Artificial Neural Networks Model for Predicting Type 2 Diabetes Mellitus Based on VDR Gene FokI Polymorphism, Lipid Profile and Demographic Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Population
2.2. Blood Collection and Chemistry Tests
2.3. DNA Extraction and Quantification
2.4. Polymerase Chain Reaction (PCR)
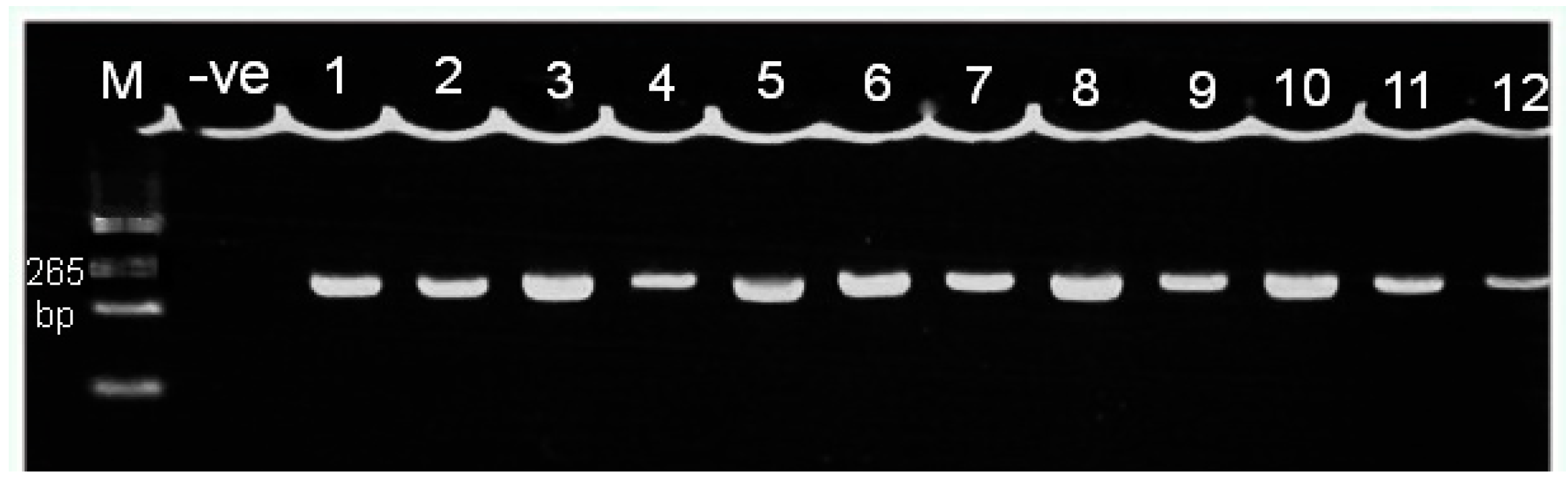
2.5. Agarose Gel Electrophoresis
2.6. DNA Sequencing
2.7. Statistical Analysis
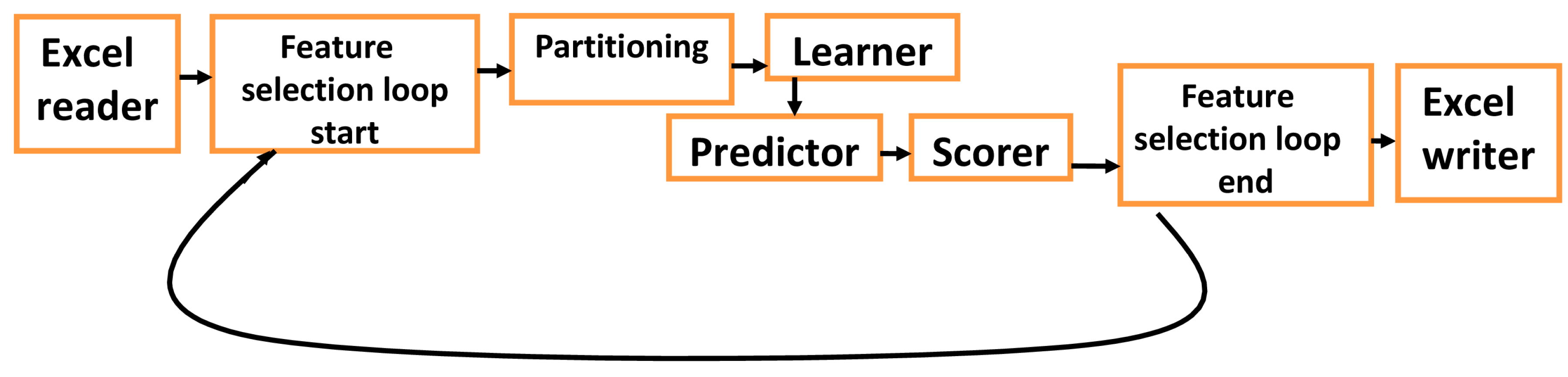
2.8. Multilayer FNN
3. Results
3.1. Demographic and Clinical Data
3.2. Genotyping of the FokI Polymorphism
3.3. Comparison of the Genotypes and Alleles
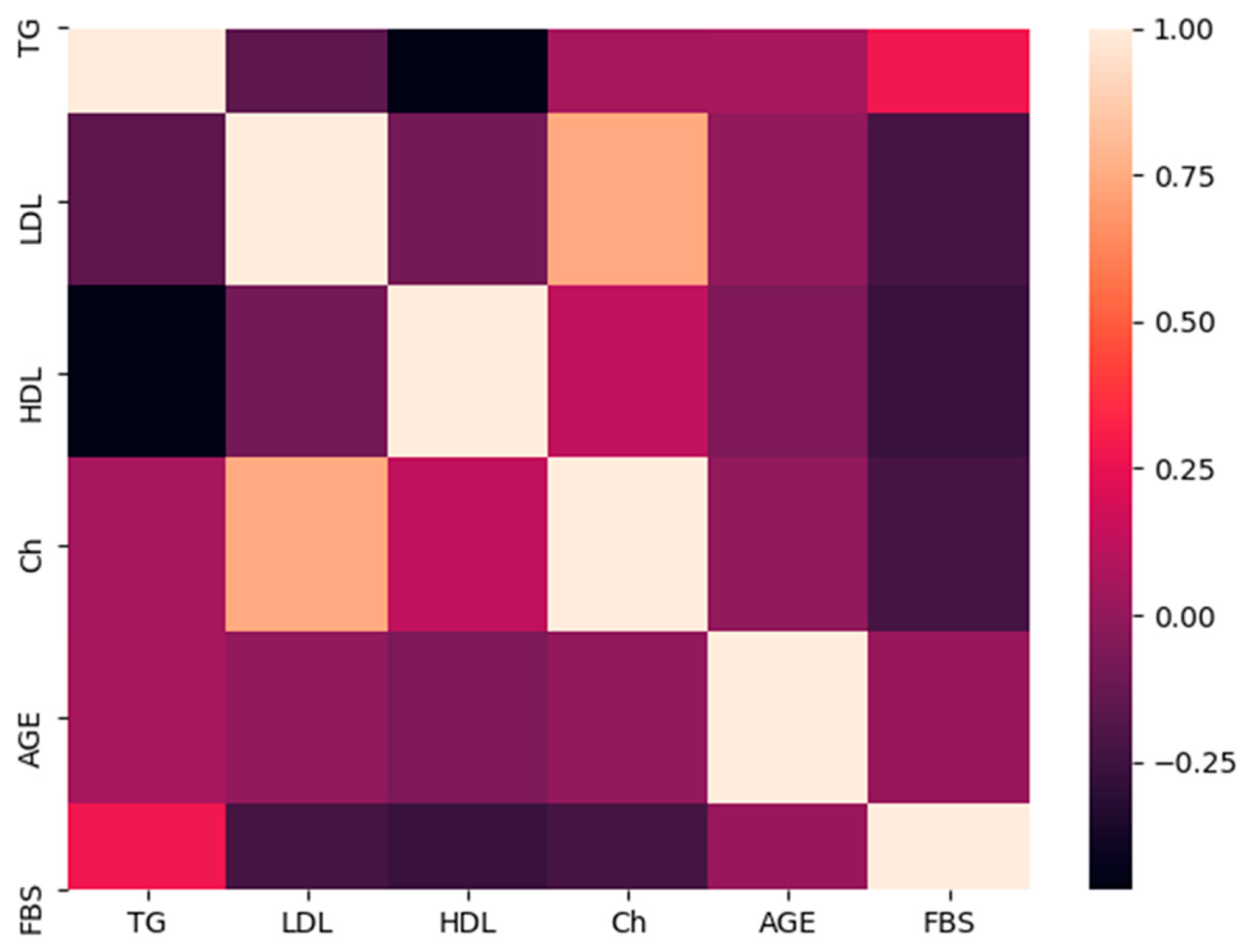
3.4. Comparative Analyses of the Lipid Profile
3.5. Comparison of the Lipid Profile and FokI Polymorphism Genotypes
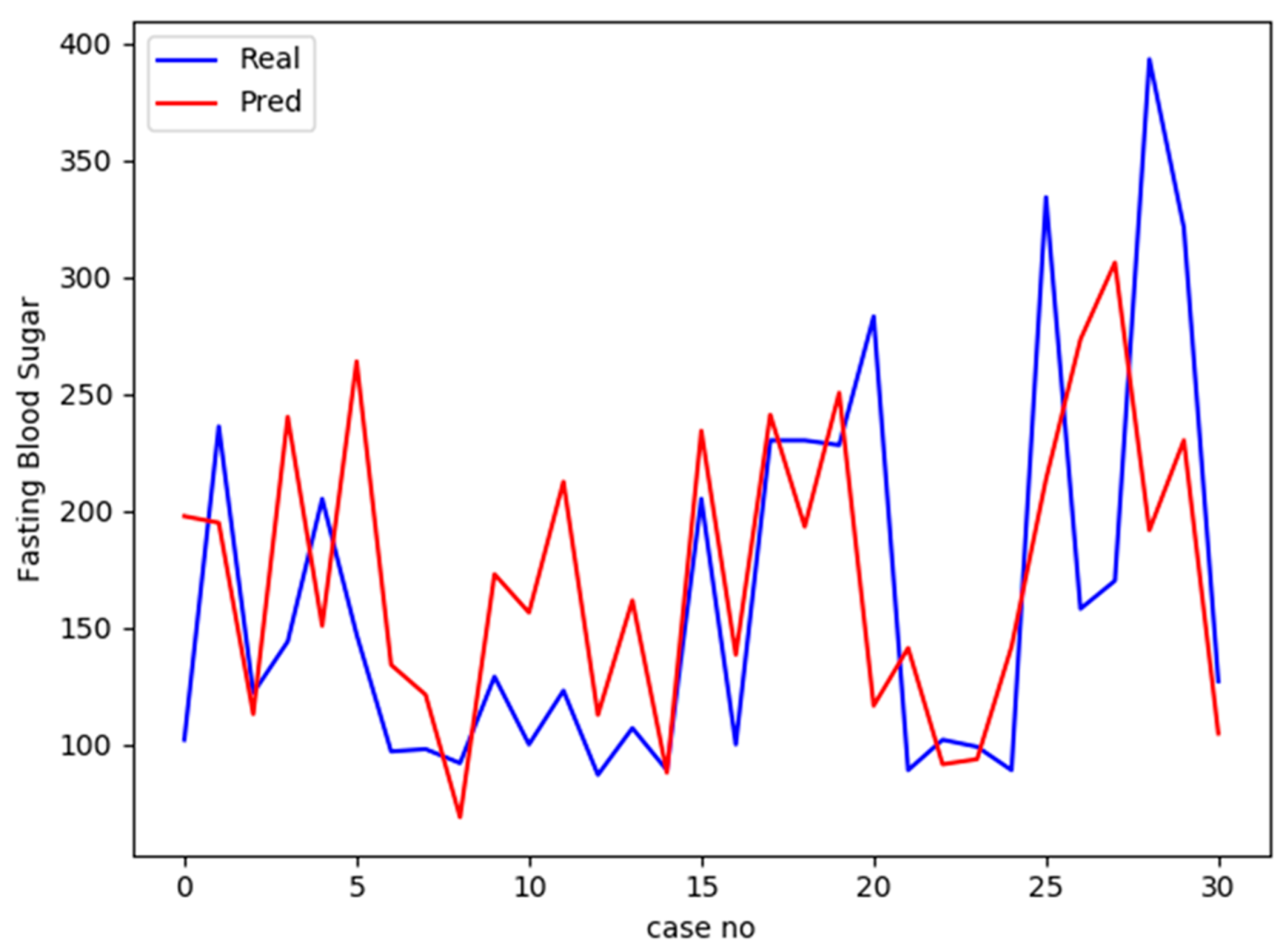
3.6. Predicting Diabetes for the Testing Set
4. Discussion
4.1. Association of FokI Polymorphism Genotypes with T2DM
4.2. Association of Lipid Profile with T2DM
4.3. Association of FokI Polymorphism Genotypes with Lipid Profile
4.4. Prediction of Diabetes Status Using FNN
5. Limitations of the Study
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Krauss, R.M. Lipids and Lipoproteins in Patients With Type 2 Diabetes. Diabetes Care 2004, 27, 1496–1504. [Google Scholar] [CrossRef] [PubMed]
