Abstract
A considerable fraction of the female workforce worldwide is making ends meet by doing various jobs informally at home or in nearby places, rather than at employers’ premises. The contribution of these female home-based workers (FHBWs) is significant to the country’s economic growth. FHBWs are often confronted with numerous occupational diseases due to a lack of awareness of occupational safety and health measures, and unhealthy living and working conditions. The informality of FHBWs prevents them from getting proper healthcare, safety, and other dispensations enjoyed by formal employees. Despite their undeniable importance, health issues of FHBWs are still overlooked. This study is an attempt to discover the frequent co-occurring occupational diseases encountered by FHBWs in Punjab, a province of Pakistan. Frequent itemset mining (FIM) or co-occurrence grouping is a technique of data science that identifies the associations among different entities in the data. Based on FIM, the D-GENE algorithm is applied in this study to efficiently discover frequent co-occurring diseases in the data obtained from the Punjab Home-based Workers Survey (2016). The far-reaching goal of the study is to bring awareness of the occupational health issues and safety risks to the health authorities as well as to the FHBWs.
1. Introduction
Led by the search of low-cost inputs by businesses [1] with the increase in poverty levels, the informal economy is expanding at a fast pace in developing countries. Female home-based workers (FHBWs) are categorized as informal workers who carry out remunerative work at home or in adjacent premises, rather than at employers’ premises. They are usually engaged in manufacturing and post-manufacturing tasks such as embroidery/stitching, carpet weaving, paper products, handicrafts, football making, and others. These workers are often confronted with unhealthy living and working conditions. Thus, the home-cum-workplace environment puts them at higher risk of suffering from occupational diseases.
Falling sick results in below-par performance, low-quality output, and significant delays in product delivery. Eventually, a dreadful impact is witnessed on the earnings of FHBWs, which further reduces their ability to seek appropriate medical treatment. The irony is that, due to their invisibility, policymakers and health practitioners cannot understand the occupational safety and health problems faced by FHBWs.
According to the UN-Women report [2], home-based workers contributed almost 400 billion PKR (nearly 3.8% of the total GDP) through their wages to the economy of Pakistan in 2013–2014. However, they are excluded from the mainstream market economy, health protection, occupational safety, and social insurance. In Punjab, 44.29% of the population is engaged with an informal economy in which female and male labor force participation rates are 6.77% and 37.52%, respectively. The majority of the female labor force is home-based workers [3].
According to the Punjab Home-based Workers Survey (2016) [4], FHBWs face the following challenges while working.
- Poor lighting situation in the workplace.
- Lack of ventilation and protective equipment.
- Workplaces are congested, hot, and suffocated.
- Workplaces contain dust and fumes.
- Subject to chemical hazards.
- Working with sharp machines/tools.
- Bad posture.
- Long working hours.
These challenges pose massive health risks; thus, FHBWs are often confronted with numerous occupational diseases. A healthy FHBW not only generates household income but also plays a pivotal role in enhancing productivity that leads towards sustainable economic growth. Despite their undeniable importance, pertinent government organizations and health practitioners are ignorant of the occupational health issues of FHBWs. Undoubtedly, the ignorance is due to the disguised and isolated way of working of home-based workers. To the best of our knowledge, no comprehensive scientific study exists that has identified the frequent occupational diseases faced by FHBWs. The discovery of frequent occupational diseases could help in identifying and eradicating the inadequacies faced by the FHBWs. Furthermore, robust medical surveillance is required for the early detection of occupational diseases and injuries [5].
This study is an attempt to analyze FHBWs occupational disease data from a data science perspective. Data science is defined as a set of principles to extract knowledge from data. Techniques based on data science principles are increasingly being applied in medical diagnostics and informatics [6,7]. Today’s sophisticated tools based on data science principles are capable of discovering significant underlying associations or co-occurring groups in the data. Thus, a data science insight can help in exploring the frequent co-occurring diseases faced by FHBWs. Frequent itemset mining (FIM) is an indispensable data science technique for exploring frequent co-occurring items in voluminous databases [8]. The co-occurrence reveals strong associations among the items.
The role of FIM has become even more important in the era of big data, where abundant co-occurrence grouping exists. FIM has been applied primarily to perform market basket analysis, in which it explores the items frequently bought together by customers [8]. FIM techniques are also used to perform classification [9], clustering [10], and mining association rules [11]. At present, along with other applications, FIM is also being applied in medical applications.
In this study, FIM is applied to discover the frequent co-occurring diseases faced by FHBWs. The FHBW data were obtained from the Punjab Home-based Workers Survey (2016), conducted by the Bureau of Statistics Punjab, Pakistan [4]. The far-reaching objectives of identifying frequent co-occurring occupational diseases in this study are the following.
- Promoting occupational safety and health.
- Making public health authorities well aware of the co-occurring occupational diseases faced by the FHBWs. Ample awareness can help the authorities to adopt the correct policies, so that appropriate medical treatment and compensation can be provided to FHBWs.
- Providing awareness to the FHBWs of the health and safety risks they are exposed to every day while working.
