Selectable Markers and Reporter Genes for Engineering the Chloroplast of Chlamydomonas reinhardtii
Abstract
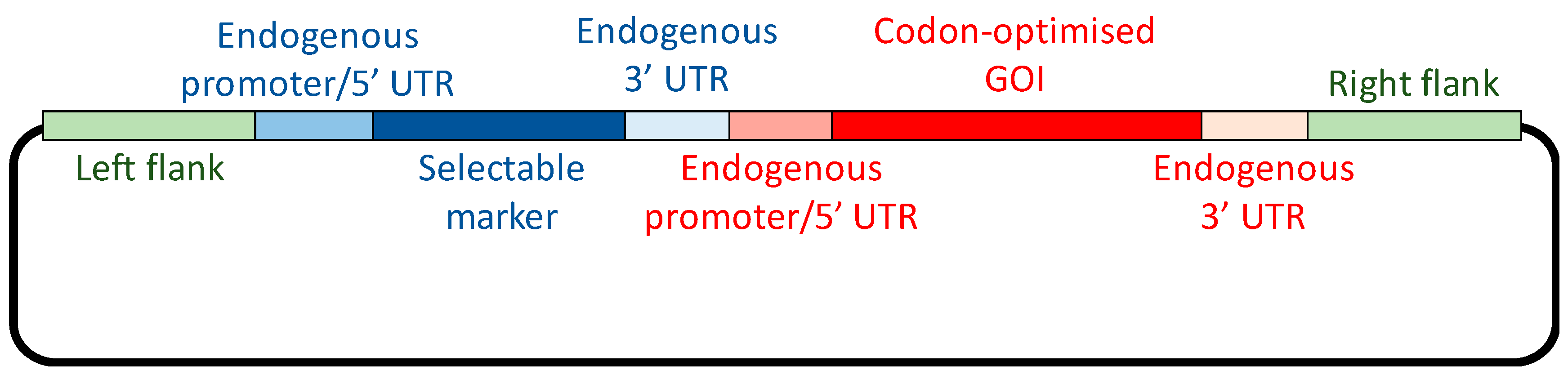
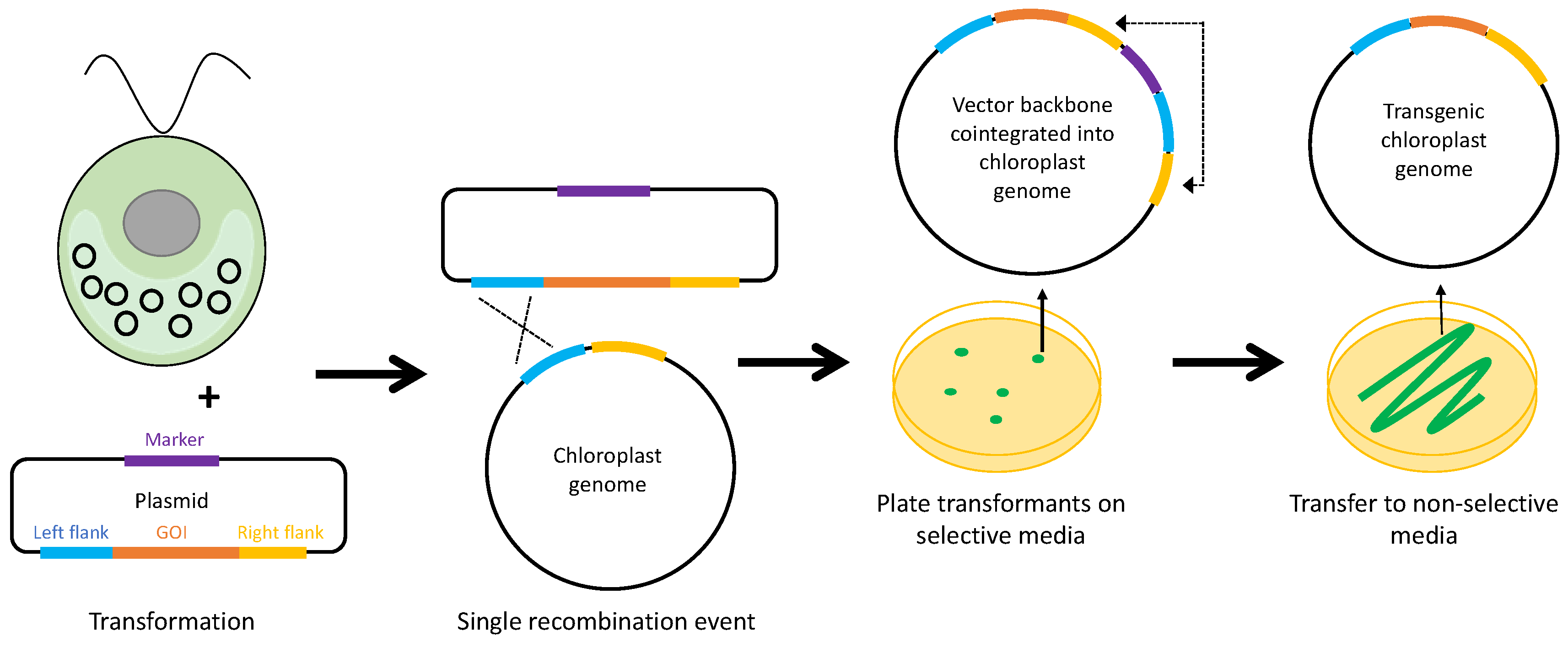
1. Introduction
2. Selectable Markers
2.1. Selection Based on Antibiotic Resistance
2.2. Selection Based on Herbicide Resistance
2.3. Selection Based on Restoration of Photosynthesis
2.4. Other Positive Selection Markers
2.5. Negative Selectable Markers
2.6. Removal of Selectable Markers
3. Reporters
3.1. Resistance Markers or Endogenous Genes as Reporters
3.2. β-glucuronidase as a Reporter
3.3. Fluorescence-Based Reporters
3.4. Luciferase-Based Reporters
4. Future Directions
5. Conclusion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Scaife, M.A.; Nguyen, G.T.D.T.; Rico, J.; Lambert, D.; Helliwell, K.E.; Smith, A.G. Establishing Chlamydomonas reinhardtii as an industrial biotechnology host. Plant J. 2015, 82, 532–546. [Google Scholar] [CrossRef] [PubMed]
- Rasala, B.A.; Mayfield, S.P. Photosynthetic biomanufacturing in green algae; Production of recombinant proteins for industrial, nutritional, and medical uses. Photosynth. Res. 2015, 123, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Hempel, F.; Maier, U.G. Microalgae as solar-powered protein factories. Adv. Exp. Med. Biol. 2016, 896, 241–262. [Google Scholar] [CrossRef] [PubMed]
- Harris, E.H. Chlamydomonas as a model organism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 363–406. [Google Scholar] [CrossRef] [PubMed]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Maréchal-Drouard, L.; et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Gallaher, S.D.; Fitz-Gibbon, S.T.; Strenkert, D.; Purvine, S.O.; Pellegrini, M.; Merchant, S.S. High-throughput sequencing of the chloroplast and mitochondrion of Chlamydomonas reinhardtii to generate improved de novo assemblies, analyze expression patterns and transcript speciation, and evaluate diversity among laboratory strains and wild isolates. Plant J. 2018, 93, 545–565. [Google Scholar] [CrossRef] [PubMed]
- Mussgnug, J.H. Genetic tools and techniques for Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol. 2015, 99, 5407–5418. [Google Scholar] [CrossRef] [PubMed]
- Purton, S.; Szaub, J.B.; Wannathong, T.; Young, R.; Economou, C.K. Genetic engineering of algal chloroplasts: Progress and prospects. Rus. J. Plant Physiol. 2013, 60, 521–528. [Google Scholar] [CrossRef]
- Mayfield, S.P.; Franklin, S.E.; Lerner, R.A. Expression and assembly of a fully active antibody in algae. Proc. Natl. Acad. Sci. USA 2003, 100, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Tran, M.; Van, C.; Barrera, D.J.; Pettersson, P.L.; Peinado, C.D.; Bui, J.; Mayfield, S.P. Production of unique immunotoxin cancer therapeutics in algal chloroplasts. Proc. Natl. Acad. Sci. USA 2013, 110, E15–E22. [Google Scholar] [CrossRef] [PubMed]
- Murbach, T.S.; Glávits, R.; Endres, J.R.; Hirka, G.; Vértesi, A.; Béres, E.; Szakonyiné, I.P. A toxicological evaluation of Chlamydomonas reinhardtii, a green algae. Int. J. Toxicol. 2018, 37, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Purton, S. Tools and techniques for chloroplast transformation of Chlamydomonas. Adv. Exp. Med. Biol. 2007, 616, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Dyo, Y.M.; Purton, S. The algal chloroplast as a synthetic biology platform for production of therapeutic proteins. Microbiology 2018, 164, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Qian, K.; Su, N.; Chang, H.; Liu, J.; Shen, G. Foot-and-mouth disease virus VP1 protein fused with cholera toxin B subunit expressed in Chlamydomonas reinhardtii chloroplast. Biotechnol. Lett. 2003, 25, 1087–1092. [Google Scholar] [CrossRef] [PubMed]
- Castellanos-Huerta, I.; Bañuelos-Hernández, B.; Téllez, G.; Rosales-Mendoza, S.; Brieba, L.G.; Esquivel-Ramos, E.; Beltrán-López, J.I.; Velazquez, G.; Fernandez-Siurob, I.; Bañuelos-Hernández, A.B.; et al. Recombinant hemagglutinin of avian influenza virus H5 expressed in the chloroplast of Chlamydomonas reinhardtii and evaluation of its immunogenicity in chickens. Avian Dis. 2016, 60, 784–791. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.S.; Luong, T.; Hannon, M.; Tran, M.; Gregory, J.A.; Shen, Z.; Briggs, S.P.; Mayfield, S.P. Heterologous expression of the C-terminal antigenic domain of the malaria vaccine candidate Pfs48/45 in the green algae Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol. 2013, 97, 1987–1995. [Google Scholar] [CrossRef] [PubMed]
- Dreesen, I.A.J.; Hamri, G.C.-E.; Fussenegger, M. Heat-stable oral alga-based vaccine protects mice from Staphylococcus aureus infection. J. Biotechnol. 2010, 145, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Demurtas, O.C.; Massa, S.; Ferrante, P.; Venuti, A.; Franconi, R.; Giuliano, G. A Chlamydomonas-Derived Human papillomavirus 16 E7 vaccine induces specific tumor protection. PLoS ONE 2013, 8, e61473. [Google Scholar] [CrossRef] [PubMed]