- Al-Daghri, N.M.; Al-Attas, O.; Alokail, M.S.; Alkharfy, K.M.; Draz, H.M.; Agliardi, C.; Mohammed, A.K.; Guerini, F.R.; Clerici, M. Vitamin D Receptor Gene Polymorphisms and HLA DRB1*04 Cosegregation in Saudi Type 2 Diabetes Patients. J. Immunol. 2012, 188, 1325–1332. [Google Scholar] [CrossRef] [PubMed]
- Taneja, N.; Khadagawat, R.; Mani, S. BsmI and TaqI Polymorphisms in Vitamin D Receptor Gene of Type 2 Diabetes Mellitus Patients From North India. Asian J. Pharm. Clin. Res. 2016, 9, 186–189. [Google Scholar] [CrossRef][Green Version]
- Al-Darraji, S.; Al-Azzawie, H.; Al-Kharsani, A. Vitamin D Status and its Receptor Genes BsmI, FokI, ApaI, TaqI Polymorphism in Relation to Glucose Metabolism in Obese Iraqi Type 2 Diabetes Mellitus Patients. J. Mol. Genet. Med. 2017, 11, 1000260. [Google Scholar] [CrossRef]
- Iyer, A.P.; Lanham New, S.; Khoja, S.; AL-Ghamdi, M.A.; Bahlas, S. Association of Apa I polymorphism of vitamin d receptor gene with type 2 diabetes mellitus in Saudi population. J. Exp. Biol. Agric. Sci. 2017, 5, 271–276. [Google Scholar] [CrossRef]