To efficiently mine frequent co-occurring occupational diseases, the recently proposed FIM algorithm, Deferring the Generation of Power sets for Discovering Frequent Itemsets from Sparse Big data (D-GENE) is used in this study. D-GENE was chosen due to its superior runtime efficiency as well as having the least memory consumption, especially for sparse data. D-GENE has shown its superiority over other state-of-the-art methods like Apriori and FP-growth [12]. Considering the sparse nature of the data used in this study, D-GENE was believed to be the optimal choice to discover frequent co-occurring diseases from them.
The organization of this paper is as follows. A review of related work is introduced in Section 2. Section 3 describes the problem of FIM. Experimental details followed by a complete example are given in Section 4. Detailed results are given in Section 5. A detailed discussion is given in Section 6. Section 7 concludes this study.
2. Related Work
Medical data were analyzed by using the Apriori algorithm [8] to discover frequent diseases in diverse geographical areas at a given time [13]. Another technique was proposed for predicting the risk level of patients suffering from heart disease [14]. Disease symptoms were identified and used as input. Frequent itemsets were generated based on a pre-defined minimum support threshold. It was envisaged that the resultant frequent itemsets would help in making better diagnostics as well as determining the disease risks at the early stages.
A modified version of the K-means algorithm was proposed to analyze medical datasets to discover early on and monthly periodic frequent patterns [15]. Periodical frequent patterns between the years 2012 and 2013 and month-wise frequent patterns were discovered. An idea of temporal view was used to adapt K-means for this purpose. A prototype was made on the proposed methodology that yielded useful results in finding diseases.
To aid in effectively diagnosing polycystic ovarian syndrome (PCOS), FIM was applied to discover recurring patterns among the symptoms of PCOS patients [16]. The Apriori algorithm was incorporated for predicting susceptible cases as well. A software system was built to produce well-timed forecasting of diabetes as well as cardiovascular diseases [17]. The system aims to predict whether the patient is at risk of suffering from a non-communicable disease or not. Most frequent disease patterns were identified for prediction.
Furthermore, innovative techniques were proposed to mine associations of infrequent diseases from electronic patient datasets [18]. A novel measure of special casual leverage and a model of the fuzzy recognition-primed decision was developed. These models mined the casual associations between drug and their reactions. A study on the data mining process in health care recommended association rule-based Apriori for generating a frequency of diseases [19]. EMR data belonging to hospitals located in different geographical areas with various time periods were used for the study. The research output identified four different diseases that occurred frequently in a different geographical area in a particular year.
Techniques based on associative classification including CBA and CMAR were applied to predict tuberculosis in a database containing twelve preliminary symptoms and one class attribute [20]. The class label of an unidentified sample was predicted as tuberculosis (TB) associated with HIV based on a higher confidence rule. Most of the resulting classifier rules might assist doctors in making better disease diagnoses.
A framework for the early assessment of rheumatoid arthritis (RA) was proposed by integrating efficient associative classifiers [21]. RA risk patterns from a large clinical database were identified. Frequently occurring risk patterns classifying the disease status were extracted. A weighted approach was incorporated for the integration of correlation and popularity information into the measure to prevent the objectivity of the results.
A technique based on association rule learning was developed to automatically detect ECG beats during myocardial ischemia [22]. Furthermore, a novel technique was proposed to identify common symptoms of patients suffering from depression [23]. FIM and association rules were used on a depression database containing 5954 records. The Apriori predictive approach was used to generate the rules for patients suffering from heart disease [24]. Based on the rules, factors were discovered that caused heart problems in both males and females. The study revealed that females are less prone to coronary heart disease as compared to males.
Recently, a new FIM algorithm, Deferring the Generation of Power sets for Mining Frequent Itemsets in Sparse Big data (D-GENE), was proposed [12]. D-GENE uses the concept of power set from set theory to generate an Iterative Trimmed Transaction Lattice (ITTL) of each transaction. However, before generating the ITTL, D-GENE cuts off the infrequent part of each transaction. Thus, the generated ITTLs consume the least memory. Moreover, D-GENE generates the ITTL of similar trimmed transactions once by introducing a deferred ITTL generation and compression strategy. These mechanisms help D-GENE enormously in getting maximal efficiency both in terms of runtime and consumption of memory, particularly for sparse data. Therefore, the D-GENE algorithm was chosen in this study to find frequent co-occurring occupational diseases among FHBWs.
3. Frequent Itemset Mining Problem
It was assumed that I = {i1, i2, ...in} denotes a set containing all items present in a transactional database; T denotes a transaction containing some items of I, such that T ⊆ I and having a distinctive identifier TID; and D denotes a database, which is a set of transactions = { T1, T2,…, Tn }. S denotes an itemset where S ⊆ I. S is also called a k-itemset, where | S | = k. T contains S if and only if S ⊆ T; the support of S, denoted as sup(S), is the percentage of transactions in D in which S exists. Assume that minsup is the user-specified minimum support threshold. S is regarded as a frequent itemset if and only if minsup ≤ sup(S). For a database D and at a particular minsup value, the task is to explore all frequent itemsets with their supports [8].
4. Mining Frequent Co-Occurring Diseases
This section covers the working principle and the implementation details of the D-GENE algorithm for mining frequent co-occurring diseases [12]. In the first place, the detailed explanation of the dataset, preprocessing, and transformation steps is provided. Afterward, the whole implementation procedure is provided in view of the dataset used.