- Gregory, J.A.; Shepley-McTaggart, A.; Umpierrez, M.; Hurlburt, B.K.; Maleki, S.J.; Sampson, H.A.; Mayfield, S.P.; Berin, M.C. Immunotherapy using algal-produced Ara h 1 core domain suppresses peanut allergy in mice. Plant Biotechnol. J. 2016, 14, 1541–1550. [Google Scholar] [CrossRef] [PubMed]
- Hirschl, S.; Ralser, C.; Asam, C.; Gangitano, A.; Huber, S.; Ebner, C.; Bohle, B.; Wolf, M.; Briza, P.; Ferreira, F.; et al. Expression and characterization of functional recombinant Bet v 1.0101 in the chloroplast of Chlamydomonas reinhardtii. Int. Arch. Allergy Immunol. 2017, 173, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Odom, O.W.; Malone, C.L.; Thangamani, S.; Herrin, D.L. Expression of a synthetic gene for the major cytotoxin (Cyt1Aa) of Bacillus thuringiensis subsp. israelensis in the chloroplast of wild-type Chlamydomonas. Biology 2018, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Gimpel, J.A.; Henríquez, V.; Mayfield, S.P. In metabolic engineering of eukaryotic microalgae: Potential and challenges come with great diversity. Front Microbiol. 2015, 6, 1376. [Google Scholar] [CrossRef] [PubMed]
- Taunt, H.N.; Stoffels, L.; Purton, S. Green biologics: The algal chloroplast as a platform for making biopharmaceuticals. Bioengineered 2018, 9, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Bock, R. Engineering plastid genomes: Methods, tools, and applications in basic research and biotechnology. Annu. Rev. Plant Biol. 2015, 66, 211–241. [Google Scholar] [CrossRef] [PubMed]
- Wannathong, T.; Waterhouse, J.C.; Young, R.E.B.; Economou, C.K.; Purton, S. New tools for chloroplast genetic engineering allow the synthesis of human growth hormone in the green alga Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol. 2016, 100, 5467–5477. [Google Scholar] [CrossRef] [PubMed]
- Pourmir, A.; Noor-Mohammadi, S.; Johannes, T.W. Production of xylitol by recombinant microalgae. J. Biotechnol. 2013, 165, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Rasala, B.A.; Muto, M.; Sullivan, J.; Mayfield, S.P. Improved heterologous protein expression in the chloroplast of Chlamydomonas reinhardtii through promoter and 5′ untranslated region optimization. Plant Biotechnol. J. 2011, 9, 674–683. [Google Scholar] [CrossRef] [PubMed]
- Franklin, S.; Ngo, B.; Efuet, E.; Mayfield, S.P. Development of a GFP reporter gene for Chlamydomonas reinhardtii chloroplast. Plant J. 2002, 30, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.; Taunt, H.; Purton, S. Codon Usage Optimizer. Available online: https://github.com/khai-/CUO (accessed on 27 July 2018).
- Goldschmidt-Clermont, M. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: A selectable marker for site-directed transformation of Chlamydomonas. Nucleic Acids Res. 1991, 19, 4083–4089. [Google Scholar] [CrossRef] [PubMed]
- Doetsch, N.A.; Favreau, M.R.; Kuscuoglu, N.; Thompson, M.D.; Hallick, R.B. Chloroplast transformation in Euglena gracilis: Splicing of a group III twintron transcribed from a transgenic psbK operon. Curr. Genet. 2001, 39, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Galarza, J.I.; Gimpel, J.A.; Rojas, V.; Arredondo-Vega, B.O.; Henríquez, V. Over-accumulation of astaxanthin in Haematococcus pluvialis through chloroplast genetic engineering. Algal Res. 2018, 31, 291–297. [Google Scholar] [CrossRef]
- Svab, Z.; Maliga, P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl. Acad. Sci. USA 1993, 90, 913–917. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Lutz, K.A.; Maliga, P. Efficient plastid transformation in Arabidopsis. Plant Physiol. 2017, 175, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Chiyoda, S.; Yamato, K.T.; Kohchi, T. Plastid transformation of sporelings and suspension-cultured cells from the liverwort Marchantia polymorpha L. Methods Mol. Biol. 2014, 1132, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Bateman, J.M.; Purton, S. Tools for chloroplast transformation in Chlamydomonas: Expression vectors and a new dominant selectable marker. Mol. Gen. Genet. 2000, 263, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.-C.; Klaus, A.S.M.J.; Herz, A.S.; Zou, A.Z.; Koop, H.-U.; Golds, A.T.J. Efficient plastid transformation in tobacco using the aphA-6 gene and kanamycin selection. Mol. Genet. Genom. 2002, 268, 19–27. [Google Scholar] [CrossRef]