- Mahjoubi, I.; Kallel, A.; Sbaï, M.H.; Ftouhi, B.; Ben Halima, M.; Jemaa, Z.; Feki, M.; Slimane, H.; Jemaa, R.; Kaabachi, N. Lack of association between FokI polymorphism in vitamin D receptor gene (VDR) & type 2 diabetes mellitus in the Tunisian population. Indian J. Med. Res. 2016, 144, 46–51. [Google Scholar] [CrossRef]
- Malik, R.; Farooq, R.; Mehta, P.; Ishaq, S.; Din, I.; Shah, P.; Majid, S. Association of Vitamin D Receptor Gene Polymorphism in Adults With Type 2 Diabetes in the Kashmir Valley. Can. J. Diabetes 2018, 42, 251–256. [Google Scholar] [CrossRef]
- Angel, B.; Lera, L.; Sánchez, H.; Oyarzún, A.; Albala, C. FokI polymorphism in vitamin D receptor gene: Differential expression of TNFα in peripheral mononuclear cells of type 2 diabetic subjects. Meta Gene 2016, 7, 1–6. [Google Scholar] [CrossRef]
- Abdeltif, E.; Benrahma, H.; Charoute, H.; Barakat, H.; Kandil, M.; Rouba, H. Vitamin D receptor gene polymorphisms and vitamin D status and susceptibility to type 2 diabetes mellitus in Moroccans patients. Int. J. Sci. Res. Publ. 2014, 4, 1–8. [Google Scholar]
- Mackawy, A.M.H.; Badawi, M.E.H. Association of vitamin D and vitamin D receptor gene polymorphisms with chronic inflammation, insulin resistance and metabolic syndrome components in type 2 diabetic Egyptian patients. Meta Gene 2014, 2, 540–556. [Google Scholar] [CrossRef]
