Nanomaterial-Based Sensor Array Signal Processing and Tuberculosis Classification Using Machine Learning
Abstract
:1. Introduction
- We utilize multiple data preprocessing techniques, such as sensor signal extraction, data normalization, data calibration, and sensor selection, which apply to similar tasks involving MTS sensor signals.
- We propose various ML-based models, namely long short-term memory (LSTM), convolutional neural network (CNN), Gramian angular field-CNN (GAF-CNN), and multivariate time series with MinCutPool (MT-MinCutPool), to classify the small TB dataset, where the proposed low-power model features a simplified and shallow network architecture, incorporating a limited number of parameters. This design results in lowered computational complexity and effectively reduces power consumption. We then compare the performance of our proposed models with several state-of-the-art methods commonly used in MTSC tasks.
- To encourage further research on MTSC with small-dataset problems, we provide an open-source of our work, which is accessible on 5 March 2023 at: https://github.com/ChenxiLiu6/TB-Classification.git.
2. Related Work
3. Background
3.1. Time Series Classification
3.2. Encoding Time Series as Images by Gramian Angular Field (GAF)
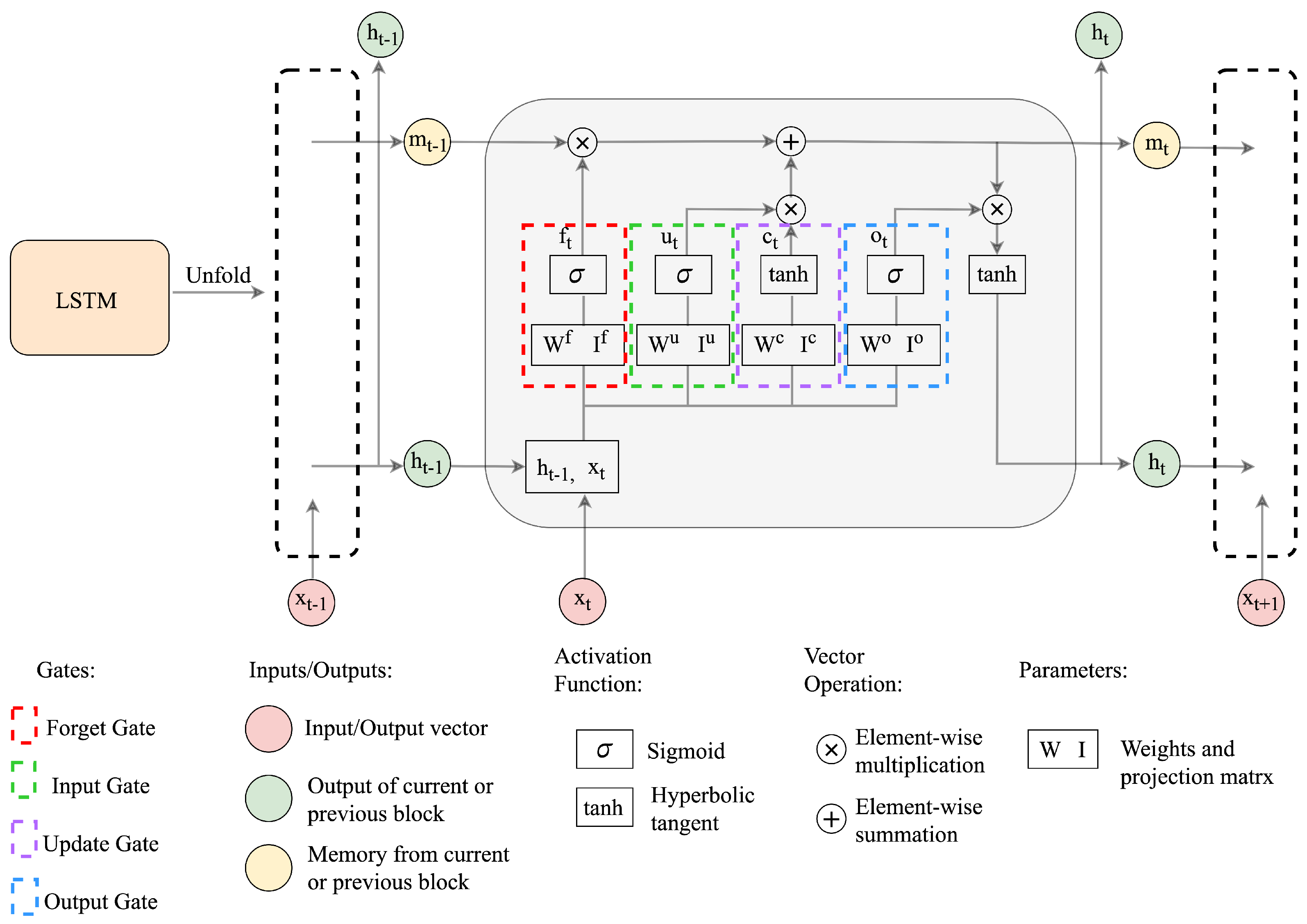
3.3. The Long Short-Term Memory (LSTM) Network
3.4. The Graph Neural Network Model
Spectral Clustering and MinCutPool
4. Data Preprocessing
4.1. Dataset Description
4.2. Middle Part Signal Extraction
4.3. Data Normalization
4.4. Data Calibration
4.5. Sensor Selection by Using the Pearson Correlation Coefficient Matrix
5. Proposed Methods for Multivariate Time Series Classification
5.1. Proposed LSTM Network
5.2. Convolution Neural Network (CNN)
5.3. GAF-CNN
5.4. MTSC with Graph Laplacian and MinCutPool
Graph Structure Learning Using Laplacian Matrix
| Algorithm 1: Build the Laplacian adjacency matrix. |
