Molecular Characterization of Twenty-Five Marine Cyanobacteria Isolated from Coastal Regions of Ireland
Abstract
:1. Introduction
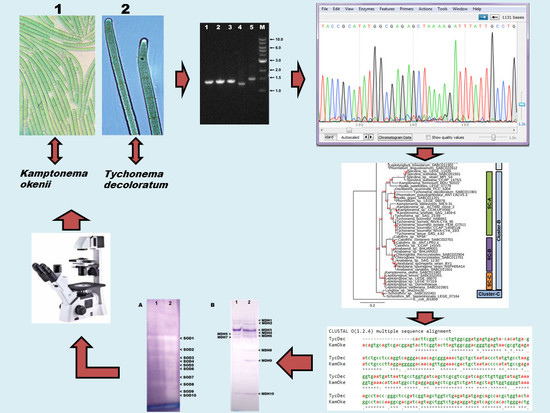
2. Materials and Methods
2.1. Cyanobacterial Isolates
2.2. Morphological Characterization and Identification
2.3. DNA Isolation and PCR Amplification
2.4. DNA Sequencing, Editing and BLAST Analysis
2.5. Construction of Phylogenetic Tree
2.6. Preparation of Isoenzyme Samples and Native-PAGE
2.7. Native-PAGE Gel Staining for SOD (EC 1.15.1.1)
2.8. Native-PAGE Gel Staining for MDH (EC 1.1.1.37)
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pham, H.T.L.; Nguyen, L.T.T.; Duong, T.A.; Bui, D.T.T.; Doan, Q.T.; Nguyen, H.T.T.; Mundt, S. Diversity and bioactivities of nostocacean cyanobacteria isolated from paddy soil in Vietnam. Syst. Appl. Microbiol. 2017, 40, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.P.; Häder, D.-P. Photobiology and ecophysiology of rice field cyanobacteria. Photochem. Photobiol. 1996, 64, 887–896. [Google Scholar] [CrossRef]
- Whitton, B.A.; Potts, M. (Eds.) Introduction to the Cyanobacteria. In The Ecology of Cyanobacteria, Their Diversity in Time and Space; Springer: Berlin, Germany, 2000; pp. 1–9. [Google Scholar]
- Stanier, R.Y.; Sistrom, W.R.; Hansen, T.A.; Whitton, B.A.; Castenholz, R.W.; Pfennig, N.; Gorlenko, V.N.; Kondratieva, E.N.; Eimhjellen, K.E.; Whittenbury, R.; et al. Proposal to place the nomenclature of the cyanobacteria (blue-green algae) under the rules of the International Code of Nomenclature of Bacteria. Int. J. Syst. Bacteriol. 1978, 28, 335–336. [Google Scholar] [CrossRef]
- Anand, N. Culture studies and taxonomy of blue-green algae-certain identification problems. Algol. Stud. Arch. Hydrobiol. 1988, 80, 141–147. [Google Scholar]
- Chang, T.P. Sheath formation in Oscillatoria aghardhii Gomot. Schweiz. Z. Hydrobiology 1977, 39, 178–181. [Google Scholar]
- Hoffmann, L. Quelques remarques sur la classification des Oscillatoriaceae. Cryptogam. Algol. 1985, 6, 71–79. [Google Scholar]
- Hoffmann, L.; Demoulin, V. Morphological variability of some species of Scytonemataceae (Cyanophyceae) under different culture conditions. Bull. K. Belg. Bot. Ver. 1985, 118, 189–197. [Google Scholar]
- Pearson, J.E.; Kingsbury, J.M. Culturally induced variation in four morphologically diverse blue-green algae. Am. J. Bot. 1966, 53, 192–200. [Google Scholar] [CrossRef]
- Wilmotte, A. Growth and morphological variability of six strains of Phormidium cf. ectocarpi Gomont (Cyanophyceae) cultivated under different temperatures and light intensities. Algol. Stud. Arch. Hydrobiol. 1988, 80, 35–46. [Google Scholar]
- Komárek, J. Genetic identity of the ‘Anacystis nidulans’ strain Kratz-Allen/Bloom 625 with Synechococcus Näg. 1849. Arch. Protistenkd. 1970, 112, 343–364. [Google Scholar]
- Laamanen, M.J.; Forsström, L.; Sivonen, K. Diversity of Aphanizomenon flos-aquae (Cyanobacterium) populations along a Baltic Sea salinity gradient. Appl. Environ. Microbiol. 2002, 68, 5296–5303. [Google Scholar] [CrossRef] [PubMed]