- Vélayoudom-Céphise, F.L.; Larifla, L.; Donnet, J.P.; Maimaitiming, S.; Deloumeaux, J.; Blanchet, A.; Massart, C.; Munoz-Bellili, N.; Merle, S.; Chout, R.; et al. Vitamin D deficiency, vitamin D receptor gene polymorphisms and cardiovascular risk factors in Caribbean patients with type 2 diabetes. Diabetes Metab. 2011, 37, 540–545. [Google Scholar] [CrossRef] [PubMed]
- Alghadir, A.; Alghwiri, A.A.; Awad, H.; Anwer, S. Ten-year Diabetes Risk Forecast in the Capital of Jordan: Arab Diabetes Risk Assessment Questionnaire Perspective—A Strobe-Complaint Article. Medicine 2016, 95, e3181. [Google Scholar] [CrossRef] [PubMed]
- Ajlouni, K.; Khader, Y.S.; Batieha, A.; Ajlouni, H.; El-Khateeb, M. An increase in prevalence of diabetes mellitus in Jordan over 10 years. J. Diabetes Complicat. 2008, 22, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.W.; Mokdad, A.H.; Walke, H.; As’ad, M.; Al-Nsour, M.; Zindah, M.; Belbeisi, A. Projected Burden of Chronic, Noncommunicable Diseases in Jordan. Prev. Chronic. Dis. 2009, 6, A78. [Google Scholar]
- International Diabetes Federation. Available online: https://idf.org/our-network/regions-members/middle-east-and-north-africa/members/37-jordan.html (accessed on 3 August 2020).