4.1. Data Collection
The Bureau of Statistics, Punjab, Pakistan conducted “The Punjab Home-based Workers Survey (2016)” from which the data of female participants were extracted for this study [4]. The ages of female participants ranged from 15 to more than 60 years. The survey contained a questionnaire composed of six diseases, where each of them was represented by a checkbox. Participants were asked to check the checkboxes relevant to their diseases. The diseases mentioned in the questionnaire are shown in Table 1.

Table 1.
Diseases mentioned in the survey.
Some example records collected from the participants are given in Table 2. It was found that the collected data were sparse in nature, because each participant was confronted with fewer diseases out of the total 6 diseases mentioned in the survey.

Table 2.
Responded questionnaire.
4.2. Data Preprocessing
To conduct experiments, the data, containing 6791 records, were split into 5 datasets, where each dataset belonged to a distinct age group. Table 3 shows the features of the datasets. Experiments were performed on each dataset to find the frequent co-occurring diseases in all age groups.

Table 3.
Datasets for experiments.
4.3. Data Transformation
The collected data needed to be transformed into a specific format to apply the D-GENE algorithm. Like any other FIM algorithm, D-GENE takes a transactional database as an input to explore frequent itemsets. Therefore, each record in the database was regarded as a transaction containing items, where each item represented a disease. Table 4 shows the diseases included in the questionnaire along with the item symbol attached to them.

Table 4.
Item symbols corresponding to the diseases.
Table 5 shows the transformed version of Table 2. All the datasets were transformed likewise to be used as input to the algorithm.

Table 5.
A transformed version of Table 2.
4.4. Implementation
After converting the data into a suitable form, the D-GENE algorithm was applied to every dataset mentioned in Table 3 to find the frequent co-occurring itemsets (diseases). For a given dataset, at any user-specified minsup value, D-GENE works in three phases. To understand the underlying procedure, this section illustrates how D-GENE works at minsup 23% for the dataset of age group 25–34.
4.4.1. D-GENE Phase 1
In the first phase, the following tasks are performed to get frequent 1-itemsets.
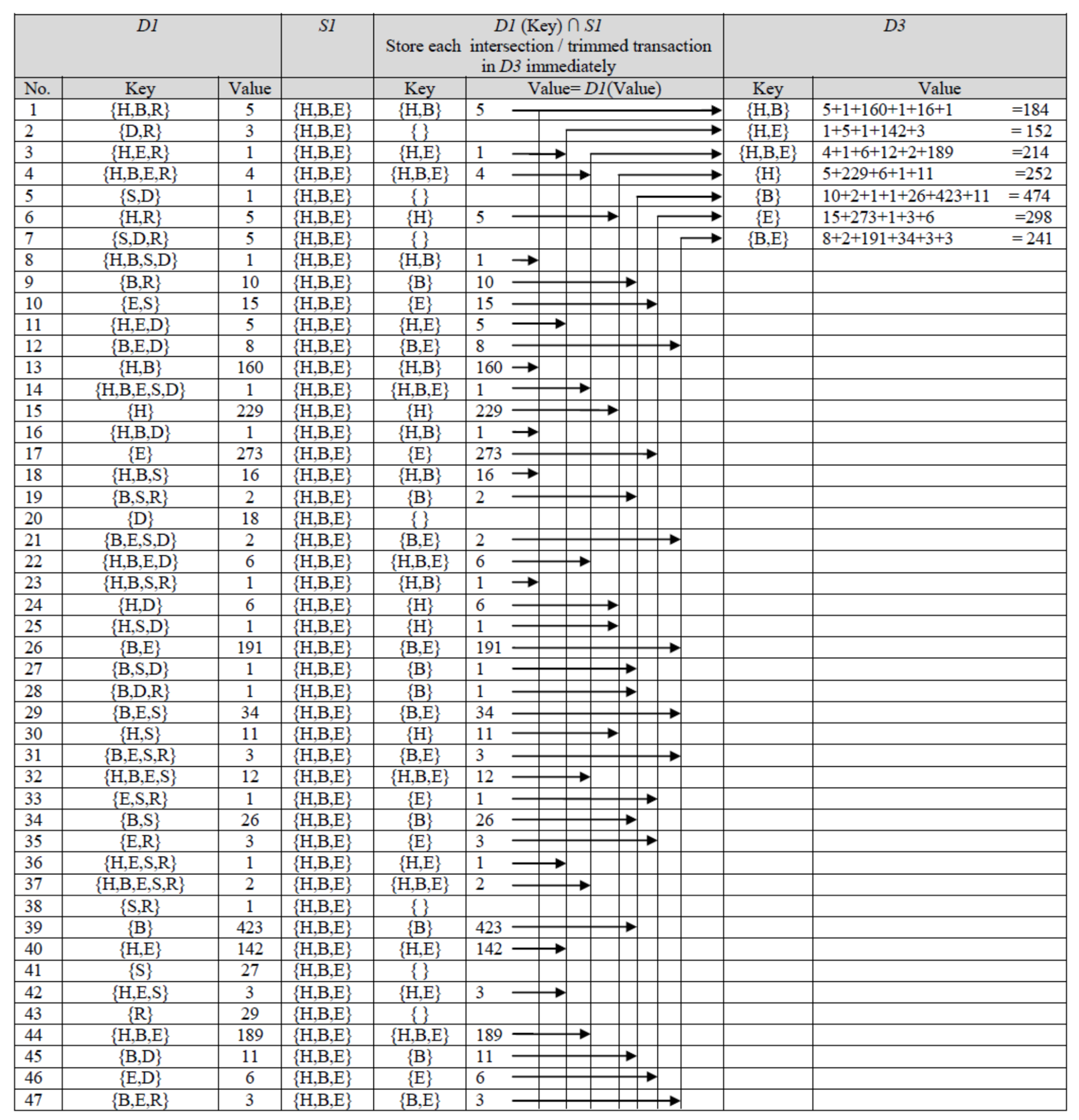