- Laloui, W.; Palinska, K.A.; Rippka, R.; Partensky, F.; Tandeau de Marsac, N.; Herdman, M.; Iteman, I. Genotyping of axenic and non-axenic isolates of the genus Prochlorococcus and the OMF-‘Synechococcus’ clade by size, sequence analysis or RFLP of the internal transcribed spacer of the ribosomal operon. Microbiology 2002, 148, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.; Huang, T.-C.; Chaw, S.-M. Molecular phylogeny of nitrogen-fixing unicellular cyanobacteria. Bot. Bull. Acad. Sin. 2001, 42, 181–186. [Google Scholar]
- Wilmotte, A. Molecular evolution and taxonomy of the cyanobacteria. In The Molecular Biology of Cyanobacteria; Bryant, D.A., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1994; pp. 1–25. [Google Scholar]
- Zheng, W.; Song, T.; Bao, X.; Bergman, B.; Rasmussen, U. High cyanobacterial diversity in coralloid roots of cycads revealed by PCR fingerprinting. FEMS Microbiol. Ecol. 2002, 40, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Ryan, U.M.; Monis, P.; McGregor, G.B.; Bath, A.; Gordon, C.; Paparini, A. Polyphasic identification of cyanobacterial isolates from Australia. Water Res. 2014, 59, 248–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biemelt, S.; Keetman, U.; Mock, H.-P.; Grimm, B. Expression and activity of isoenzymes of superoxide dismutase in wheat roots in response to hypoxia and anoxia. Plant Cell Environ. 2000, 23, 135–144. [Google Scholar] [CrossRef]
- Kim, E.J.; Kim, H.-P.; Hah, Y.C.; Roe, J.H. Differential expression of superoxide dismutase containing Ni and Fe/Zn in Streptomyces coelicolor. Eur. J. Biochem. 1996, 241, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.K.; Uma, L.; Subramanian, G. Nitrogen stress induced changes in the marine cyanobacterium Oscillatoria willei BDU 130511. FEMS Microbiol. Ecol. 2003, 45, 263–272. [Google Scholar] [CrossRef]
- Tripathi, S.N.; Srivastava, P. Presence of stable active oxygen scavenging enzymes superoxide dismutase, ascorbate peroxidase and catalase in a desiccation-tolerant cyanobacterium Lyngbya arboricola under dry state. Curr. Sci. 2001, 81, 197–200. [Google Scholar]
- Okada, S.; Kanematsu, S.; Asada, K. Intracellular distribution of manganese and ferric superoxide dismutase in blue-green algae. FEBS Lett. 1979, 103, 106–110. [Google Scholar] [CrossRef]
- Priya, B.; Premanandh, J.; Dhanalakshmi, T.R.; Uma, L.; Prabaharan, D.; Subramanian, G. Comparative analysis of cyanobacterial superoxide dismutases to discriminate canonical forms. BMC Genom. 2007, 8, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, F.A.; Lazoski, C.; Noireau, F.; Solé-Cava, A.M. Allozyme relationships among ten species of Rhodniini, showing paraphyly of Rhodnius including Psammolestes. Med. Vet. Entomol. 2002, 16, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Lia, V.; Comas, C.I.; Cortés, M.C.; Hunziker, J.H. Isozyme variation in Larrea ameghinoi and Larrea nitida (Zygophyllaceae): Genetic diversity and its bearing on their relationship. Genetica 1999, 106, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Pasquet, R.S. Genetic relationships among subspecies of Vigna unguiculata (L.) Walp. based on allozyme variation. Theor. Appl. Genet. 1999, 98, 1104–1119. [Google Scholar] [CrossRef]