 |
5.4.1. Temporal Feature Extraction
5.4.2. Spatial–Temporal Modeling
5.4.3. Graph Coarsening by MinCutPool
5.4.4. Graph-Level Embedding Classification
6. Experiments
6.1. Evaluation Metrics
- Accuracy: It measures the proportion of the correct predictions among all of the predictions made by the models.
- Sensitivity (true positive rate, TPR): It measures the proportion of true positive models made by the model out of all actual positive samples. It indicates the ability of the model to correctly identify the positive samples, which is very important for clinical settings. It indicates the ability of a model to identify negative samples correctly.
- Specificity (true negative rate, TNR): It measures the proportion of true negative predictions made by the model out of all actual negative samples.
- AUC: (Area under the ROC curve): It considers the performance of a classifier over all possible threshold values, taking into account both the true positive rate (sensitivity) and the false positive rate (1—specificity).
6.2. Experimental Setup
7. Results and Discussion
7.1. Main Results
7.2. Discussion
- Medical diagnosis: GAF image conversion could further assist in identifying a range of medical conditions, from electrocardiogram (ECG) rhythms to Alzheimer’s signals. GAF image conversion in these fields may allow for better clinical decision-making and enhance the accuracy of machine learning models.
- Financial time-series analysis: GAF image conversion could be leveraged to predict stock prices and fluctuations in the currency market and further enhance the effectiveness of ML algorithms in predicting trends and changes.
- Speech recognition: GAF-based image classification could enable more accurate identification of speech patterns, phonemes, and speech denoising. The use of images could be particularly effective in noisy environments where traditional audio inputs may be challenged.