- Dataset is compressed by storing repeated (similar) transactions once. D-GENE uses D1, a dictionary ADT for this purpose. The procedure is shown in Figure 1. If a transaction is read for the first time, it is kept as a key in D1 with value 1. The value of a key shows its support count (frequency). If a transaction is read again, it is not stored in D1; instead, its value is incremented. Thus, D1 contains distinct transactions by storing similar transactions once. It is interesting to see that the number of distinct transactions stored in D1 was 47 only, revealing that numerous similar transactions exist in the dataset. The most repeated transaction in D1 was {B} with value 423. This states that 423 participants checked the checkbox representing the disease of backache. By compressing the dataset, the algorithm performed the forthcoming actions on 47 transactions only, whereas the original dataset contained 1899 transactions. In Figure 1, the first transaction stored as a key in D1 was {H, B, R} with value 5. This means that out of 1899 participants, 5 participants were suffering from three co-occurring diseases denoted by H, B, and R.
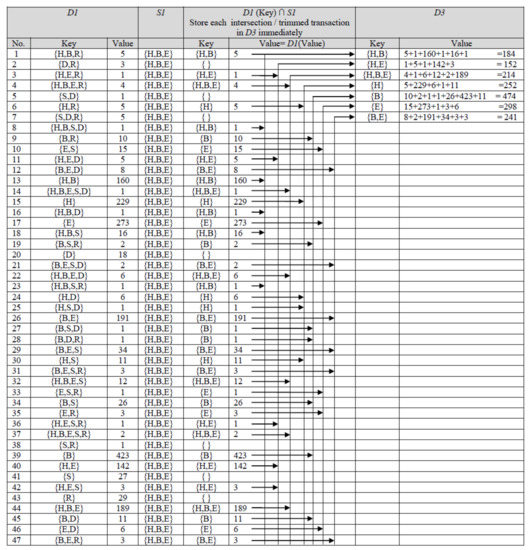 Figure 1. First and second phase of D-GENE for age group 25–34 at minsup 23%.
Figure 1. First and second phase of D-GENE for age group 25–34 at minsup 23%. - Moreover, the support count of every single item in each transaction is calculated. D-Gene uses another dictionary ADT, D2, in which every single item is stored as a key whose value is set to 1 for its first arrival. On subsequent arrivals of the same single item, its value is incremented accordingly. Table 6 shows the state of D2. The dataset contained 1899 transactions; thus, 23% minsup means that the single items whose support count was not less than 436.77 will be believed to be frequent. Support count of items {H}, {B}, and {E} were greater than minsup; therefore, they were selected as frequent 1-itemsets. However, {S}, {D}, and {R} were infrequent. According to the anti-monotone property [6], a superset of an infrequent subset cannot be frequent; thus, co-occurring itemsets containing {S}, {D}, and {R} cannot be frequent. {H}, {B}, and {E}, were stored in S1, a set ADT shown in Figure 1. Discovered frequent 1-itemsets were used in the subsequent phases to discover frequent co-occurring itemsets.
 Table 6. Support of single itemsets in age group 25–34 at minsup 23%.
Table 6. Support of single itemsets in age group 25–34 at minsup 23%.
4.4.2. D-GENE Phase 2
In the second phase, D-GENE reads one key from D1 at a time, takes its intersection with S1, and stores the intersection in another dictionary ADT namely, D3, with the value equal to the value of the key currently read. The procedure is shown in Figure 1. The intersection shows that the infrequent part of each key is neglected; thus, a trimmed transaction is stored in D3. If the same trimmed transaction occurs again, its value in D3 is increased by the value of the key read from D1 at that particular instant of time. The state of D3 in Figure 1 shows each trimmed transaction, stored as a key, with its value showing its cumulative support count.