- Kumar, S.; Dhingra, A.; Daniell, H. Stable transformation of the cotton plastid genome and maternal inheritance of transgenes. Plant Mol. Biol. 2004, 56, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Tabatabaei, I.; Ruf, S.; Bock, R. A bifunctional aminoglycoside acetyltransferase/phosphotransferase conferring tobramycin resistance provides an efficient selectable marker for plastid transformation. Plant Mol. Biol. 2017, 93, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ruf, S.; Bock, R. Chloramphenicol acetyltransferase as selectable marker for plastid transformation. Plant Mol. Biol. 2011, 76, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Zienkiewicz, M.; Krupnik, T.; Drożak, A.; Golke, A.; Romanowska, E. Transformation of the Cyanidioschyzon merolae chloroplast genome: Prospects for understanding chloroplast function in extreme environments. Plant Mol. Biol. 2017, 93, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Gan, Q.; Jiang, J.; Han, X.; Wang, S.; Lu, Y. Engineering the chloroplast genome of oleaginous marine microalga Nannochloropsis oceanica. Front. Plant Sci. 2018, 9, 439. [Google Scholar] [CrossRef] [PubMed]
- Georgianna, D.R.; Hannon, M.J.; Marcuschi, M.; Wu, S.; Botsch, K.; Lewis, A.J.; Hyun, J.; Mendez, M.; Mayfield, S.P. Production of recombinant enzymes in the marine alga Dunaliella tertiolecta. Algal Res. 2013, 2, 2–9. [Google Scholar] [CrossRef]
- Carrer, H.; Hockenberry, T.N.; Svab, Z.; Maliga, P. Kanamycin resistance as a selectable marker for plastid transformation in tobacco. Mol. Gen. Genet. 1993, 241, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Newman, S.M.; Boynton, J.E.; Gillham, N.W.; Randolph-Anderson, B.L.; Johnson, A.M.; Harris, E.H. Transformation of chloroplast ribosomal RNA genes in Chlamydomonas: Molecular and genetic characterization of integration events. Genetics 1990, 126, 875–888. [Google Scholar] [PubMed]
- Przibilla, E.; Heiss, S.; Johanningmeier, U.; Trebst, A. Site-specific mutagenesis of the D1 subunit of photosystem II in wild-type Chlamydomonas. Plant Cell 1991, 3, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Newman, S.M.; Harris, E.H.; Johnson, A.M.; Boynton, J.E.; Gillham, N.W. Nonrandom distribution of chloroplast recombination events in Chlamydomonas reinhardtii: Evidence for a hotspot and an adjacent cold region. Genetics 1992, 132, 413–429. [Google Scholar] [PubMed]
- Lutz, K.A.; Knapp, J.E.; Maliga, P. Expression of bar in the plastid genome confers herbicide resistance. Plant Physiol. 2001, 125, 1585–1590. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Qin, S.; Jiang, P. Chloroplast transformation of Platymonas (Tetraselmis) subcordiformis with the bar gene as selectable marker. PLoS ONE 2014, 9, e98607. [Google Scholar] [CrossRef] [PubMed]
- Bellucci, M.; De Marchis, F.; Ferradini, N.; Pompa, A.; Veronesi, F.; Rosellini, D. A mutant Synechococcus gene encoding glutamate 1-semialdehyde aminotransferase confers gabaculine resistance when expressed in tobacco plastids. Plant Cell Rep. 2015, 34, 2127–2136. [Google Scholar] [CrossRef] [PubMed]
- Lapidot, M.; Raveh, D.; Sivan, A.; Arad, S.; Shapira, M. Stable chloroplast transformation of the unicellular red alga Porphyridium species. Plant Physiol. 2002, 129, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.C.; Melis, A. Marker-free genetic engineering of the chloroplast in the green microalga Chlamydomonas reinhardtii. Plant Biotechnol. J. 2013, 11, 818–828. [Google Scholar] [CrossRef] [PubMed]
- Bertalan, I.; Munder, M.C.; Weiß, C.; Kopf, J.; Fischer, D.; Johanningmeier, U. A rapid, modular and marker-free chloroplast expression system for the green alga Chlamydomonas reinhardtii. J. Biotechnol. 2015, 195, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Boynton, J.; Gillham, N.; Harris, E.; Hosler, J.; Johnson, A.; Jones, A.; Randolph-Anderson, B.; Robertson, D.; Klein, T.; Shark, K.; et al. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 1988, 240, 1534–1538. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.; Boynton, J.E.; Gillham, N.W. Cotranscription of the wild-type chloroplast atpE gene encoding the CF1/CF0 epsilon subunit with the 3′ half of the rps7 gene in Chlamydomonas reinhardtii and characterization of frameshift mutations in atpE. Mol. Gen. Genet. 1990, 221, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Bingham, S.E.; Xu, R.; Webber, A.N. Transformation of chloroplasts with the psaB gene encoding a polypeptide of the photosystem I reaction center. FEBS Lett. 1991, 292, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Day, A.; Dowson-Day, M.; Shen, G.F.; Dixon, R. The Klebsiella pneumoniae nitrogenase Fe protein gene (nifH) functionally substitutes for the chlL gene in Chlamydomonas reinhardtii. Biochem. Biophys. Res. Commun. 2005, 329, 966–975. [Google Scholar] [CrossRef] [PubMed]