- Schifino-Wittmann, M.T. Germplasm characterization of some Lathyrus species native to Rio Grande do Sul (Southern Brazil). Lathyrus Lathyrism Newsl. 2001, 2, 89–90. [Google Scholar]
- Schenk, H.E.A.; Hofer, I.; Metzner, H. Malate-dehydrogenase isoenzyme enden als potentielles chemotaxonomisches kriterium für cyanophyceen-species. Biochem. Syst. 1973, 1, 179–184. [Google Scholar] [CrossRef]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Desikachary, T.V. Cyanophyta; Indian Council of Agricultural Research: New Delhi, India, 1959; pp. 1–686. [Google Scholar]
- Komárek, J.; Anagnostidis, K. Cyanoprokaryota, 1. Teil/1st Part: Chroococcales; Süsswasserflora von Mitteleuropa; Gustav Fischer: Jena, Germany, 1998. [Google Scholar]
- Komárek, J.; Anagnostidis, K. Cyanoprokaryota, 2. Teil/2nd Part: Oscillatoriales; Süsswasserflora von Mitteleuropa; Elsevier/Spektrum Akademischer: München, Germany, 2005. [Google Scholar]
- Saha, S.K.; Uma, L.; Subramanian, G. An improved method for marine cyanobacterial DNA isolation. World J. Microbiol. Biotechnol. 2005, 21, 877–881. [Google Scholar] [CrossRef]
- Donovan, F. Establishment of Irish Marine Cyanobacterial Germplasm and Characterisation of Selected Cyanobacteria for Industrially Potential Biomolecules. Master’s Thesis, Limerick Institute of Technology, Limerick, UK, 2014. [Google Scholar]
- Nübel, U.; Garcia-Pichel, F.; Muyzer, G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 1997, 63, 3327–3332. [Google Scholar] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Beauchamp, C.; Fridovich, I. Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971, 44, 276–287. [Google Scholar] [CrossRef]
- Wendel, J.F.; Weeden, N.F. Visualization and interpretation of plant isozymes. In Isozymes in Plant Biology; Soltis, D.E., Soltis, P.S., Eds.; Chapman and Hall: London, UK, 1989; pp. 5–45. [Google Scholar]
- Brito, Â.; Ramos, V.; Mota, R.; Lima, S.; Santos, A.; Vieira, J.; Vieira, C.P.; Kaštovský, J.; Vasconcelos, V.M.; Tamagnini, P. Description of new genera and species of marine cyanobacteria from the Portuguese Atlantic coast. Mol. Phylogenet. Evol. 2017, 111, 18–34. [Google Scholar] [CrossRef] [PubMed]
- Komárek, J.; Kaštovský, J.; Mareš, J.; Johansen, J.R. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia 2014, 86, 295–335. [Google Scholar]
- Strunecky, O.; Komarek, J.; Šmarda, J. Kamptonema (Microcoleaceae, Cyanobacteria), a new genus derived from the polyphyletic Phormidium on the basis of combined molecular and cytomorphological markers. Preslia 2014, 86, 193–207. [Google Scholar]
- Bravakos, P.; Kotoulas, G.; Skaraki, K.; Pantazidou, A.; Economou-Amilli, A. A polyphasic taxonomic approach in isolated strains of cyanobacteria from thermal springs of Greece. Mol. Phylogenet. Evol. 2016, 98, 147–160. [Google Scholar] [CrossRef] [PubMed]



