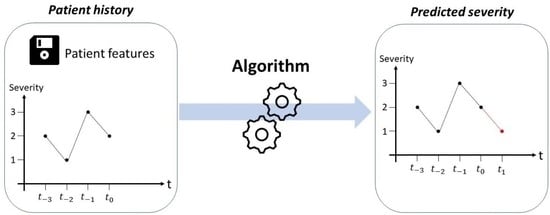
Deep Learning for Predicting Congestive Heart Failure
Abstract
:1. Introduction
- High prevalence (1–3% in adult population).
- An incidence of 1–20 cases per 1000 population.
- High mortality: from 30 days mortality of 2–3% to five years mortality of 50–75%.
- Increased treatment costs. Mainly due to the increase of people over 65 years of age.
2. Related Work
3. Materials and Methods
3.1. Dataset
3.2. Prediction System Development
3.2.1. Data Pre-Processing
- BNP interpolation
- Cleaning
- Scaling
- Structuring data
- Class Mild: BNP < 500
- Class Moderate: 500 < BNP < 1000
- Class Severe: BNP > 1000
3.2.2. Models Design
- Temporal data: Weight, ejection fraction, BNP, NYHA class, age, systolic arterial pressure, diastolic arterial pressure, cardiac frequency, ACEblockers dose, Betablockers dose, diuretics dose.
- Boolean data: Ischemic heart disease, hypertension, dyslipidemia, sinus rhythm, atrial fibrillation, diabetes, BPCO, nitrates.
- Ordinal data: Sarta dose level, Betablockers dose level, compliance.
4. Results
- True Positives (TP): Positive instances correctly classified
- False Positives (FP): Negative instances classified as positive
- True Negatives (TN): Negative instances correctly classified
- False Negatives (FN): Positive instances classified as negative
- Accuracy: Represents the number of correct predictions with respect to the total number of samples.
- Recall or Sensitivity: Ratio of positive instances correctly classified to total (actual) positives in the dataset .
- Precision: Accuracy of the positive predictions .
- False positive rate (FPR): Ratio of false positives to the total number of actual negative events .
- Area Under Receiver Operating Characteristic (AUROC) curve: Area under the Receiver Operator Characteristic (ROC) curve, which plots the true positive rate (recall) against the FPR. To plot the entire curve, these two metrics are evaluated many times after the variation of a classification threshold. The best value for the AUROC is 1.
- Area Under Precision Recall Curve (AUPRC): This metric is particularly used for binary responses. It is appropriate for rare events and is not dependent on model specificity. In this case, the axes are defined as precision and recall, respectively.
- Confusion matrix: This matrix is the most complete way of representing results. It is shown as a table containing true values in the rows and predicted values in the columns. A perfect confusion matrix is diagonal; values in the diagonal are predicted correctly and the others are not.
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| ANN | Artificial Neural network |
| AUPRC | Area Under Precision Recall Curve |
| AUROC or AUC | Area Under ROC |
| BNP | Brain Natriuretic Peptide |
| CDSS | Clinical Decision Support System |
| CHF | Congestive Heart Failure |
| CNN | Convolutional Neural network |
| COPD | Chronic Obstructive Pulmonary Disease |
| DL | Deep Learning |
| ECG | Electrocardiogram |
| EDV | End Diastolic Volume |
| EF | Ejection Fraction |
| EHR | Electronic Health Records |
| ESC | European Society of Cardiology |
| ESV | End Systolic Volume |
| FP | False Positive |
| FN | False Negative |
| HRV | Heart Rate Variability |
| HF | Heart Failure |
| ICD | Implantable Cardioverter Defibrillator |
| ICDCRT | Implantable Cardioverter Defibrillator Cardiac Resynchronization Therapy |
| KNN | K-Nearest Neighbors |
| LMT | Logistic Model Tree |
| ML | Machine Learning |
| NB | Naive Bayes |
| NN | Neural Network |
| NYHA | New York Heart Association |
| PPM | Pulse Per Minute |
| RF | Random Forest |
| RNN | Recurrent Neural network |
| ROC | Receiver Operating Characteristic |
| ROT | Rotation Tree |
| SVM | Support Vector machine |
| TP | True Positive |