- Young, R.E.B.; Purton, S. Codon reassignment to facilitate genetic engineering and biocontainment in the chloroplast of Chlamydomonas reinhardtii. Plant Biotechnol. J. 2016, 14, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Remacle, C.; Cline, S.; Boutaffala, L.; Gabilly, S.; Larosa, V.; Barbieri, M.R.; Coosemans, N.; Hamel, P.P. The ARG9 gene encodes the plastid-resident N-acetyl ornithine aminotransferase in the green alga Chlamydomonas reinhardtii. Eukaryot. Cell 2009, 8, 1460–1463. [Google Scholar] [CrossRef] [PubMed]
- Changko, S.; Young, R.E.B.; Purton, S. Development of ptxD as a chloroplast selectable marker and crop protection tool for Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol. 2018. submitted for publication. [Google Scholar]
- Barone, P.; Zhang, X.H.; Widholm, J.M. Tobacco plastid transformation using the feedback-insensitive anthranilate synthase α-subunit of tobacco (ASA2) as a new selectable marker. J. Exp. Bot. 2009, 60, 3195–3202. [Google Scholar] [CrossRef] [PubMed]
- Gisby, M.F.; Mudd, E.A.; Day, A. Growth of transplastomic cells expressing D-amino acid oxidase in chloroplasts is tolerant to D-alanine and inhibited by D-valine. Plant Physiol. 2012, 160, 2219–2226. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, R.; Hu, Z.; Li, H.; Lu, S.; Zhang, J.; Lin, Y.; Zhou, F. Expression of a codon-optimized dsdA gene in tobacco plastids and rice nucleus confers D-serine tolerance. Front. Plant Sci. 2016, 7, 640. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Muthukumar, B.; Lee, S.B. Marker free transgenic plants: Engineering the chloroplast genome without the use of antibiotic selection. Curr. Genet. 2001, 39, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Serino, G.; Maliga, P. A negative selection scheme based on the expression of cytosine deaminase in plastids. Plant J. 1997, 12, 697–701. [Google Scholar] [CrossRef] [PubMed]
- Young, R.E.B.; Purton, S. Cytosine deaminase as a negative selectable marker for the microalgal chloroplast: A strategy for the isolation of nuclear mutations that affect chloroplast gene expression. Plant J. 2014, 80, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Goldschmidt-Clermont, M.; Soen, S.Y.; Franzén, L.G.; Rochaix, J.D. Directed chloroplast transformation in Chlamydomonas reinhardtii: Insertional inactivation of the psaC gene encoding the iron sulfur protein destabilizes photosystem I. EMBO J. 1991, 10, 2033–2040. [Google Scholar] [CrossRef] [PubMed]
- Reifschneider-Wegner, K.; Kanygin, A.; Redding, K.E. Expression of the [FeFe] hydrogenase in the chloroplast of Chlamydomonas reinhardtii. Int. J. Hydrogen Energy 2014, 39, 3657–3665. [Google Scholar] [CrossRef]
- Dejtisakdi, W.; Miller, S.M. Overexpression of Calvin cycle enzyme fructose 1,6-bisphosphatase in Chlamydomonas reinhardtii has a detrimental effect on growth. Algal Res. 2016, 14, 116–126. [Google Scholar] [CrossRef]
- Wani, S.H.; Sah, S.K.; Sági, L.; Solymosi, K. Transplastomic plants for innovations in agriculture. A review. Agron. Sustain. Dev. 2015, 35, 1391–1430. [Google Scholar] [CrossRef]
- Muto, M.; Henry, R.E.; Mayfield, S.P. Accumulation and processing of a recombinant protein designed as a cleavable fusion to the endogenous Rubisco LSU protein in Chlamydomonas chloroplast. BMC Biotechnol. 2009, 9, 26. [Google Scholar] [CrossRef] [PubMed]
- Esland, L.; Young, R.; Purton, S. Bacterial genes encoding rifampicin and tetracycline resistance determinants as potential selectable markers for transformation of the C. reinhardtii chloroplast. Unpublished work. 2018. [Google Scholar]
- Ninlayarn, T. Chloroplast Genetic Engineering in the Microalga Chlamydomonas reinhardtii: Molecular Tools and Applications. PhD Thesis, University College London, London, UK, February 2012. [Google Scholar]
- Boynton, J.E.; Gillham, N.W. Chloroplast transformation in Chlamydomonas. Methods Enzymol. 1993, 217, 510–536. [Google Scholar] [CrossRef] [PubMed]
- Gillham, N.W.; Boynton, J.E.; Harris, E.H. Specific elimination of mitochondrial DNA from Chlamydomonas by intercalating dyes. Curr. Genet. 1987, 12, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Spicer, A.; Purton, S. Genetic engineering of microalgae: Current status and future prospects. In Microalgal Production for Biomass and High-Value Products; Slocombe, S.P., Benemann, J.R., Eds.; CRC Press: Boca Raton, FL, USA, 2016; pp. 139–164. [Google Scholar]
- Gatignol, A.; Durand, H.; Tiraby, G. Bleomycin resistance conferred by a drug-binding protein. FEBS Lett. 1988, 230, 171–175. [Google Scholar] [CrossRef]
- Melnyk, A.H.; Wong, A.; Kassen, R. The fitness costs of antibiotic resistance mutations. Evol. Appl. 2015, 8, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.J.; Movva, N.R.; Tizard, R.; Crameri, R.; Davies, J.E.; Lauwereys, M.; Botterman, J. Characterization of the herbicide-resistance gene bar from Streptomyces hygroscopicus. EMBO J. 1987, 6, 2519–2523. [Google Scholar] [CrossRef] [PubMed]