| No | Primer Name | Sequence (5′–3′) | Reference |
|---|---|---|---|
| 1 | CYA106F | CGGACGGGTGAGTAACGCGTGA | [35] |
| 2 | FDSKS_CyaF1 | AGAGTTTGATCCTGGCTCAGGATG | Present study |
| 3 | FDSKS_CyaF2 | TGCTTAACACATGCAAGTCGAACG | Present study |
| 4 | FDSKS_CyaF3 | TAGTGGCGGACGGGTGAGTAAC | Present study |
| 5 | FDSKS_CyaR1 | CACCTTCCGGTACGGCTACCTTG | Present study |
| 6 | FDSKS_CyaR2 | TACAAGGCCCGGGAACGTATTCACC | Present study |
| 7 | FDSKS_CyaR3 | GCATTGTAGTACGTGTGTAGCCCA | Present study |
| Species | Strain | Sampling Site | Primer Pairs | Edited Sequence Size (bp) | GenBank Accession Numbers | |
|---|---|---|---|---|---|---|
| 1 | Anabaena variabilis | SABC011501 | Ballybunion (Lat 52.511389, Lon 9.677496) | 2, 5 | 1385 | KX765290 |
| 2 | Calothrix contarenii | SABC022701 | Kilkee (Lat 52.685969, Lon 9.652605) | 2, 5 | 1388 | KT740998 |
| 3 | Chlorogloea microcystoides | SABC011701 | Ballybunion (Lat 52.511389, Lon 9.677496) | 4, 5 | 1308 | KY807916 |
| 4 | Chlorogloea microcystoides | SABC022904 | Kilkee (Lat 52.685969, Lon 9.652605) | 3, 5 | 1345 | KY807917 |
| 5 | Hyella gigas | SABC011201 | Ballybunion (Lat 52.511389, Lon 9.677496) | 2, 5 | 1381 | KX818207 |
| 6 | Kamptonema okenii | SABC011902 | Ballybunion (Lat 52.511389, Lon 9.677496) | 2, 5 | 1369 | KY807915 |
| 7 | Leptolyngbya africana | SABC021601 | Kilkee (Lat 52.685969, Lon 9.652605) | 2, 5 | 1392 | KT740999 |
| 8 | Leptolyngbya africana | SABC011801 | Ballybunion (Lat 52.511389, Lon 9.677496) | 2, 5 | 1371 | KX818206 |
| 9 | Leptolyngbya ectocarpi | SABC012402 | Ballybunion (Lat 52.511389, Lon 9.677496) | 2, 5 | 1376 | KX765291 |
| 10 | Leptolyngbya foveolarum | SABC011302 | Ballybunion (Lat 52.511389, Lon 9.677496) | 2, 5 | 1384 | KX818208 |
| 11 | Leptolyngbya fragilis | SABC012503 | Ballybunion (Lat 52.511389, Lon 9.677496) | 2, 5 | 1377 | KX818209 |
| 12 | Leptolyngbya fragilis | SABC031801 | Tralee (Lat 52.306668, Lon 9.857311) | 3, 5 | 1352 | KX818202 |
| 20 | Leptolyngbya norvegica | SABC031702 | Tralee (Lat 52.306668, Lon 9.857311) | 2, 5 | 1385 | KX818211 |
| 13 | Leptolyngbya tenuis | SABC010201 | Ballybunion (Lat 52.511389, Lon 9.677496) | 3, 5 | 1335 | KX765288 |
| 14 | Leptolyngbya valderiana | SABC022801 | Kilkee (Lat 52.685969, Lon 9.652605) | 3, 5 | 1349 | KY807918 |
| 15 | Phormidium angustissimum | SABC011301 | Ballybunion (Lat 52.511389, Lon 9.677496) | 1, 5 | 1317 | KX818204 |
| 16 | Phormidium angustissimum | SABC020801 | Kilkee (Lat 52.685969, Lon 9.652605) | 2, 5 | 1382 | KT740997 |
| 17 | Phormidium angustissimum | SABC022612 | Kilkee (Lat 52.685969, Lon 9.652605) | 1, 5 | 1312 | KX765287 |
| 18 | Phormidium angustissimum | SABC022901 | Kilkee (Lat 52.685969, Lon 9.652605) | 2, 5 | 1396 | KX818205 |
| 19 | Phormidium angustissimum | SABC030403 | Tralee (Lat 52.306668, Lon 9.857311) | 1, 5 | 1317 | KX818203 |
| 21 | Phormidium sp. | SABC022903 | Kilkee (Lat 52.685969, Lon 9.652605) | 3, 5 | 1362 | KT741000 |
| 22 | Pseudanabaena minima | SABC031701 | Tralee (Lat 52.306668, Lon 9.857311) | 2, 5 | 1379 | KX818210 |