- Taylor, R. Insulin resistance and type 2 diabetes. Diabetes 2012, 61, 778–779. [Google Scholar] [CrossRef]
- Zheng, S.; Xu, H.; Zhou, H.; Ren, X.; Han, T.; Chen, Y.; Qiu, H.; Wu, P.; Zheng, J.; Wang, L.; et al. Associations of lipid profiles with insulin resistance and β cell function in adults with normal glucose tolerance and different categories of impaired glucose regulation. PLoS ONE 2017, 12, e0172221. [Google Scholar] [CrossRef]
- Kavakiotis, I.; Tsave, O.; Salifoglou, A.; Maglaveras, N.; Vlahavas, I.; Chouvarda, I. Machine learning and data mining methods in diabetes research. Comput. Struct. Biotechnol. J. 2017, 15, 104–116. [Google Scholar] [CrossRef]
- Lee, B.J.; Kim, J.Y. Identification of type 2 diabetes risk factors using phenotypes consisting of anthropometry and triglycerides based on machine learning. IEEE J. Biomed. Health Inform. 2016, 20, 39–46. [Google Scholar] [CrossRef]
- Alghamdi, M.; Al-Mallah, M.; Keteyian, S.; Brawner, C.; Ehrman, J.; Sakr, S. Predicting diabetes mellitus using SMOTE and ensemble machine learning approach: The henry ford exercise testing (FIT) project. PLoS ONE 2017, 12, e0179805. [Google Scholar] [CrossRef]
- Faruqui, S.; Du, Y.; Meka, R.; Alaeddini, A.; Li, C.; Shirinkam, S.; Wang, J. Development of a Deep Learning Model for Dynamic Forecasting of Blood Glucose Level for Type 2 Diabetes Mellitus: Secondary Analysis of a Randomized Controlled Trial. JMIR mHealth uHealth 2019, 7, e14452. [Google Scholar] [CrossRef]
- Amar, Y.; Shilo, S.; Oron, T.; Amar, E.; Phillip, M.; Segal, E. Clinically Accurate Prediction of Glucose Levels in Patients with Type 1 Diabetes. Diabetes Technol. Ther. 2020. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Nikolayeva, O.; Luo, J.; Li, D. Building Risk Prediction Models for Type 2 Diabetes Using Machine Learning Techniques. Prev. Chronic Dis. 2019, 16, E130. [Google Scholar] [CrossRef] [PubMed]
- Sanal, M.G.; Paul, K.; Kumar, S.; Ganguly, N.K. Artificial Intelligence and Deep Learning: The Future of Medicine and Medical Practice. J. Assoc. Physicians India 2019, 67, 71–73. [Google Scholar] [PubMed]
- Nguyen, B.P.; Pham, H.N.; Tran, H.; Nghiem, N.; Nguyen, Q.H.; Do, T.; Tran, C.T.; Simpson, C.R. Predicting the onset of type 2 diabetes using wide and deep learning with electronic health records. Comput. Methods Programs Biomed. 2019, 182, 105055. [Google Scholar] [CrossRef]
- Raman, R.; Srinivasan, S.; Virmani, S.; Sivaprasad, S.; Rao, C.; Rajalakshmi, R. Fundus photograph-based deep learning algorithms in detecting diabetic retinopathy. Eye 2019, 33, 97–109. [Google Scholar] [CrossRef]
- Woldaregay, A.Z.; Årsand, E.; Walderhaug, S.; Albers, D.; Mamykina, L.; Botsis, T.; Hartvigsen, G. Data-driven modeling and prediction of blood glucose dynamics: Machine learning applications in type 1 diabetes. Artif. Intell. Med. 2019, 98, 109–134. [Google Scholar] [CrossRef]
- The World Health Organization. Definition and Diagnosis of Diabetes Mellitus and Intermediate Hyperglycaemia: Report of a WHO/IDF Consultation. 2006. Available online: https://www.who.int/diabetes/publications/diagnosis_diabetes2006/en/ (accessed on 8 July 2020).