- Ye, G.-N.; Colburn, S.M.; Xu, C.W.; Hajdukiewicz, P.T.J.; Staub, J.M. Persistence of unselected transgenic DNA during a plastid transformation and segregation approach to herbicide resistance. Plant Physiol. 2003, 133, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Maughan, S.C.; Cobbett, C.S. Methionine sulfoximine, an alternative selection for the bar marker in plants. J. Biotechnol. 2003, 102, 125–128. [Google Scholar] [CrossRef]
- Franco, A.R.; Javier, F.; Pez-Siles, L.; Rdenas, J.C. Resistance to phosphinothricin (glufosinate) and its utilization as a nitrogen source by Chlamydomonas reinhardtii. Appl. Environ. Microbiol. 1996, 62, 3834–3839. [Google Scholar] [PubMed]
- Fernández-San Millán, A.; Obregón, P.; Veramendi, J. Over-expression of peptide deformylase in chloroplasts confers actinonin resistance, but is not a suitable selective marker system for plastid transformation. Transgenic Res. 2011, 20, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Kahn, A.; Kannangara, C.G. Gabaculine-resistant mutants of Chlamydomonas reinhardtii with elevated glutamate 1-semialdehyde aminotransferase activity. Carlsberg Res. Commun. 1987, 52, 73–81. [Google Scholar] [CrossRef]
- Ye, G.N.; Hajdukiewicz, P.T.; Broyles, D.; Rodriguez, D.; Xu, C.W.; Nehra, N.; Staub, J.M. Plastid-expressed 5-enolpyruvylshikimate-3-phosphate synthase genes provide high level glyphosate tolerance in tobacco. Plant J. 2001, 25, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Bruggeman, A.J.; Kuehler, D.; Weeks, D.P. Evaluation of three herbicide resistance genes for use in genetic transformations and for potential crop protection in algae production. Plant Biotechnol. J. 2014, 12, 894–902. [Google Scholar] [CrossRef] [PubMed]
- Kanda, K.; Ishida, T.; Hirota, R.; Ono, S.; Motomura, K.; Ikeda, T.; Kitamura, K.; Kuroda, A. Application of a phosphite dehydrogenase gene as a novel dominant selection marker for yeasts. J Biotechnol. 2014, 182–183, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Vallon, O.; Spalding, M.H. Amino acid metabolism. In The Chlamydomonas Sourcebook, 2nd ed.; Stern, D., Ed.; Academic Press: Oxford, UK, 2009; Volume 2, pp. 116–158. [Google Scholar]
- Nishimura, K.; Tomoda, Y.; Nakamoto, Y.; Kawada, T.; Ishii, Y.; Nagata, Y. Alanine racemase from the green alga Chlamydomonas reinhardtii. Amino Acids 2007, 32, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Maliga, P. Plastid transformation in higher plants. Annu. Rev. Plant Biol. 2004, 55, 289–313. [Google Scholar] [CrossRef] [PubMed]
- Whitney, S.M.; Sharwood, R.E. Construction of a tobacco master line to improve Rubisco engineering in chloroplasts. J. Exp. Bot. 2008, 59, 1909–1921. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.J.; Duff, R.J. The aldehyde dehydrogenase (ALDH) gene superfamily of the moss Physcomitrella patens and the algae Chlamydomonas reinhardtii and Ostreococcus tauri. Bryologist 2009, 112, 1–11. [Google Scholar] [CrossRef]
- Miki, B.; McHugh, S. Selectable marker genes in transgenic plants: Applications, alternatives and biosafety. J. Biotechnol. 2004, 107, 193–232. [Google Scholar] [CrossRef] [PubMed]
- Fischer, N.; Stampacchia, O.; Redding, K.; Rochaix, J.D. Selectable marker recycling in the chloroplast. Mol. Gen. Genet. 1996, 251, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Iamtham, S.; Day, A. Removal of antibiotic resistance genes from transgenic tobacco plastids. Nat. Biotechnol. 2000, 18, 1172–1176. [Google Scholar] [CrossRef] [PubMed]
- Kode, V.; Mudd, E.A.; Iamtham, S.; Day, A. Isolation of precise plastid deletion mutants by homology-based excision: A resource for site-directed mutagenesis, multi-gene changes and high-throughput plastid transformation. Plant J. 2006, 46, 901–909. [Google Scholar] [CrossRef] [PubMed]
- Klaus, S.M.J.; Huang, F.-C.; Golds, T.J.; Koop, H.-U. Generation of marker-free plastid transformants using a transiently cointegrated selection gene. Nat. Biotechnol. 2004, 22, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Lutz, K.A.; Maliga, P. Construction of marker-free transplastomic plants. Curr. Opin. Biotechnol. 2007, 18, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Kasai, Y.; Harayama, S. Construction of marker-free transgenic strains of Chlamydomonas reinhardtii using a Cre/loxP-mediated recombinase system. PLoS ONE 2016, 11, e0161733. [Google Scholar] [CrossRef] [PubMed]