| 23 | Schizothrix sp. | SABC022401 | Kilkee (Lat 52.685969, Lon 9.652605) | 4, 6 | 1198 | KX765289 |
| 24 | Spirulina subsalsa | SABC051501 | Cork (Lat 51.793171, Lon 8.268673) | 3, 5 | 1313 | KY807919 |
| 25 | Tychonema decoloratum | SABC011901 | Ballybunion (Lat 52.511389, Lon 9.677496) | 3, 5 | 1343 | KY807914 |
| Forms | Description | Taxonomic Identification | Strains/Remarks |
|---|---|---|---|
| A | Thallus gelatinous, dark green, trichome without sheath, flexuous, heterocystous, cells barrel shaped to spherical, 3–5 µm broad, 5–6 µm long, heterocysts oval, 6 µm broad, sometimes up to 8 µm long, spores colorless or yellowish brown. | Anabaena variabilis | 1 |
| B | Thallus crustaceous, compact, firm, dull green, filaments densely arranged, filaments up to 1 mm long, trichome swollen at the base, 9–15 µm broad at base, sheath thick, colorless, trichome 6–8 µm broad in the middle, trichomes with basal, more or less spherical or hemispherical heterocysts, filament ending in a long hair. | Calothrix contarenii | 1 |
| C | Small but macroscopically visible thallus, wart-like, made up by union of a number of daughter dark-green colonies. Cells spherical, ellipsoidal or polygonal-rounded, 1–5 µm in diameter. Sometimes cells form Anabaena-like rows of barrel shaped cells with very irregularly shaped trichomes. | Chlorogloea microcystoides | 2/Sheath of isolate SABC011701 is often yellowish brown. |
| D | Thallus microscopic to dot-like on agar plate, often gelatinous, pseudofilaments bent or irregularly branched with cells in one or in many rows forming a chroococcacean stage; pseudofilaments 100–125 µm long, base cells 5–7.5 µm broad, cells in pseudofilament up to 17 µm long, cells at tip of pseudofilament 5 µm long, mucilaginous sheath usually thin, cells content highly granular to homogenous, spherical to slightly angular sporangia found in proximal end producing endospores. | Hyella gigas | 1 |
| E | Thallus dark blue-green, trichomes mostly straight to slightly curved, trichomes 3.5–5 µm wide, cells1.5–4.5 µm long, end cell rounded and often bent without calyptra, cells shorter than broad, trichome attenuated, cell content partially minutely granular to homogenous, not distinctly constricted at cross walls. | Kamptonema okenii | 1 |
| F | Thallus green, mucilaginous, filaments long, mostly entangled, straight to irregularly curved, trichome 1–1.2 µm broad, 1.5–2 µm long, cells quadrate or slightly rectangular, indistinctly constricted, very thin sheath, apical cell conical round. | Leptolyngbya africana | 2/SABC011801 filaments loosely entangled, cells 1–1.5 µm broad, 1.5–3–4 µm long, constrictions in old filaments are distinct, end cell round |
| G | Thallus yellowish brown to yellowish green, filaments densely entangled, curved or bent, fragile, hormogonia present (usually 2–5 cells long), sheath distinct in older cells, no evident sheath in hormogonia. Cells 1–1.2 µm broad, 1–1.5–2.5 µm long, apical cell rounded to conical rounded, without calyptra. | Leptolyngbya ectocarpi | 1 |