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BCE | binary cross entropy |
| CNN | convolution neural network |
| CV | computer vision |
| DL | deep learning |
| DTW | dynamic time warping |
| GAF | Gramian angular field |
| GCN | graph convolution network |
| GNN | graph neural network |
| LSTM | long short-term memory |
| MTF | Markov transition field |
| ML | Machine Learning |
| MLSTM-FCN | multivariate LSTM fully convolutional network |
| MT-MinCutPool | multivariate time series with MinCutPool |
| MTPool | multivariate time series classification with variational graph pooling |
| MTS | multivariate time series |
| MTSC | multivariate time series classification |
| NLP | natural language processing |
| NNs | neural networks |
| SC | spectral clustering |
| TB | tuberculosis |
| TSC | time series classification |
| VOC | volatile organic compound |
| WHO | World Health Organization |
References
- Zaman, K. Tuberculosis: A global health problem. J. Health Popul. Nutr. 2010, 28, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Vishinkin, R.; Busool, R.; Mansour, E.; Fish, F.; Esmail, A.; Kumar, P.; Gharaa, A.; Cancilla, J.C.; Torrecilla, J.S.; Skenders, G.; et al. Profiles of volatile biomarkers detect tuberculosis from skin. Adv. Sci. 2021, 8, 2100235. [Google Scholar] [CrossRef] [PubMed]
- Nakhleh, M.K.; Amal, H.; Jeries, R.; Broza, Y.Y.; Aboud, M.; Gharra, A.; Ivgi, H.; Khatib, S.; Badarneh, S.; Har-Shai, L.; et al. Diagnosis and classification of 17 diseases from 1404 subjects via pattern analysis of exhaled molecules. ACS Nano 2017, 11, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Broza, Y.Y.; Vishinkin, R.; Barash, O.; Nakhleh, M.K.; Haick, H. Synergy between nanomaterials and volatile organic compounds for non-invasive medical evaluation. Chem. Soc. Rev. 2018, 47, 4781–4859. [Google Scholar] [CrossRef]
- Röck, F.; Barsan, N.; Weimar, U. Electronic nose: Current status and future trends. Chem. Rev. 2008, 108, 705–725. [Google Scholar] [CrossRef] [PubMed]
- Huynh, T.P.; Khatib, M.; Srour, R.; Plotkin, M.; Wu, W.; Vishinkin, R.; Hayek, N.; Jin, H.; Gazit, O.M.; Haick, H. Composites of polymer and carbon nanostructures for self-healing chemical sensors. Adv. Mater. Technol. 2016, 1, 1600187. [Google Scholar] [CrossRef]
- Brigato, L.; Iocchi, L. A Close Look at Deep Learning with Small Data. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021. [Google Scholar]
- Shaikhina, T.; Lowe, D.; Daga, S.; Briggs, D.; Higgins, R.; Khovanova, N. Machine Learning for Predictive Modelling based on Small Data in Biomedical Engineering. IFAC-PapersOnLine 2015, 48, 469–474. [Google Scholar] [CrossRef]
- Hirata, Y.; Katori, Y.; Shimokawa, H.; Suzuki, H.; Blenkinsop, T.A.; Lang, E.J.; Aihara, K. Testing a neural coding hypothesis using surrogate data. J. Neurosci. Methods 2008, 172, 312–322. [Google Scholar] [CrossRef]
- Barton, T.; Yu, H.; Rogers, K.; Fulda, N.; Chiang, S.h.W.; Yorgason, J.; Warnick, K.F. Towards Low-Power Machine Learning Architectures Inspired by Brain Neuromodulatory Signalling. J. Low Power Electron. Appl. 2022, 12, 59. [Google Scholar] [CrossRef]
- Musa, A.; Hassan, M.; Hamada, M.; Aliyu, F. Low-Power Deep Learning Model for Plant Disease Detection for Smart-Hydroponics Using Knowledge Distillation Techniques. J. Low Power Electron. Appl. 2022, 12, 24. [Google Scholar] [CrossRef]
- Chopra, K.; Singh, S. Newer diagnostic tests for tuberculosis, their utility, and their limitations. Curr. Med. Res. Pract. 2020, 10, 8–11. [Google Scholar] [CrossRef]
- MacGregor-Fairlie, M.; Wilkinson, S.; Besra, G.S.; Goldberg Oppenheimer, P. Tuberculosis diagnostics: Overcoming ancient challenges with modern solutions. Emerg. Top. Life Sci. 2020, 4, 435–448. [Google Scholar]