4.4.3. D-GENE Phase 3
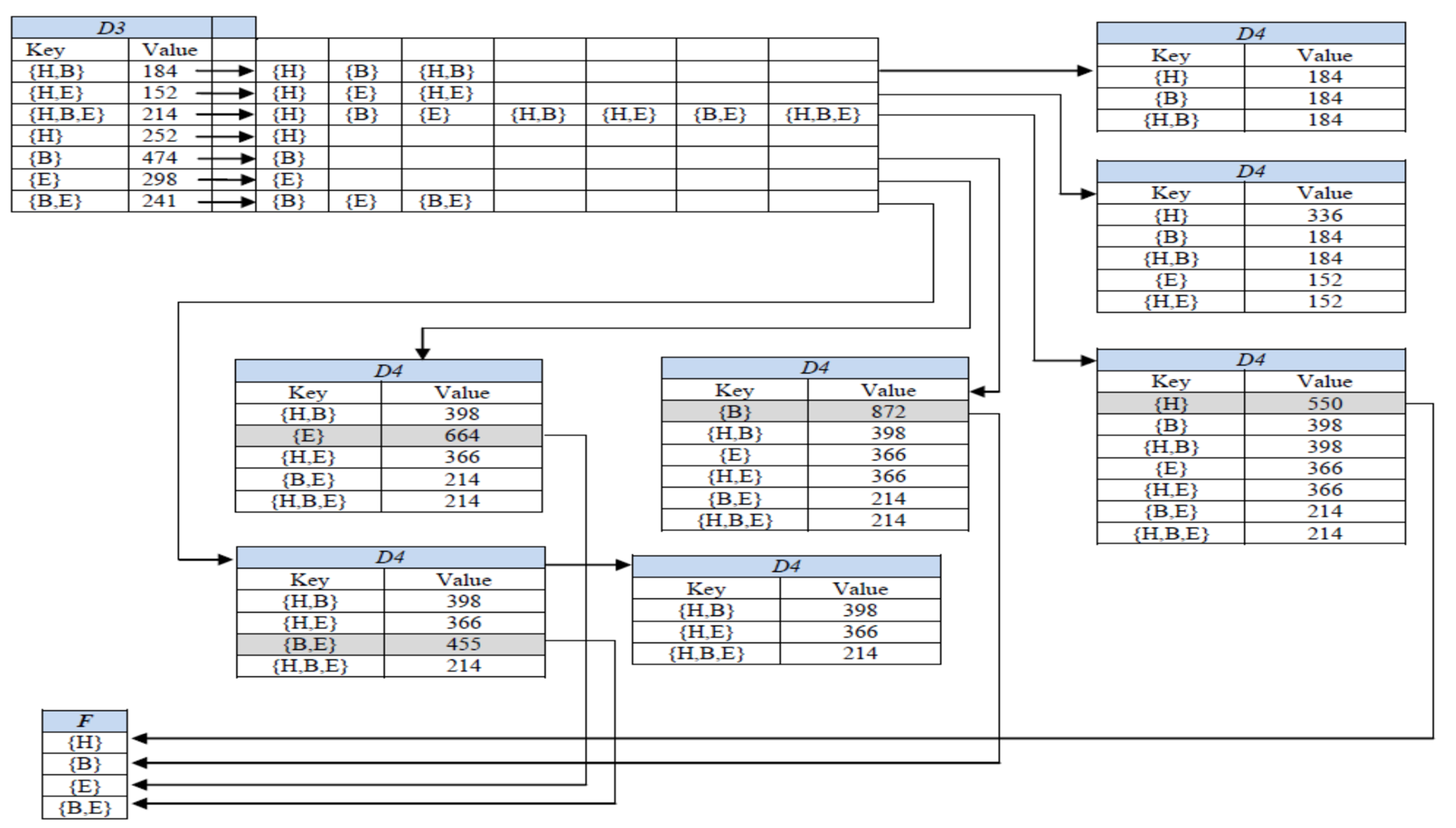
In the last phase, D-GENE reads one trimmed transaction (key) from D3 at a time, makes its ITTL by generating its power set, and stores each subset into a dictionary ADT namely, D4, as a key with the value equal to the value of the currently read key from D3. The process is shown in Figure 2. Afterward, the value of the subset is compared with the minsup. If the value of the subset is greater than or equal to minsup, it is regarded as a frequent itemset, deleted from D4, and stored in F, which is a set ADT to store all frequent itemsets. Once a subset is stored in F, it cannot be stored again in D4.
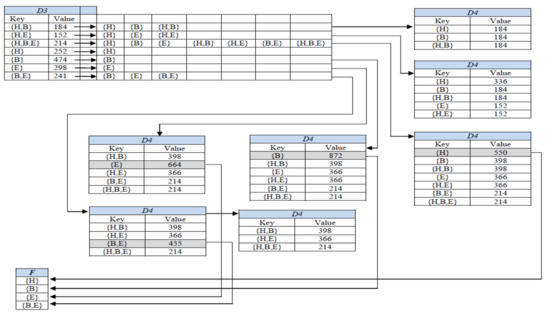
Figure 2.
Generating ITTLs in the last phase of D-GENE for age group 25–34 at minsup 23%.
However, if a subset is not frequent yet, on its occurrence in any subsequent ITTL, its existing value is increased by the value of the key read at that particular instant of time from D3. For instance, in Figure 2, itemset {H} in D4 was a subset of ITTLs of {H,B}, {H,E} {H,B,E}, and {H} having values of 184, 152, 214, and 252, respectively. After storing {H} in D4, as a subset of ITTLs of {H,B}, {H,E}, and {H,B,E}, its cumulative support count became greater than minsup; thus {H} became frequent, popped out from D4 and was stored in F. Later on, as a subset of ITTL of key {H}, the subset {H} will not be entered into D4 again as it has become frequent already.