- Friedewald, W.T.; Levy, R.I.; Fredrickson, D.S. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 1972, 18, 499–502. [Google Scholar] [CrossRef] [PubMed]
- Heather, J.M.; Chain, B. The sequence of sequencers: The history of sequencing DNA. Genomics 2016, 107, 1–8. [Google Scholar] [CrossRef]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Wheeler, D.L. GenBank. Nucleic Acids Res. 2007, 36, D25–D30. [Google Scholar] [CrossRef]
- Krishnan, S.; Hendriks, H.; Hartvigsen, M.; Graaf, A. Feed-forward neural network model for hunger and satiety related VAS score prediction. Theor. Biol. Med. Model. 2016, 13, 17. [Google Scholar] [CrossRef]
- Patel, J.L.; Goyal, R.K. Applications of Artificial Neural Networks in Medical Science. Curr. Clin. Pharmacol. 2007, 2, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Györffy, B.; Vásárhelyi, B.; Krikovszky, D.; Madácsy, L.; Tordai, A.; Tulassay, T.; Szabó, A. Gender-specific association of vitamin D receptor polymorphism combinations with type 1 diabetes mellitus. Eur. J. Endocrinol. 2003, 147, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Panierakis, C.; Goulielmos, G.; Mamoulakis, D.; Petraki, E.; Papavasiliou, E.; Galanakis, E. Vitamin D receptor gene polymorphisms and susceptibility to type 1 diabetes in Crete, Greece. Clin. Immunol. 2009, 133, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Uitterlinden, A.G.; Fang, Y.; van Meurs, J.B.J.; Pols, H.A.P.; van Leeuwen, J.P.T.M. Genetics and biology of vitamin D receptor polymorphisms. Gene 2004, 338, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Haykin, S. Neural Networks: A Comprehensive Foundation; Prentice Hall PTR: Delhi, India, 1994. [Google Scholar]
- Svozil, D.; Kvasnicka, V.; Pospichal, J.í. Introduction to multi-layer feed-forward neural networks. Chemometr. Intell. Lab. 1997, 39, 43–62. [Google Scholar] [CrossRef]
- Sijiang, D.; Vuskovic, M. Temporal vs. spectral approach to feature extraction from prehensile EMG signals. In Proceedings of the 2004 IEEE International Conference on Information Reuse and Integration, Las Vegas, NV, USA, 8–10 November 2004; pp. 3443–3450. [Google Scholar]
- Malecki, M.T.; Frey, J.; Moczulski, D.; Klupa, T.; Kozek, E.; Sieradzki, J. Vitamin D Receptor Gene Polymorphisms and Association with Type 2 Diabetes Mellitus in a Polish Population. Exp. Clin. Endocrinol. Diabetes 2003, 111, 505–509. [Google Scholar] [CrossRef]
- Bid, H.K.; Konwar, R.; Aggarwal, C.G.; Gautam, S.; Saxena, M.; Nayak, V.L.; Banerjee, M. Vitamin D receptor (FokI, BsmI and TaqI) gene polymorphisms and type 2 diabetes mellitus: A North Indian study. Indian J. Med. Sci. 2009, 63, 187–194. [Google Scholar] [CrossRef]
- Rawoof, A.M.; Promela, M.; Arif, A.B.; SheikhIshaq; Rabia, F. Vitamin D Receptor (VDR) Gene Polymorphismand Type 2 Diabeticmellitus (T2DM)-A Case Control Study in Ethnic Population of Kashmir Valley. Adv. Res. Gastroentero. Hepatol. 2017, 4, 555648. [Google Scholar] [CrossRef]
- Sriram, R.D.; Reddy, S.S.K. Artificial Intelligence and Digital Tools: Future of Diabetes Care. Clin. Geriatr. Med. 2020, 36, 513–525. [Google Scholar] [CrossRef]
- Pradhan, N.; Rani, G.; Dhaka, V.S.; Poonia, R.C. Diabetes Prediction Using Artificial Neural Network. In Deep Learning Techniques for Biomedical and Health Informatics; Agarwal, B., Balas, V.E., Jain, L.C., Poonia, R.C., Manisha, Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 327–339. [Google Scholar]
- Guo, Y.; Hao, Z.; Zhao, S.; Gong, J.; Yang, F. Artificial Intelligence in Health Care: Bibliometric Analysis. J. Med. Internet Res. 2020, 22, e18228. [Google Scholar] [CrossRef]