| H | Thallus mucilaginous, blue-green to dark green, filaments variously curved to parallelly arranged, sheath thin but firmly attached to the trichome, cells somewhat shorter than broad, 1–1.5 µm broad and 0.8–1.8 µm long, cross walls constricted in old filaments, cell content homogenous, apical cells flat rounded, no calyptra. | Leptolyngbya foveolarum | 1 |
| I | Thallus light blue green, mucilaginous, filaments entangled and variously curved, cells slightly longer than broad, sometimes quadrate, constricted at cross-walls, filaments easily broken apart, sheath diffluent, end cell conical, very long open-ended sheath, | Leptolyngbya fragilis | 2/SABC012503 cells 1.5–2 µm broad, 2–3 µm long; SABC031801 cells 1.5 µm broad, 2–2.5–3 µm long. |
| J | Thallus brown or brownish green, some pseudobranching, sheaths initially thin or absent, cells ~1.5 µm broad, 1.5–2 (4) µm long, cells sometime quadrate, distinctly constricted at the cross walls, sheath later thick, older filament with sheath 2–3 µm, filaments often wavy within sheath, apical cell slightly elongated and tapered. | Leptolyngbya norvegica | 1 |
| K | Thallus membranous, irregularly expanded, very thin diffluent sheath, trichome olive-green or blue-green, straight or slightly bent but densely entangled, cells 1.2–1.5 μm broad, 1.5–2 μm long, cross wall constricted, cell content homogenous to slightly granular, end cells attenuated, mostly acute conical or round, no calyptra. | Leptolyngbya tenuis | 1 |
| L | Thallus mucilaginous, blue green to dark green, filaments long and strong, not easily breaking into short fragments, sheath thin, mucilaginous, cells longer than broad, 1.5 μm broad, 2.5–3 μm long, cross wall not distinctly constricted, granule on cross-wall, cell content homogenous to slightly granular, end cell acute conical to round. | Leptolyngbya valderiana | 1 |
| M | Thallus pale green to blue green, mucilaginous, filaments with thin but firm sheaths, cells longer than broad, trichomes bent and entangled densely or loosely, end cell mostly conical round, no calyptra, cross walls distinct in old filaments, cell content homogenous. | Phormidium angustissimum | 5/SABC011301 cells 1 µm broad, 1.5 µm long; SABC020801 cells 0.8–1 µm broad, 1 µm long; SABC022612 cells 1–1.5 µm broad, 1.5–2 µm long; SABC022901 cells 1–1.2 µm broad, 1.5–2 µm long; SABC030403 filaments brownish, cells 0.8–1 µm broad, 1–1.5 µm long. |
| N | Thallus mucilaginous, distinct and firm sheath all through the trichome, cells longer than broad, 0.8–1 µm broad, 3–4 µm long, cross wall distinctly constricted, cell content homogenous, apical cell acute conical or round, no calyptra. | Phormidium sp. | 1 |
| O | Trichomes solitary or crowded in clusters, straight or almost straight, pale blue-green, cells barrel shaped, 1.2–1.5 µm broad, 1.2–1.5 µm long, intensely constricted at cross walls, no heterocysts or sheath, end cells round | Pseudanabaena minima | 1 |