Similarly, the cumulative support count of {B, E} in D4 is the sum of the values of both of its supersets, {H, B, E} and {B, E} in D3. Figure 2 shows the state of D4 containing all subsets as keys with their cumulative support counts as values. D4 in Figure 2 shows that the supports of {H}, {B}, {E}, and {B,E} were greater than minsup, thus they were regarded as frequent itemsets. However, the supports of {H,B}, {H,E}, and {H,B,E} were less than minsup; thus, they were not frequent. Frequent itemsets after reading each key of D2 are stored in F. The algorithm stops after this step and prints F.
Thus, it was concluded that 23% of participants of the age group 25–34 were suffering from the following frequent diseases.
- Headache.
- Backache.
- Affected eyesight.
- Backache and affected eyesight together.
5. Results
The D-GENE algorithm was run for each dataset to discover frequent itemsets at different minsup values. Table 7 shows the state of dictionary ADT, D1, for each dataset during the first phase of D-GENE execution. The state of D1 depicts that distinct transactions were far less in number compared to the total transactions in each dataset. Instead of generating ITTLs of all transactions, D-GENE generates ITTLs of distinct transactions in the last step; thus, it gets better runtime efficiency and consumes the least memory. An illustration of ITTL generation is shown in Figure 2.

Table 7.
Distinct transactions of each dataset stored in D1.
Table 8, Table 9, Table 10, Table 11 and Table 12 contain the frequent co-occurring diseases for each age group at different minsup thresholds. Table 8 represents frequent co-occurring diseases for the age group 15–24. It shows that itemset {B, E} was frequent when minsup was 21%, which means that 21% of participants belonging to the age group 15–24 were suffering from backache and affected eyesight diseases jointly. At minsup 9%, itemsets, {B,E}, {H,B}, {H,E}, and {H,B,E} were frequent, which means that 9% of participants were suffering from the following co-occurring diseases:

Table 8.
Frequent co-occurring diseases for age group 15–24.

Table 9.
Frequent co-occurring diseases for age group 25–34.

Table 10.
Frequent co-occurring diseases for age group 35–44.

Table 11.
Frequent co-occurring diseases for age group 45–54.

Table 12.
Frequent co-occurring diseases for age group >54.
- Backache and affected eyesight.
- Headache and backache.
- Headache and affected eyesight.
- Headache, backache, and affected eyesight.
The rest of the records in Table 8 can be inferred accordingly.
Table 9 represents frequent co-occurring diseases for the age group 25–34. It shows that itemset {B, E} was frequent when minsup was 23%, which means that 23% of participants belonging to the age group 25–34 were suffering from both backache and affected eyesight diseases. At minsup 11%, itemsets {B,E}, {H,B}, {H,E}, and {H,B,E} were frequent, which means that 11% of participants were facing the following co-occurring diseases:
- Backache and affected eyesight.
- Headache and backache.
- Headache and affected eyesight.
- Headache, backache, and affected eyesight.
The rest of the records in Table 9 can be analyzed in the same fashion.
Table 10 represents frequent co-occurring diseases for the age group 35–44. It shows that itemset {B, E} was frequent when minsup was 26%, which means that 26% of participants belonging to the age group 35–44 were confronting backache and affected eyesight diseases jointly. At minsup 12%, itemsets {B,E}, {H,B}, {H,E}, and {H,B,E} were frequent, which means that 12% of participants were suffering from the following co-occurring diseases:
- Backache and affected eyesight.
- Headache and backache.
- Headache and affected eyesight.
- Headache, backache, and affected eyesight.
Similarly, interpretation of the rest of the records in Table 10 can be done.
Table 11 represents frequent co-occurring diseases for the age group 45–54. It shows that itemset {B,E} was frequent when minsup was 31%, which means that 31% of participants belonging to age group 45–54 were facing backache and affected eyesight diseases jointly. At minsup 23%, itemset {B,E} and {H,E} were frequent, which means that 23% of participants of age group 45–54 were facing backache and affected eyesight jointly and headache and affected eyesight jointly. Itemsets {B,E}, {H,B}, {H,E}, and {H,B,E} were frequent at minsup 14%, which means that 14% of the participants were facing the following co-occurring diseases:
- Backache and affected eyesight.
- Headache and backache.
- Headache and affected eyesight.
- Headache, backache, and affected eyesight.
The rest of the records in Table 11 can be inferred accordingly.
Table 12 represents frequent co-occurring diseases for the age group (>54). It shows that itemset {B,E} was frequent when minsup was 33%, which means that 33% of participants belonging to age group (>54) were suffering from backache and affected eyesight diseases jointly. At minsup 27%, itemset {B, E} and {H, E} were frequent, which means that 27% of participants of the age group >54 were facing backache and affected eyesight and headache and affected eyesight jointly. Itemsets {B,E}, {H,B}, {H,E}, and {H,B,E} were frequent at minsup 16%, which means that 16% of the participants were facing the following co-occurring diseases:
- Backache and affected eyesight.
- Headache and backache.
- Headache and affected eyesight.
- Headache, backache, and affected eyesight.
Other records in Table 12 can be interpreted likewise.