- Xie, L.; Yang, S.; Squirrell, D.; Vaghefi, E. Towards implementation of AI in New Zealand national diabetic screening program: Cloud-based, robust, and bespoke. PLoS ONE 2020, 15, e0225015. [Google Scholar] [CrossRef] [PubMed]
- Rahimloo, P.; Jafarian, A. Prediction of Diabetes by Using Artificial Neural Network, Logistic Regression Statistical Model and Combination of Them. Bull. Société R. Sci. Liège 2016, 85, 1148–1164. [Google Scholar]
- Zou, Q.; Qu, K.; Luo, Y.; Yin, D.; Ju, Y.; Tang, H. Predicting Diabetes Mellitus With Machine Learning Techniques. Front. Genet. 2018, 9, 515. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, Y.; Niu, M.; Wang, C.; Wang, Z. Machine learning for characterizing risk of type 2 diabetes mellitus in a rural Chinese population: The Henan Rural Cohort Study. Sci. Rep. 2020, 10, 4406. [Google Scholar] [CrossRef]
- Luque-Baena, R.M.; Urda, D.; Subirats, J.L.; Franco, L.; Jerez, J.M. Application of genetic algorithms and constructive neural networks for the analysis of microarray cancer data. Theor. Biol. Med. Model. 2014, 11, S7. [Google Scholar] [CrossRef]
- Nikooyeh, B.; Neyestani, T.R.; Farvid, M.; Alavi-Majd, H.; Houshiarrad, A.; Kalayi, A.; Shariatzadeh, N.; Gharavi, A.; Heravifard, S.; Tayebinejad, N.; et al. Daily consumption of vitamin D- or vitamin D + calcium-fortified yogurt drink improved glycaemic control in patients with type 2 diabetes: A randomized clinical trial. Am. J. Clin. Nutr. 2011, 93, 764–771. [Google Scholar] [CrossRef]
- Buchanan, T.A.; Watanabe, R.M.; Xiang, A.H. Limitations in surrogate measures of insulin resistance. J. Clin. Endocrinol. Metab. 2010, 95, 4874–4876. [Google Scholar] [CrossRef]
- Chen, X.; Wang, J.; Sun, D.; Liang, J. A Novel Hybrid Evolutionary Algorithm Based on PSO and AFSA for Feedforward Neural Network Training. In Proceedings of the 4th International Conference on Wireless Communications, Networking and Mobile Computing, Dalian, Liaoning, China, 12–14 October 2008; pp. 1–5. [Google Scholar]
| Parameter | Category | T2DM Patients (n = 82) | Healthy Controls (n = 82) | χ2 | p-Value |
|---|---|---|---|---|---|
| Age (years) | 20–60 | 67 | 67 | - | - |
| >60 | 15 | 15 | - | - | |
| Gender | Male | 37 | 37 | - | - |
| Female | 45 | 45 | - | - | |
| Lipid profile (mg/dL) | |||||
| HDL | <40 | 46 | 21 | 13.75 | 0.0002 * |
| ≥60 | 6 | 18 | |||
| LDL | ≤130 | 55 | 48 | 0.25 | 0.61 |
| >160 | 5 | 6 | |||
| TC | ≤200 | 58 | 39 | 1.43 | 0.23 |
| >240 | 6 | 8 | |||
| TG | ≤150 | 24 | 52 | 24.06 | <0.00001 * |
| >200 | 40 | 13 | |||
| Genotype | Healthy Controls (n = 82) | T2DM Patients (n = 82) |
|---|---|---|
| Mutated homozygous (CC) | 49 | 44 |
| Heterozygous (CT) | 27 | 31 |
| Wild homozygous (TT) | 6 | 7 |
| Genotype | Observed Value | Expected Value | χ2 | p-Value |
|---|---|---|---|---|
| CC | 49 | 47.36 | 0.69 | 0.4 |
| TT | 6 | 4.72 | ||
| CT | 27 | 29.91 |
| FokI Polymorphism | T2DM Patients n (%) | Healthy Controls n (%) | Odds Ratio (OR) | 95% CI | p-Value |
|---|---|---|---|---|---|
| Genotypes | |||||
| CC | 44 (53.7) | 49 (59.8) | CC vs. (CT and TT) 0.78 | 0.42–1.45 | 0.4 |
| CT | 31 (37.8) | 27 (32.9) | CT vs. (CC and TT) 1.24 | 0.65–2.35 | 0.5 |
| TT | 7 (8.5) | 6 (7.3) | TT vs. (CC and CT) 1.18 | 0.38–3.68 | 0.7 |
| Alleles | |||||
| C | 119 (72.6) | 125 (76.2) | 0.82 | 0.50–1.36 | 0.4 |
| T | 45 (27.4) | 39 (23.8) | 1.21 | 0.74–1.99 |
| Parameter (mg/dL) | T2DM Patients | Healthy Controls | p-Value | ||
|---|---|---|---|---|---|
| D-Value | Mean ± SD (mg/dL) | D-Value | Mean ± SD (mg/dL) | ||
| TC | 0.06 (0.91) | 173.9 ± 47.2 | 0.08 (0.64) | 201.4 ± 37.7 | <0.001 * |