| TN | True Negative |
References
- Malik, A.; Brito, D.; Vaqar, S.; Chhabra, L. Congestive heart failure. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Groenewegen, A.; Rutten, F.H.; Mosterd, A.; Hoes, A.W. Epidemiology of heart failure. Eur. J. Heart Fail. 2020, 22, 1342–1356. Available online: https://onlinelibrary.wiley.com/doi/pdf/10.1002/ejhf.1858 (accessed on 1 November 2022). [CrossRef] [PubMed]
- Jones, N.R.; Roalfe, A.K.; Adoki, I.; Hobbs, F.R.; Taylor, C.J. Survival of patients with chronic heart failure in the community: A systematic review and meta-analysis. Eur. J. Heart Fail. 2019, 21, 1306–1325. [Google Scholar] [CrossRef] [Green Version]
- McDonagh, T.A.; Metra, M.; Adamo, M.; Gardner, R.S.; Baumbach, A.; Böhm, M.; Burri, H.; Butler, J.; Čelutkienė, J.; Chioncel, O.; et al. 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: Developed by the Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC) With the special contribution of the Heart Failure Association (HFA) of the ESC. Eur. Heart J. 2021, 42, 3599–3726. [Google Scholar] [PubMed]
- Bragazzi, N.L.; Zhong, W.; Shu, J.; Abu Much, A.; Lotan, D.; Grupper, A.; Younis, A.; Dai, H. Burden of heart failure and underlying causes in 195 countries and territories from 1990 to 2017. Eur. J. Prev. Cardiol. 2021, 28, 1682–1690. Available online: https://academic.oup.com/eurjpc/advance-article-pdf/doi/10.1093/eurjpc/zwaa147/36239071/zwaa147.pdf (accessed on 1 November 2022). [CrossRef]
- Cook, C.; Cole, G.; Asaria, P.; Jabbour, R.; Francis, D.P. The annual global economic burden of heart failure. Int. J. Cardiol. 2014, 171, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Urbich, M.; Globe, G.; Pantiri, K.; Heisen, M.; Bennison, C.; Wirtz, H.S.; Di Tanna, G.L. A systematic review of medical costs associated with heart failure in the USA (2014–2020). Pharmacoeconomics 2020, 38, 1219–1236. [Google Scholar] [CrossRef] [PubMed]
- Lesyuk, W.; Kriza, C.; Kolominsky-Rabas, P. Cost-of-illness studies in heart failure: A systematic review 2004–2016. BMC Cardiovasc. Disord. 2018, 18, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Shafie, A.A.; Tan, Y.P.; Ng, C.H. Systematic review of economic burden of heart failure. Heart Fail. Rev. 2018, 23, 131–145. [Google Scholar] [CrossRef]
- Ambrosy, A.P.; Fonarow, G.C.; Butler, J.; Chioncel, O.; Greene, S.J.; Vaduganathan, M.; Nodari, S.; Lam, C.S.; Sato, N.; Shah, A.N.; et al. The Global Health and Economic Burden of hospitalizations for Heart Failure. J. Am. Coll. Cardiol. 2014, 63, 1123–1133. Available online: https://www.jacc.org/doi/pdf/10.1016/j.jacc.2013.11.053 (accessed on 1 November 2022). [CrossRef]
- Savarese, G.; Becher, P.M.; Lund, L.H.; Seferovic, P.; Rosano, G.M.C.; Coats, A.J.S. Global burden of heart failure: A comprehensive and updated review of epidemiology. Cardiovasc. Res. 2022, cvac013. Available online: https://academic.oup.com/cardiovascres/advance-article-pdf/doi/10.1093/cvr/cvac013/43972759/cvac013.pdf (accessed on 1 November 2022). [CrossRef]
- El Naqa, I.; Murphy, M.J. What is machine learning? In Machine Learning in Radiation Oncology; Springer: Cham, Switzerland, 2015; pp. 3–11. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Mcculloch, W.S.; Pitts, W. A logical calculus nervous activity. Bull. Math. Biol. 1990, 52, 99–115. [Google Scholar] [CrossRef] [PubMed]
- You, S.; Lei, B.; Wang, S.; Chui, C.K.; Cheung, A.C.; Liu, Y.; Gan, M.; Wu, G.; Shen, Y. Fine perceptive gans for brain mr image super-resolution in wavelet domain. IEEE Trans. Neural Netw. Learn. Syst. 2022; Online ahead of print. [Google Scholar]
- Yu, W.; Lei, B.; Ng, M.K.; Cheung, A.C.; Shen, Y.; Wang, S. Tensorizing GAN with high-order pooling for Alzheimer’s disease assessment. IEEE Trans. Neural Netw. Learn. Syst. 2021, 33, 4945–4959. [Google Scholar] [CrossRef]