- Gill, C.M.; Dolan, L.; Piggott, L.M.; McLaughlin, A.M. New developments in tuberculosis diagnosis and treatment. Breathe 2022, 18. [Google Scholar] [CrossRef]
- Santos, J.A.; Leite, A.; Soares, P.; Duarte, R.; Nunes, C. Delayed diagnosis of active pulmonary tuberculosis-potential risk factors for patient and healthcare delays in Portugal. BMC Public Health 2021, 21, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Broza, Y.Y.; Haick, H. Nanomaterial-based sensors for detection of disease by volatile organic compounds. Nanomedicine 2013, 8, 785–806. [Google Scholar] [CrossRef]
- Konvalina, G.; Haick, H. Sensors for breath testing: From nanomaterials to comprehensive disease detection. Acc. Chem. Res. 2014, 47, 66–76. [Google Scholar] [CrossRef]
- Tisch, U.; Billan, S.; Ilouze, M.; Phillips, M.; Peled, N.; Haick, H. Volatile organic compounds in exhaled breath as biomarkers for the early detection and screening of lung cancer. Int. J. Clin. Rev. 2012. [Google Scholar] [CrossRef]
- Turner, A.P.; Magan, N. Electronic noses and disease diagnostics. Nat. Rev. Microbiol. 2004, 2, 161–166. [Google Scholar] [CrossRef]
- Nweke, H.F.; Teh, Y.W.; Al-garadi, M.A.; Alo, U.R. Deep learning algorithms for human activity recognition using mobile and wearable sensor networks: State of the art and research challenges. Expert Syst. Appl. 2018, 105, 233–261. [Google Scholar] [CrossRef]
- Rajkomar, A.; Oren, E.; Chen, K.; Dai, A.M.; Hajaj, N. Scalable and accurate deep learning with electronic health records. NPJ Digit. Med. 2018, 1, 18. [Google Scholar] [CrossRef]
- Nwe, T.L.; Dat, T.H.; Ma, B. Convolutional neural network with multi-task learning scheme for acoustic scene classification. In Proceedings of the 2017 Asia-Pacific Signal and Information Processing Association Annual Summit and Conference (APSIPA ASC), Aloft Kuala Lumpur Sentral, Malaysia, 12–15 December 2017; pp. 1347–1350. [Google Scholar]
- Xiao, D.; Su, J.; Che, H. Research on Stock Price Time Series Prediction Based on Deep Learning and Autoregressive Integrated Moving Average. Sci. Program. 2022, 2022, 4758698. [Google Scholar] [CrossRef]
- Wang, Z.; Oates, T. Encoding Time Series as Images for Visual Inspection and Classification Using Tiled Convolutional Neural Networks. In Proceedings of the Workshops at the Twenty-Ninth AAAI Conference on Artificial Intelligence, Austin, TX, USA, 25–30 January 2015; Volume 1. [Google Scholar]
- Pascanu, R.; Gulcehre, C.; Cho, K.; Bengio, Y. How to construct deep recurrent neural networks. arXiv 2013, arXiv:1312.6026. [Google Scholar]
- Karim, F.; Majumdar, S.; Darabi, H.; Chen, S. LSTM Fully Convolutional Networks for Time Series Classification. IEEE Access 2018, 6, 1662–1669. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, K. Supervised Sequence Labelling with Recurrent Neural Networks. Ph.D. Thesis, Technical University of Munich, Munich, Germany, 2008. [Google Scholar]
- Bianchi, F.M.; Grattarola, D.; Alippi, C. Spectral clustering with graph neural networks for graph pooling. In Proceedings of the 37th International Conference on Machine Learning, Virtual, 13–18 July 2020; pp. 874–883. [Google Scholar]
- Ng, A.; Jordan, M.; Weiss, Y. On spectral clustering: Analysis and an algorithm. Adv. Neural Inf. Process. Syst. 2001, 14. [Google Scholar]
- Broza, Y.Y.; Zhou, X.; Yuan, M.; Qu, D.; Zheng, Y.; Vishinkin, R.; Khatib, M.; Wu, W.; Haick, H. Disease detection with molecular biomarkers: From chemistry of body fluids to nature-inspired chemical sensors. Chem. Rev. 2019, 119, 11761–11817. [Google Scholar] [CrossRef]