| P | Thallus green in young stage and then red, mucilaginous, filaments densely entangled to radial fascicles, trichomes with fine sheath, sheath diffluent and colorless, pseudobranching present, trichomes 2 µm broad, cells 2–3 µm long, often quadratic, filaments long and randomly intertwined, cells constricted at cross walls but indistinct towards apices, end cells without calyptra, end cell rounded conical. | Schizothrix sp. | 1 |
| Q | Thallus mucilaginous, blackish green, trichomes solitary to entangled with other trichomes, regularly screw-like coils, 4.2–4.8 µm broad, 2.1–2.6 µm long, coils dextral, regularly tightly joined to one another, arranged nearly in parallel, rapidly motile with screw like rotation, end cells rounded. | Spirulina subsalsa | 1 |
| R | Thallus light blue-green, trichomes free-moving, mostly straight to slightly curved, cell content homogenous, cell constrictions not distinct, cells somewhat shorter than wide, 3.9–5.3 µm long, 6.5–8 µm wide, no heterocysts or sheath, end cells slightly bend round at one end, acute conical on the other end. | Tychonema decoloratum | 1 |
| Morphotype | Fragment Size (bp) | Query Coverage (%) | Identity (%) | Closest Match in Genbank (Accession Number) |
|---|---|---|---|---|
| Anabaena variabilis SABC011501 | 1385 | 98 | 99 | Anabaena sp. SAG 12.82 (KT290322.1) |
| 98 | 99 | Nodularia spumigena strain BY1 (AF268004.1) | ||
| 99 | 98 | Nodularia spumigena strain NSPH05A14 (AF268017.1) | ||
| Calothrix contarenii SABC022701 | 1388 | 98 | 98 | Calothrix sp. CCAP 1410/5 (HF678513.1) |
| 98 | 97 | Calothrix sp. ANT.LPR2.4 (AY493597.1) | ||
| 98 | 97 | Calothrix sp. XP9A (AM230670.1) | ||
| Chlorogloea microcystoides SABC022904 | 1345 | 95 | 99 | Chlorogloea microcystoides SABC011701 (KY807916.1) |
| 99 | 97 | Anabaena sp. BHUAR002 (HM235817.1) | ||
| 98 | 97 | Anabaena sp. BHUAR001 (HM235816.1) | ||
| Hyella gigas SABC011201 | 1381 | 94 | 91 | Hyella patelloides LEGE 07179 (HQ832901.1) |
| 98 | 99 | Phormidium sp. LEGE 06078 (HM217075.1) | ||
| 98 | 99 | Phormidium pseudopriestleyi ANT.LACV5.3 (AY493600.1) | ||
| Kamptonema okenii SABC011902 | 1369 | 97 | 91 | Kamptonema animale SAG 1459-6 (EF654087.1) |
| 97 | 91 | Kamptonema sp. CCM-UFV066 (MG563377.1) | ||
| 97 | 91 | Kamptonema sp. ACT692 clone 2 (MK247996.1) | ||
| Leptolyngbya africana SABC021601 | 1392 | 99 | 97 | Phormidium angustissimum SABC020801 (KT740997.1) |
| 99 | 95 | Leptolyngbya sp. CCAP 1442/1 (HE975019.1) | ||
| 99 | 95 | Leptolyngbya sp. HBC1 (EU249120.1) | ||
| Leptolyngbya ectocarpi SABC012402 | 1376 | 99 | 98 | Phormidesmis priestleyi ANT.LACV5.1 (AY493586.1) |
| 99 | 97 | Leptolyngbya sp. ANT.L52.1 (AY493584.1) | ||
| 99 | 97 | LPP-group cyanobacterium QSSC8cya (AF170758.1) | ||
| Leptolyngbya foveolarum SABC 011302 | 1384 | 100 | 99 | Leptolyngbya sp. LEGE 07296 (HM217082.1) |
| 99 | 99 | Leptolyngbya sp. LEGE 07298 (HM217044.1) | ||
| 99 | 99 | Leptolyngbya norvegica SABC031702 (KX818211.1) | ||
| Leptolyngbya fragilis SABC012503 | 1377 | 99 | 99 | Leptolyngbya foveolarum SABC011302 (KX818208.1) |