- Corneille, S.; Lutz, K.; Svab, Z.; Maliga, P. Efficient elimination of selectable marker genes from the plastid genome by the CRE-lox site-specific recombination system. Plant J. 2001, 27, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Kittiwongwattana, C.; Lutz, K.; Clark, M.; Maliga, P. Plastid marker gene excision by the phiC31 phage site-specific recombinase. Plant Mol. Biol. 2007, 64, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Kumar, S.; Thomson, J.G. Precise excision of plastid DNA by the large serine recombinase Bxb1. Plant Biotechnol. J. 2014, 12, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Hajdukiewicz, P.T.; Gilbertson, L.; Staub, J.M. Multiple pathways for Cre/lox-mediated recombination in plastids. Plant J. 2001, 27, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Xiong, A.S.; Peng, R.H.; Zhuang, J.; Davies, J.; Zhang, J.; Yao, Q.H. Advances in directed molecular evolution of reporter genes. Crit. Rev. Biotechnol. 2012, 32, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Zerges, W.; Rochaix, J.-D. The 5′ leader of a chloroplast mRNA mediates the translational requirements for two nucleus-encoded functions in Chlamydomonas reinhardtii. Mol. Cell. Biol. 1994, 14, 5268–5277. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre-Legendre, L.; Choquet, Y.; Kuras, R.; Loubéry, S.; Douchi, D.; Goldschmidt-Clermont, M. A nucleus-encoded chloroplast protein regulated by iron availability governs expression of the photosystem I subunit PsaA in Chlamydomonas reinhardtii. Plant Physiol. 2015, 167, 1527–1540. [Google Scholar] [CrossRef] [PubMed]
- Franklin, J.L.; Zhang, J.; Redding, K. Use of aminoglycoside adenyltransferase translational fusions to determine topology of thylakoid membrane proteins. FEBS Lett. 2003, 536, 97–100. [Google Scholar] [CrossRef]
- Li, J.; Goldschmidt-Clermont, M.; Timko, M.P. Chloroplast-encoded chlB is required for light-independent protochlorophyllide reductase activity in Chlamydomonas reinhardtii. Plant Cell 1993, 5, 1817–1829. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, R.A. The GUS reporter gene system. Nature 1989, 342, 837–838. [Google Scholar] [CrossRef] [PubMed]
- Hull, G.A.; Devic, M. The β-glucuronidase (gus) reporter gene system. Gene fusions; spectrophotometric, fluorometric, and histochemical detection. Methods Mol. Biol. 1995, 49, 125–141. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, W.; Kindle, K.L.; Stern, D.B. In vivo analysis of Chlamydomonas chloroplast petD gene expression using stable transformation of β-glucuronidase translational fusions. Proc. Natl Acad. Sci. USA 1993, 90, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Ishikura, K.; Takaoka, Y.; Kato, K.; Sekine, M.; Yoshida, K.; Shinmyo, A. Expression of a foreign gene in Chlamydomonas reinhardtii chloroplast. J. Biosci. Bioeng. 1999, 87, 307–314. [Google Scholar] [CrossRef]
- Kato, K.; Ishikura, K.; Kasai, S.; Shinmyo, A. Efficient translation destabilizes transcripts in chloroplasts of Chlamydomonas reinhardtii. J. Biosci. Bioeng. 2006, 101, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Day, R.N.; Davidson, M.W. The fluorescent protein palette: Tools for cellular imaging. Chem. Soc. Rev. 2009, 38, 2887–2921. [Google Scholar] [CrossRef] [PubMed]
- Komine, Y.; Kikis, E.; Schuster, G.; Stern, D. Evidence for in vivo modulation of chloroplast RNA stability by 3′-UTR homopolymeric tails in Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA 2002, 99, 4085–4090. [Google Scholar] [CrossRef] [PubMed]
- Reed, M.L.; Wilson, S.K.; Sutton, C.A.; Hanson, M.R. High-level expression of a synthetic red-shifted GFP coding region incorporated into transgenic chloroplasts. Plant J. 2001, 27, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Rasala, B.A.; Barrera, D.J.; Ng, J.; Plucinak, T.M.; Rosenberg, J.N.; Weeks, D.P.; Oyler, G.A.; Peterson, T.C.; Haerizadeh, F.; Mayfield, S.P. Expanding the spectral palette of fluorescent proteins for the green microalga Chlamydomonas reinhardtii. Plant J. 2013, 74, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Braun-Galleani, S.; Baganz, F.; Purton, S. Improving recombinant protein production in the Chlamydomonas reinhardtii chloroplast using vivid Verde Fluorescent Protein as a reporter. Biotechnol. J. 2015, 10, 1289–1297. [Google Scholar] [CrossRef] [PubMed]
- Boehm, C.R.; Ueda, M.; Nishimura, Y.; Shikanai, T.; Haseloff, J. A cyan fluorescent reporter expressed from the chloroplast genome of Marchantia polymorpha. Plant Cell Physiol. 2016, 57, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Larrea-Alvarez, M.; Purton, S. Expression of a cyan fluorescent reporter in the chloroplast of the green alga Chlamydomonas reinhardtii. Unpublished work. 2018. [Google Scholar]
- Lauersen, K.J.; Kruse, O.; Mussgnug, J.H. Targeted expression of nuclear transgenes in Chlamydomonas reinhardtii with a versatile, modular vector toolkit. Appl. Microbiol. Biotechnol. 2015, 99, 3491–3503. [Google Scholar] [CrossRef] [PubMed]