6. Discussion
Table 13 summarizes Table 8, Table 9, Table 10, Table 11 and Table 12, depicting the frequent co-occurring diseases in each age group.

Table 13.
A summary of frequent co-occurring diseases in all age groups.
Therefore, it can be observed that:
- The most devastating co-occurring diseases faced by FHBWs are backache and affected eyesight.
- Headache and backache are the second most common co-occurring diseases.
- Headache and affected eyesight together are the third most frequent.
- Headache, backache, and affected eyesight together are the fourth most frequent.
Furthermore, the intensity increased with the increase in the age of workers. It can be inferred that the working conditions of numerous FHBWs are not per their job specifications. Ironically, they are unaware of this fact, thus, compromising their health. It is imperative to educate FHBWs, the complete requirements of their work, and relevant health and safety risks. This is essential because FHBWs are unregistered; thus, the state does not treat them as registered workers with benefits like health care and others.
The resulting disease patterns suggest that FHBWs should take essential precautionary measures while at work. FHBWs could never know the pertinent occupational diseases ahead of time due to the absence of any scientific study highlighting such risk factors. Thus, this study is novel and the foremost attempt to identify the leading co-occurring occupational diseases. By applying frequent itemset mining, a fundamental data science technique, this study has revealed insights that would not be possible with conventional statistical techniques. It is envisioned that the discovered disease patterns, if adequately communicated by relevant NGOs and health practitioners, would make FHBWs aware of the diseases that they are susceptible to in future. Eventually, they would take essential precautionary measures, such as the installation of proper lighting in the working area and the adoption appropriate working postures along with others.
To spread awareness, relevant organizations [25,26] and competent health authorities might adopt the following strategies:
- Effective use of all types of media.
- Organizing work-specific training programs.
7. Conclusions
The majority of female workers in Pakistan are categorized as Female Home-Based Workers (FHBWs). They usually work in unhealthy conditions and are unaware of pertinent health and safety risks. Therefore, FHBWs often suffer from numerous occupational diseases. The biggest downside is the informal nature of their job due to which FHBWs cannot get adequate health care, safety, and other fringe benefits from the government. Additionally, their work-related health issues are usually overlooked by the relevant health authorities. Therefore, there is a dire need to identify the most common diseases faced by FHBWs.
In this study, D-GENE, a recent FIM algorithm, was applied to data obtained from the Punjab Home-based Workers Survey (2016) to find co-occurring frequent diseases faced by FHBWs. Discovered frequent disease patterns (affected eyesight and backache) clearly signify that inadequate lighting and incorrect working postures are the dominant factors affecting FHBWs’ health. It is envisioned that the results of the study will be useful for both the relevant health authorities and FHBWs. Health authorities will be able to make more favorable health policies after getting a clear insight. Eventually, FHBWs would take essential precautionary measures, such as the installation of proper lighting in the working area and the adoption of appropriate working postures along with others. A healthy and well aware FHBW will become more productive and a useful contributor to the sustainable economic growth of the country.
Author Contributions
Conceptualization, M.Y., A.A., and M.U.C.; methodology, M.Y., A.A., and M.J.; software, M.Y., M.U.C., and F.H.; formal analysis, M.Y., J.-H.L., and E.J.; writing—original draft preparation, M.Y., A.A., and M.U.C.; writing—review and editing, F.H., M.J., and Z.L.; supervision, J.-H.L. and Z.L.; project administration, M.U.C., funding acquisition, Z.L. All authors have read and agreed to the published version of the manuscript.
Funding
Publication of this article was financially supported by the Chair of Electrical Engineering, Wroclaw University of Science and Technology.
Data Availability Statement
Data that were used in this article were obtained from the Punjab Home-based Workers Survey (2016).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ashraf, A.; Herzer, D.; Nunnenkamp, P. Greenfield FDI, cross-border M&As, and government size. J. Int. Trade Econ. Dev. 2017, 26, 566–584. [Google Scholar] [CrossRef]
- Women’ Economic Participation and Empowerment in Pakistan Status Report. 2016. Available online: https://asiapacific.unwomen.org/en/digital-library/publications/2016/05/status-report-on-womens-economic-participation-and-empowerment (accessed on 1 January 2021).
- Labor Force Survey 2017-18, Pakistan Bureau of Statistics. 2018. Available online: http://www.pbs.gov.pk/content/labour-force-survey-2017-18-annual-report (accessed on 1 January 2021).
- The Punjab Home Based Workers Survey 2016, Bureau of Statistics Punjab. 2016. Available online: http://bos.gop.pk/system/files/PHBWSFinalReport_1.pdf (accessed on 10 December 2020).
- Protecting Workers’ Health, World Health Organization. 2017. Available online: https://www.who.int/news-room/fact-sheets/detail/protecting-workers’-health (accessed on 1 January 2021).