| 98 | 99 | Leptolyngbya sp. LEGE 07080 (HM217085.1) | ||
| 99 | 99 | Leptolyngbya norvegica SABC031702 (KX818211.1) | ||
| Leptolyngbya norvegica SABC031702 | 1385 | 99 | 99 | Leptolyngbya foveolarum SABC011302 (KX818208.1) |
| 99 | 99 | Leptolyngbya sp. LEGE 07314 (HM217061.1) | ||
| 99 | 99 | Leptolyngbya sp. LEGE 07298 (HM217044.1) | ||
| Leptolyngbya tenuis SABC010201 | 1335 | 99 | 99 | Leptolyngbya valderiana SABC022801 (KY807918.1) |
| 99 | 99 | Leptolyngbya sp. LEGE 06070 (HM217074.1) | ||
| 98 | 93 | Phormidium tenue BDU 92362 (KU958134.1) | ||
| Leptolyngbya valderiana SABC022801 | 1349 | 100 | 100 | Leptolyngbya sp. LEGE 06070 (HM217074.1) |
| 100 | 99 | Leptolyngbya sp. LEGE 07319 (HM217045.1) | ||
| 98 | 100 | Leptolyngbya sp. Doroninskoye (KT753316.1) | ||
| Phormidium angustissimum SABC022901 | 1396 | 99 | 99 | Leptolyngbya norvegica SABC031702 (KX818211.1) |
| 99 | 98 | Leptolyngbya sp. LEGE 07298 (HM217044.1) | ||
| 99 | 97 | Phormidium sp. 195-A12 (EU282429.1) | ||
| Phormidium sp. SABC022903 | 1362 | 98 | 99 | Leptolyngbya fragilis SABC031801 (KX818202.1) |
| 98 | 99 | Nodosilinea sp. CENA183 (KC695874.1) | ||
| 98 | 98 | Oscillatoria sp. BS594 (KM019975.1) | ||
| Pseudanabaena minima SABC031701 | 1379 | 99 | 99 | Plectonema sp. F3 (AF091110.1) |
| 99 | 97 | Leptolyngbya sp. ANT.L52.1 (AY493584.1) | ||
| 99 | 96 | Phormidesmis sp. CENA339 (KT731156.1) | ||
| Schizothrix sp. SABC022401 | 1198 | 99 | 96 | Schizothrix aff. septentrionalis LEGE 07164 (KU951800.1) |
| 93 | 96 | Lyngbya sp. aka2mo30 (AB863114.1) | ||
| 97 | 92 | Leptolyngbya sp. RS03 (JF518829.1) | ||
| Spirulina subsalsa SABC051501 | 1313 | 99 | 99 | Spirulina sp. LEGE 11,439 (KU951804.1) |
| 99 | 99 | Spirulina sp. strain MPI S4 (Y18792.1) | ||
| 99 | 98 | Spirulina subsalsa CCAP 1475/1 (HF678502.1) | ||
| Tychonema decoloratum SABC011901 | 1343 | 98 | 84 | Tychonema sp. SAG 23.89 (KM019964.1) |
| 97 | 84 | Tychonema bourrellyi HAB663 (FJ184385.1) | ||
| 98 | 90 | Oscillatoria acuminata strain PCC 6304 (NR_102463.1) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shiels, K.; Browne, N.; Donovan, F.; Murray, P.; Saha, S.K. Molecular Characterization of Twenty-Five Marine Cyanobacteria Isolated from Coastal Regions of Ireland. Biology 2019, 8, 59. https://doi.org/10.3390/biology8030059
Shiels K, Browne N, Donovan F, Murray P, Saha SK. Molecular Characterization of Twenty-Five Marine Cyanobacteria Isolated from Coastal Regions of Ireland. Biology. 2019; 8(3):59. https://doi.org/10.3390/biology8030059
Chicago/Turabian StyleShiels, Katie, Norma Browne, Fiona Donovan, Patrick Murray, and Sushanta Kumar Saha. 2019. "Molecular Characterization of Twenty-Five Marine Cyanobacteria Isolated from Coastal Regions of Ireland" Biology 8, no. 3: 59. https://doi.org/10.3390/biology8030059
APA StyleShiels, K., Browne, N., Donovan, F., Murray, P., & Saha, S. K. (2019). Molecular Characterization of Twenty-Five Marine Cyanobacteria Isolated from Coastal Regions of Ireland. Biology, 8(3), 59. https://doi.org/10.3390/biology8030059