| TG | 0.08 (0.58) | 214.6 ±90.4 | 0.13 (0.09) | 141.3 ± 77.7 | <0.001 * |
| HDL | 0.13 (0.13) | 39.4 ± 19.7 | 0.07 (0.74) | 51.0 ± 14.0 | <0.001 * |
| LDL | 0.11 (0.25) | 87.8 ± 50.1 | 0.07 (0.73) | 121.3 ± 34.6 | <0.001 * |
| Parameter (mg/dL) | Category (n) | CC | CT | TT | χ2 | p-Value |
|---|---|---|---|---|---|---|
| TC | ≤200 (58) | 31 | 22 | 5 | 0.73 | 0.6 |
| >240 (6) | 3 | 3 | 0 | |||
| TG | ≤150 (23) | 14 | 7 | 2 | 1.82 | 0.4 |
| >200 (40) | 22 | 17 | 1 | |||
| HDL | <40 (46) | 23 | 19 | 4 | 1.47 | 0.4 |
| ≥60 (6) | 4 | 1 | 1 | |||
| LDL | ≤130 (55) | 32 | 20 | 3 | 1.21 | 0.5 |
| >160 (5) | 2 | 3 | 0 |
| Parameter (mg/dL) | Category (n) | CC | CT | TT | χ2 | p-Value |
|---|---|---|---|---|---|---|
| TC | ≤200 (39) | 26 | 9 | 4 | 1.38 | 0.5 |
| >240 (8) | 5 | 3 | 0 | |||
| TG | ≤150 (53) | 34 | 13 | 6 | 3.35 | 0.1 |
| >200 (13) | 7 | 6 | 0 | |||
| HDL | ≥60 (18) | 10 | 6 | 2 | 2.81 | 0.2 |
| <40 (22) | 12 | 10 | 0 | |||
| LDL | ≤130 (48) | 33 | 11 | 4 | 2.29 | 0.3 |
| >160 (6) | 3 | 3 | 0 |
| Diabetes | No Diabetes | |
|---|---|---|
| Predicted diabetes | a | b |
| Predicted no diabetes | c | d |
| a + c | b + d |
| Prediction | Trial 1 | Trial 2 | Trial 3 | Trial 4 | ||||
|---|---|---|---|---|---|---|---|---|
| D | ND | D | ND | D | ND | D | ND | |
| D | 13 | 17 | 15 | 13 | 13 | 1 | 15 | 6 |
| ND | 4 | 3 | 1 | 6 | 6 | 12 | 1 | 11 |
| Population | Number of Controls | Number of T2DM Patients | OR (95% CI) | p-Value | Results | Reference |
|---|---|---|---|---|---|---|
| Kashmir valley | 100 | 100 | 0.32 (0.1704–0.6154) | <0.001 | Yes | [42] |
| Morocco | 177 | 176 | 0.35 (0.14–0.83) | 0.01 | Yes | [9] |
| Santiago, Metropolitan Region, Chile | 160 | 160 | 3.52 (1.53–8.09) | 0.003 | Yes | [8] |
| Tunisia | 302 | 439 | 1.19 (0.63–2.25) | 0.5 | No | [6] |
| Iraq | 75 | 200 | 0.21(0.10–0.40) | <0.0001 | Yes | [4] |
| Poland | 239 | 308 | 1.08 (0.85–1.37) | >0.05 | No | [40] |
| North India | 160 | 100 | 0.72 (0.49–1.06) | 0.098 | No | [41] |
| Saudi | 285 | 285 | 0.73 (0.56–0.95) | 0.02 | Yes | [2] |
| Egypt | 60 | 130 | 0.51 (0.37–0.72) | 0.001 | Yes | [10] |
| Caribbean | 76 | 201 | 0.52 (0.30–0.90) | 0.02 | Yes | [11] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hatmal, M.M.; Abderrahman, S.M.; Nimer, W.; Al-Eisawi, Z.; Al-Ameer, H.J.; Al-Hatamleh, M.A.I.; Mohamud, R.; Alshaer, W. Artificial Neural Networks Model for Predicting Type 2 Diabetes Mellitus Based on VDR Gene FokI Polymorphism, Lipid Profile and Demographic Data. Biology 2020, 9, 222. https://doi.org/10.3390/biology9080222
Hatmal MM, Abderrahman SM, Nimer W, Al-Eisawi Z, Al-Ameer HJ, Al-Hatamleh MAI, Mohamud R, Alshaer W. Artificial Neural Networks Model for Predicting Type 2 Diabetes Mellitus Based on VDR Gene FokI Polymorphism, Lipid Profile and Demographic Data. Biology. 2020; 9(8):222. https://doi.org/10.3390/biology9080222
Chicago/Turabian StyleHatmal, Ma’mon M., Salim M. Abderrahman, Wajeha Nimer, Zaynab Al-Eisawi, Hamzeh J. Al-Ameer, Mohammad A. I. Al-Hatamleh, Rohimah Mohamud, and Walhan Alshaer. 2020. "Artificial Neural Networks Model for Predicting Type 2 Diabetes Mellitus Based on VDR Gene FokI Polymorphism, Lipid Profile and Demographic Data" Biology 9, no. 8: 222. https://doi.org/10.3390/biology9080222
APA StyleHatmal, M. M., Abderrahman, S. M., Nimer, W., Al-Eisawi, Z., Al-Ameer, H. J., Al-Hatamleh, M. A. I., Mohamud, R., & Alshaer, W. (2020). Artificial Neural Networks Model for Predicting Type 2 Diabetes Mellitus Based on VDR Gene FokI Polymorphism, Lipid Profile and Demographic Data. Biology, 9(8), 222. https://doi.org/10.3390/biology9080222






