongshun Huang, Long Han, Mingyu Wang, and et al. 2025. "Transcriptomic Analysis Identifies Molecular Response of the Tolerant Alfalfa (Medicago sativa) Cultivar Nongjing 1 to Saline-Alkali Stress" Biology 14, no. 4: 439. https://doi.org/10.3390/biology14040439
APA StyleZhang, D., Li, J., Zhang, Y., Zhang, Y., Wang, W., Li, Z., Zhu, P., Huang, Y., Han, L., Wang, M., Zhang, Z., Shen, Z., Han, W., Mou, L., Zhuang, X., Pang, Q., Wang, J., & Li, L. (2025). Transcriptomic Analysis Identifies Molecular Response of the Tolerant Alfalfa (Medicago sativa) Cultivar Nongjing 1 to Saline-Alkali Stress. Biology, 14(4), 439. https://doi.org/10.3390/biology14040439