- Saheb, T.; Izadi, L. Paradigm of IoT big data analytics in the healthcare industry: A review of scientific literature and mapping of research trends. Telemat. Inform. 2019, 41, 70–85. [Google Scholar] [CrossRef]
- Nilashi, M.; Ibrahim, O.; Ahmadi, H.; Shahmoradi, L. A knowledge-based system for breast cancer classification using fuzzy logic method. Telemat. Inform. 2017, 34, 133–144. [Google Scholar] [CrossRef]
- Agrawal, R.; Imieliński, T.; Swami, A. Mining association rules between sets of items in large databases. ACM SIGMOD Rec. 1993, 22, 207–216. [Google Scholar] [CrossRef]
- Cheng, H.; Yan, X.; Han, J.; Yu, P.S. Direct Discriminative Pattern Mining for Effective Classification. In Proceedings of the 2008 IEEE 24th International Conference on Data Engineering, Cancun, Mexico, 7–12 April 2008; pp. 169–178. [Google Scholar]
- Wang, H.; Wang, W.; Yang, J.; Yu, P.S. Clustering by pattern similarity in large data sets. In Proceedings of the 2002 ACM SIGMOD International Conference on Management of Data—SIGMOD, Madison, WI, USA, 3–6 June 2002; pp. 394–405. [Google Scholar]
- Ceglar, A.; Roddick, J.F. Association mining. ACM Comput. Surv. 2006, 38. [Google Scholar] [CrossRef]
- Yasir, M.; Habib, M.A.; Ashraf, M.; Sarwar, S.; Chaudhry, M.U.; Shahwani, H.; Ahmad, M.; Faisal, C.M.N. D-GENE: Deferring the GENEration of Power Sets for Discovering Frequent Itemsets in Sparse Big Data. IEEE Access 2020, 8, 27375–27392. [Google Scholar] [CrossRef]
- Ilayaraja, M.; Meyyappan, T. Mining medical data to identify frequent diseases using Apriori algorithm. In Proceedings of the 2013 International Conference on Pattern Recognition, Informatics and Mobile Engineering, Salem, India, 21–22 February 2013; pp. 194–199. [Google Scholar]
- Ilayaraja, M.; Meyyappan, T. Efficient Data Mining Method to Predict the Risk of Heart Diseases through Frequent Itemsets. Procedia Comput. Sci. 2015, 70, 586–592. [Google Scholar] [CrossRef]
- Khaleel, M.A.; Dash, G.N.; Choudhury, K.S.; Khan, M.A. Medical Data Mining for Discovering Periodically Frequent Diseases from Transactional Databases. In Sustainable Design and Manufacturing 2016; Springer: Berlin/Heidelberg, Germany, 2015; pp. 87–96. [Google Scholar]
- Vikas, B.; Anuhya, B.S.; Bhargav, K.S.; Sarangi, S.; Chilla, M. Application of the Apriori Algorithm for Prediction of Polycystic Ovarian Syndrome (PCOS). In Advances in Human Factors, Business Management, Training and Education; Springer: Cham, Switzerland, 2018; pp. 934–944. [Google Scholar]
- Aldallal, A.; Al-Moosa, A.A.A. Using Data Mining Techniques to Predict Diabetes and Heart Diseases. In Proceedings of the 2018 4th International Conference on Frontiers of Signal Processing (ICFSP), Poitiers, France, 24–27 September 2018; pp. 150–154. [Google Scholar]
- Ji, Y.; Ying, H.; Tran, J.; Dews, P.; Mansour, A.; Massanari, R.M. Mining Infrequent Causal Associations in Electronic Health Databases. In Proceedings of the 2011 IEEE 11th International Conference on Data Mining Workshops, Vancouver, BC, Canada, 11 December 2011; pp. 421–428. [Google Scholar]
- Undrajavarapu, S. A Review on Data Mining Process in Healthcare Department to Identify the Frequently Occurring Diseases. IJRCCT 2015, 4, 315–318. [Google Scholar]
- Asha, T.; Natarajan, S.; Murthy, K.N.B. Associative classification in the prediction of tuberculosis. In Proceedings of the International Conference & Workshop on Emerging Trends in Technology—ICWET; ACM: New York, NY, USA, 2011. [Google Scholar]
- Chin, C.Y.; Weng, M.Y.; Lin, T.C.; Cheng, S.Y.; Yang, Y.H.K.; Tseng, V.S. Mining Disease Risk Patterns from Nationwide Clinical Databases for the Assessment of Early Rheumatoid Arthritis Risk. PLoS ONE 2015, 10, e0122508. [Google Scholar] [CrossRef] [PubMed]
- Exarchos, T.; Papaloukas, C.; Fotiadis, D.; Michalis, L. An association rule mining-based methodology for automated detection of ischemic ECG beats. IEEE Trans. Biomed. Eng. 2006, 53, 1531–1540. [Google Scholar] [CrossRef] [PubMed]
- Ghafoor, Y.; Huang, Y.-P.; Liu, S.-I. An intelligent approach to discovering common symptoms among depressed patients. Soft Comput. 2014, 19, 819–827. [Google Scholar] [CrossRef]
- Nahar, J.; Imam, T.; Tickle, K.S.; Chen, Y.-P.P. Association rule mining to detect factors which contribute to heart disease in males and females. Expert Syst. Appl. 2013, 40, 1086–1093. [Google Scholar] [CrossRef]
- Women in Informal Employment: Globalizing and Organizing (WIEGO). Available online: https://www.wiego.org/ (accessed on 20 March 2021).
- Home Based Workers. Available online: https://hnsa.org.in/home-based-workers (accessed on 20 March 2021).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).