- Welsh, D.K.; Kay, S.A. Bioluminescence imaging in living organisms. Curr. Opin. Biotechnol. 2005, 16, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Minko, I.; Holloway, S.P.; Nikaido, S.; Carter, M.; Odom, O.W.; Johnson, C.H.; Herrin, D.L. Renilla luciferase as a vital reporter for chloroplast gene expression in Chlamydomonas. Mol. Gen. Genet. 1999, 262, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Mayfield, S.P.; Schultz, J. Development of a luciferase reporter gene, luxCt, for Chlamydomonas reinhardtii chloroplast. Plant J. 2004, 37, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, T.; Onai, K.; Okamoto, K.; Minagawa, J.; Ishiura, M. Real-time monitoring of chloroplast gene expression by a luciferase reporter: Evidence for nuclear regulation of chloroplast circadian period. Mol. Cell. Biol. 2006, 26, 863–870. [Google Scholar] [CrossRef] [PubMed]
- Douchi, D.; Qu, Y.; Longoni, P.; Legendre-Lefebvre, L.; Johnson, X.; Schmitz-Linneweber, C.; Goldschmidt-Clermont, M. A nucleus-encoded chloroplast phosphoprotein governs expression of the photosystem I subunit PsaC in Chlamydomonas reinhardtii. Plant Cell 2016, 28, 1182–1199. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Zhang, B.S.; Prescher, J.A. Advances in bioluminescence imaging: New probes from old recipes. Curr. Opin. Chem. Biol. 2018, 45, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Shao, N.; Bock, R. A codon-optimized luciferase from Gaussia princeps facilitates the in vivo monitoring of gene expression in the model alga Chlamydomonas reinhardtii. Curr. Genet. 2008, 53, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Baek, K.; Kim, D.H.; Jeong, J.; Sim, S.J.; Melis, A.; Kim, J.S.; Jin, E.; Bae, S. DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci. Rep. 2016, 6, 30620. [Google Scholar] [CrossRef] [PubMed]
- Ferenczi, A.; Pyott, D.E.; Xipnitou, A.; Molnar, A. Efficient targeted DNA editing and replacement in Chlamydomonas reinhardtii using Cpf1 ribonucleoproteins and single-stranded DNA. Proc. Natl Acad. Sci. USA 2017, 114, 13567–13572. [Google Scholar] [CrossRef] [PubMed]
- Crozet, P.; Navarro, F.J.; Willmund, F.; Mehrshahi, P.; Bakowski, K.; Lauersen, K.J.; Pérez-Pérez, M.E.; Auroy, P.; Rovira, A.G.; Sauret-Gueto, S.; et al. Birth of a photosynthetic chassis: A MoClo toolkit enabling synthetic biology in the microalga Chlamydomonas reinhardtii. ACS Synth. Biol. 2018, 7, 2074–2086. [Google Scholar] [CrossRef] [PubMed]
- Barahimipour, R.; Neupert, J.; Bock, R. Efficient expression of nuclear transgenes in the green alga Chlamydomonas: Synthesis of an HIV antigen and development of a new selectable marker. Plant Mol. Biol. 2016, 90, 403–418. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Hu, H.H.; Li, Z.F.; Cheng, R.Q.; Meng, D.M.; Wang, J.; Fan, Z.C. A novel bicistronic expression system composed of the intraflagellar transport protein gene ift25 and FMDV 2A sequence directs robust nuclear gene expression in Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol. 2017, 101, 4227–4245. [Google Scholar] [CrossRef] [PubMed]
- Baier, T.; Wichmann, J.; Kruse, O.; Lauersen, K.J. Intron-containing algal transgenes mediate efficient recombinant gene expression in the green microalga Chlamydomonas reinhardtii. Nucleic Acids Res. 2018, 46, 6909–6919. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, P.; Armarego-Marriott, T.; Bock, R. Plastid transformation and its application in metabolic engineering. Curr. Opin. Biotechnol. 2018, 49, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Hahn, F.M.; Baidoo, E.; Kahlon, T.S.; Wood, D.F.; McMahan, C.M.; Cornish, K.; Keasling, J.D.; Daniell, H.; Whalen, M.C. Remodeling the isoprenoid pathway in tobacco by expressing the cytoplasmic mevalonate pathway in chloroplasts. Metab. Eng. 2012, 14, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Saxena, B.; Subramaniyan, M.; Malhotra, K.; Sarovar Bhavesh, N.; Potlakayala, S.D.; Kumar, S. Metabolic engineering of chloroplasts for artemisinic acid biosynthesis and impact on plant growth. J. Biosci. 2014, 39, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Young, R.; Purton, S. CITRIC: Cold-inducible translational readthrough in the chloroplast of Chlamydomonas reinhardtii using a novel temperature-sensitive transfer RNA. Microb. Cell Fact. 2018. submitted for publication. [Google Scholar]
- Boehm, C.R.; Bock, R. Recent advances and current challenges in synthetic biology of the plastid genetic system and metabolism. Plant Physiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Bradley, R.W.; Buck, M.; Wang, B. Tools and principles for microbial gene circuit engineering. J. Mol. Biol. 2016, 428, 862–888. [Google Scholar] [CrossRef] [PubMed]
- Triton Algae Innovations, USA. Available online: https://www.tritonai.com/ (accessed on 10 September 2018).