- Singh, D.; Singh, B. Investigating the impact of data normalization on classification performance. Appl. Soft Comput. 2020, 97, 105524. [Google Scholar] [CrossRef]
- Gravina, R.; Alinia, P.; Ghasemzadeh, H.; Fortino, G. Multi-sensor fusion in body sensor networks: State-of-the-art and research challenges. Inf. Fusion 2017, 35, 68–80. [Google Scholar] [CrossRef]
- Brena, R.; Aguileta, A.; Trejo, L.; Molino Minero Re, E.; Mayora, O. Choosing the Best Sensor Fusion Method: A Machine-Learning Approach. Sensors 2020, 20, 2350. [Google Scholar] [CrossRef]
- Benesty, J.; Chen, J.; Huang, Y.; Cohen, I. Pearson correlation coefficient. In Noise Reduction in Speech Processing; Springer: Berlin/Heidelberg, Germany, 2009; Chapter 5; pp. 37–40. [Google Scholar]
- Sainath, T.N.; Vinyals, O.; Senior, A.; Sak, H. Convolutional, Long Short-Term Memory, fully connected Deep Neural Networks. In Proceedings of the 2015 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), South Brisbane, Australia, 19–24 April 2015; pp. 4580–4584. [Google Scholar]
- Fawaz, H.I.; Forestier, G.; Weber, J.; Idoumghar, L.; Muller, P.A. Deep learning for time series classification: A review. Data Min. Knowl. Discov. 2019, 33, 917–963. [Google Scholar] [CrossRef]
- Längkvist, M.; Karlsson, L.; Loutfi, A. A review of unsupervised feature learning and deep learning for time-series modeling. Pattern Recognit. Lett. 2014, 42, 11–24. [Google Scholar] [CrossRef]
- Wei, C.; Shi, K. Multi-scale Attention Convolutional Neural Network for time series classification. Neural Netw. 2021, 136, 126–140. [Google Scholar]
- Sharma, Y.; Coronato, N.; Brown, D.E. Encoding cardiopulmonary exercise testing time series as images for classification using convolutional neural network. In Proceedings of the 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Glasgow, UK, 11–15 July 2022; pp. 1611–1614. [Google Scholar]
- Duan, Z.; Xu, H.; Wang, Y.; Huang, Y.; Ren, A.; Xu, Z.; Sun, Y.; Wang, W. Multivariate time-series classification with hierarchical variational graph pooling. Neural Netw. 2022, 154, 481–490. [Google Scholar] [CrossRef]
- Zerveas, G.; Jayaraman, S.; Patel, D.; Bhamidipaty, A.; Eickhoff, C. A transformer-based framework for multivariate time series representation learning. In Proceedings of the 27th ACM SIGKDD Conference on Knowledge Discovery & Data Mining, Singapore, 14–18 August 2021; pp. 2114–2124. [Google Scholar]
- Ruiz, A.P.; Flynn, M.; Large, J.; Middlehurst, M.; Bagnall, A. The great multivariate time series classification bake off: A review and experimental evaluation of recent algorithmic advances. Data Min. Knowl. Discov. 2021, 35, 401–449. [Google Scholar] [CrossRef] [PubMed]
- Karim, F.; Majumdar, S.; Darabi, H.; Harford, S. Multivariate LSTM-FCNs for time series classification. Neural Netw. 2019, 116, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Thanou, D.; Frossard, P.; Vandergheynst, P. Learning Laplacian Matrix in Smooth Graph Signal Representations. IEEE Trans. Signal Process. 2016, 64, 6160–6173. [Google Scholar] [CrossRef]
- Yan, S.; Xiong, Y.; Lin, D. Spatial temporal graph convolutional networks for skeleton-based action recognition. In Proceedings of the AAAI Conference on Artificial Intelligence, New Orleans, LA, USA, 2–7 February 2018; Volume 32. [Google Scholar]
- Yu, B.; Yin, H.; Zhu, Z. Spatio-temporal graph convolutional networks: A deep learning framework for traffic forecasting. In Proceedings of the International Joint Conferences on Artificial Intelligence Organization, Melbourne, Australia, 19–25 August 2017. [Google Scholar]
- Hamilton, W.; Ying, Z.; Leskovec, J. Inductive representation learning on large graphs. arXiv 2017, arXiv:1706.02216. [Google Scholar]
