- Microsynbiotix Ltd., Ireland. Available online: https://www.microsynbiotix.com/ (accessed on 10 September 2018).
- Axitan Ltd., UK. Available online: http://www.axitan.com/ (accessed on 10 September 2018).





| Gene | Phenotype | Organism | Reference |
|---|---|---|---|
| aadA | Streptomycin and spectinomycin resistance | Various, including Chlamydomonas reinhardtii, Euglena gracilis, Haematococcus pluvialis, Nicotiana tabacum, Arabidopsis thaliana, Marchantia polymorpha | [30,31,32,33,34,35] |
| aphA-6 | Kanamycin and amikacin resistance | Various, including C. reinhardtii, N. tabacum, Gossypium hirsutum | [36,37,38] |
| aac6-aph2 | Tobramycin resistance | N. tabacum | [39] |
| cat | Chloramphenicol resistance | N. tabacum, Cyanidioschyzon merolae | [40,41] |
| ble | Zeomycin resistance | Nannochloropsis oceanica | [42] |
| ereB | Erythromycin resistance | Dunaliella tertiolecta | [43] |
| nptII | Kanamycin resistance | N. tabacum, G. hirsutum | [38,44] |
| rrnS and rrnL variants | Resistance to spectinomycin, streptomycin, kanamycin, or erythromycin | C. reinhardtii | [45] |
| psbA variants | Resistance to various herbicides, e.g., metribuzin, 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), phenmedipham | C. reinhardtii | [46,47] |
| bar | Phosphinothricin resistance | N. tabacum, P. subcordiformis | [48,49] |
| hemL | Insensitivity to gabaculine | N. tabacum | [50] |
| ahaSW492S | Sulfometuron methyl resistance | Porphyridium sp. | [51] |
| Essential photosynthesis genes e.g., atpB, petB, psaB, psbA, psbH, rbcL, tscA | Restored photosynthesis in recipient strain | C. reinhardtii | [25,30,52,53,54,55,56,57] |
| trnWUCA | Restored photosynthesis by translational read-through of opal mutation in psaA-3 | C. reinhardtii | [58] |
| ARG9 | Rescued arginine prototrophy in an ARG9 mutant strain | C. reinhardtii | [59] |
| ptxD | Ability to use phosphite as a source of phosphorus | C. reinhardtii | [60] |
| ASA2 variant | Insensitivity to tryptophan analogues | N. tabacum | [61] |
| dao | Tolerance to D-alanine and sensitivity to D-valine | N. tabacum | [62] |
| dsdA | Resistance to D-serine | N. tabacum | [63] |
| BADH | Resistance to betaine aldehyde | N. tabacum | [64] |
| codA | Sensitivity to 5-fluorocytosine | N. tabacum, C. reinhardtii | [65,66] |
| Gene Name | Origin of Luciferase | Gene Details | Size of Protein (kDa) | Substrate and Co-Factor | λMAX (nm) | Reference |
|---|---|---|---|---|---|---|
| rluc | Renilla coral | Native sequence | 36 | Coelenterazine | 480 | [123] |
| luxCt | Vibrio bacterium | Codon optimised gene fusion encoding A and B subunits joined by linker. | 78 | Decanal, FMNH2 | 490 | [124] |
| lucCP | Firefly beetle | Codon optimised | 61 | Luciferin, ATP | 550–570 | [125] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esland, L.; Larrea-Alvarez, M.; Purton, S. Selectable Markers and Reporter Genes for Engineering the Chloroplast of Chlamydomonas reinhardtii. Biology 2018, 7, 46. https://doi.org/10.3390/biology7040046
Esland L, Larrea-Alvarez M, Purton S. Selectable Markers and Reporter Genes for Engineering the Chloroplast of Chlamydomonas reinhardtii. Biology. 2018; 7(4):46. https://doi.org/10.3390/biology7040046
Chicago/Turabian StyleEsland, Lola, Marco Larrea-Alvarez, and Saul Purton. 2018. "Selectable Markers and Reporter Genes for Engineering the Chloroplast of Chlamydomonas reinhardtii" Biology 7, no. 4: 46. https://doi.org/10.3390/biology7040046
APA StyleEsland, L., Larrea-Alvarez, M., & Purton, S. (2018). Selectable Markers and Reporter Genes for Engineering the Chloroplast of Chlamydomonas reinhardtii. Biology, 7(4), 46. https://doi.org/10.3390/biology7040046