| Layer (Type) | Output Shape | Activation | Parameter Number |
|---|---|---|---|
| lstm_1 (LSTM) | (bs, 32) | − | 7936 |
| dropout_1 (Dropout) | (bs, 32) | − | 0 |
| dense_1 (Dense) | (bs, 16) | ReLU | 528 |
| dense_2 (Dense) | (bs, 1) | Sigmoid | 17 |
| Layer | Stride | Activation | Kerne lSize | Input Shape | Output Shape | Parameter Number |
|---|---|---|---|---|---|---|
| Conv1D_1 | 1 | ReLU | 20 | (bs, 147, 29) | (bs, 128, 16) | 9296 |
| Max Pooling1D_1 | 2 | - | 2 | (bs, 128, 16) | (bs, 64, 16) | 0 |
| Conv1D_2 | 2 | ReLU | 2 | (bs, 64, 16) | (bs, 32, 4) | 2116 |
| Max Pooling1D_2 | 2 | - | 2 | (bs, 32, 4) | (bs, 16, 4) | 0 |
| Flatten | - | - | - | (bs, 16, 4) | (bs, 64) | 0 |
| Dense_1 | - | ReLU | - | (bs, 64) | (bs, 16) | 1040 |
| Dense_2 | - | Sigmoid | - | (bs, 16) | (bs, 1) | 17 |
| Layer | Stride | Activation | Kernel Size | Input Shape | Output Shape | Parameter Number |
|---|---|---|---|---|---|---|
| Conv2D_1 | 1 | ReLU | (5, 5) | (bs, 147, 147, 29) | (bs, 143, 143, 12) | 8712 |
| Max Pooling2D_1 | 2 | - | (2, 2) | (bs, 143, 143, 12) | (bs, 71, 71, 12) | 0 |
| Conv2D_2 | 1 | ReLU | (5, 5) | (bs, 71, 71, 12) | (bs, 67, 67,6) | 1806 |
| Max Pooling2D_2 | 2 | - | (2, 2) | (bs, 67, 67, 6) | (bs, 33, 33, 6) | 0 |
| Flatten | - | - | - | (bs, 33, 33, 6) | (bs, 6534) | 0 |
| Dense_1 | - | Sigmoid | - | (bs, 6534) | (bs, 1) | 6535 |
| LSTM | CNN | GAF-CNN | MT-MinCutPool | Model Mean | ||
|---|---|---|---|---|---|---|
| Accuracy | train | 0.646 | 0.916 | 0.659 | 0.688 | − |
| valid | 0.69 | 0.687 | 0.692 | 0.69 | − | |
| test | 0.611 | 0.606 | 0.639 | 0.604 | 0.615 | |
| Sensitivity | train | 0.618 | 0.931 | 0.766 | 0.76 | − |
| valid | 0.675 | 0.715 | 0.78 | 0.756 | − | |
| test | 0.631 | 0.694 | 0.777 | 0.728 | 0.71 | |
| Specificity | train | 0.676 | 0.901 | 0.544 | 0.612 | − |
| valid | 0.704 | 0.658 | 0.597 | 0.619 | − | |
| test | 0.594 | 0.533 | 0.532 | 0.5 | 0.54 | |
| AUC | test | 0.634 | 0.657 | 0.692 | 0.661 | 0.661 |
| MTPool [41] | GAF-Attention [40] | MLSTM-FCN [44] | Model Mean | ||
|---|---|---|---|---|---|
| Accuracy | train | 0.563 | 0.755 | 0.733 | − |
| valid | 0.577 | 0.663 | 0.693 | − | |
| test | 0.517 | 0.631 | 0.586 | 0.578 | |
| Sensitivity | train | 0.682 | 0.78 | 0.757 | − |
| valid | 0.683 | 0.688 | 0.733 | − | |
| test | 0.655 | 0.622 | 0.664 | 0.647 | |
| Specificity | train | 0.436 | 0.728 | 0.709 | − |
| valid | 0.464 | 0.636 | 0.651 | − | |
| test | 0.4 | 0.637 | 0.521 | 0.519 | |
| AUC | test | 0.538 | 0.695 | 0.648 | 0.627 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Cohen, I.; Vishinkin, R.; Haick, H. Nanomaterial-Based Sensor Array Signal Processing and Tuberculosis Classification Using Machine Learning. J. Low Power Electron. Appl. 2023, 13, 39. https://doi.org/10.3390/jlpea13020039
Liu C, Cohen I, Vishinkin R, Haick H. Nanomaterial-Based Sensor Array Signal Processing and Tuberculosis Classification Using Machine Learning. Journal of Low Power Electronics and Applications. 2023; 13(2):39. https://doi.org/10.3390/jlpea13020039
Chicago/Turabian StyleLiu, Chenxi, Israel Cohen, Rotem Vishinkin, and Hossam Haick. 2023. "Nanomaterial-Based Sensor Array Signal Processing and Tuberculosis Classification Using Machine Learning" Journal of Low Power Electronics and Applications 13, no. 2: 39. https://doi.org/10.3390/jlpea13020039
APA StyleLiu, C., Cohen, I., Vishinkin, R., & Haick, H. (2023). Nanomaterial-Based Sensor Array Signal Processing and Tuberculosis Classification Using Machine Learning. Journal of Low Power Electronics and Applications, 13(2), 39. https://doi.org/10.3390/jlpea13020039