- Wang, S.; Wang, X.; Shen, Y.; He, B.; Zhao, X.; Cheung, P.W.H.; Cheung, J.P.Y.; Luk, K.D.K.; Hu, Y. An ensemble-based densely-connected deep learning system for assessment of skeletal maturity. IEEE Trans. Syst. Man Cybern. Syst. 2020, 52, 426–437. [Google Scholar] [CrossRef]
- Mpanya, D.; Celik, T.; Klug, E.; Ntsinjana, H. Predicting mortality and hospitalization in heart failure using machine learning: A systematic literature review. IJC Heart Vasc. 2021, 34, 100773. [Google Scholar] [CrossRef] [PubMed]
- Bazoukis, G.; Stavrakis, S.; Zhou, J.; Bollepalli, S.C.; Tse, G.; Zhang, Q.; Singh, J.P.; Armoundas, A.A. Machine learning versus conventional clinical methods in guiding management of heart failure patients—A systematic review. Heart Fail. Rev. 2021, 26, 23–34. [Google Scholar] [CrossRef]
- Tripoliti, E.E.; Papadopoulos, T.G.; Karanasiou, G.S.; Naka, K.K.; Fotiadis, D.I. Heart failure: Diagnosis, severity estimation and prediction of adverse events through machine learning techniques. Comput. Struct. Biotechnol. J. 2017, 15, 26–47. [Google Scholar] [CrossRef] [Green Version]
- Krittanawong, C.; Virk, H.U.H.; Bangalore, S.; Wang, Z.; Johnson, K.W.; Pinotti, R.; Zhang, H.J.; Kaplin, S.; Narasimhan, B.; Kitai, T.; et al. Machine learning prediction in cardiovascular diseases: A meta-analysis. Sci. Rep. 2020, 10, 16057. [Google Scholar] [CrossRef]
- Olsen, C.R.; Mentz, R.J.; Anstrom, K.J.; Page, D.; Patel, P.A. Clinical applications of machine learning in the diagnosis, classification, and prediction of heart failure: Machine learning in heart failure. Am. Heart J. 2020, 229, 1–17. [Google Scholar] [CrossRef]
- Samuel, O.W.; Asogbon, G.M.; Sangaiah, A.K.; Fang, P.; Li, G. An integrated decision support system based on ANN and Fuzzy_AHP for heart failure risk prediction. Expert Syst. Appl. 2017, 68, 163–172. [Google Scholar] [CrossRef]
- Guidi, G.; Iadanza, E.; Pettenati, M.; Milli, M.; Pavone, F.; Biffi Gentili, G. Heart failure artificial intelligence-based computer aided diagnosis telecare system. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2012; Volume 7251 LNCS, pp. 278–281. [Google Scholar] [CrossRef]
- Guidi, G.; Pettenati, M.; Miniati, R.; Iadanza, E. Random forest for automatic assessment of heart failure severity in a telemonitoring scenario. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, Osaka, Japan, 3–7 July 2013; IEEE: New York, NY, USA, 2013; pp. 3230–3233. [Google Scholar] [CrossRef]
- Guidi, G.; Pettenati, M.C.; Melillo, P.; Iadanza, E. A machine learning system to improve heart failure patient assistance. IEEE J. Biomed. Health Inform. 2014, 18, 1750–1756. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Guo, X.; Abbas, H.T.; Qaraqe, K.A.; Abbasi, Q.H. Using machine learning to predict the future development of disease. In Proceedings of the 2020 International Conference on UK-China Emerging Technologies (UCET), Glasgow, UK, 20–21 August 2020; pp. 1–4. [Google Scholar]
- Schvetz, M.; Fuchs, L.; Novack, V.; Moskovitch, R. Outcomes prediction in longitudinal data: Study designs evaluation, use case in ICU acquired sepsis. J. Biomed. Inform. 2021, 117, 103734. [Google Scholar] [CrossRef] [PubMed]
- Plati, D.K.; Tripoliti, E.E.; Bechlioulis, A.; Rammos, A.; Dimou, I.; Lakkas, L.; Watson, C.; McDonald, K.; Ledwidge, M.; Pharithi, R.; et al. A machine learning approach for chronic heart failure diagnosis. Diagnostics 2021, 11, 1863. [Google Scholar] [CrossRef] [PubMed]
- Hussain, L.; Awan, I.A.; Aziz, W.; Saeed, S.; Ali, A.; Zeeshan, F.; Kwak, K.S. Detecting Congestive Heart Failure by Extracting Multimodal Features and Employing Machine Learning Techniques. BioMed Res. Int. 2020, 2020, 4281243. [Google Scholar] [CrossRef]
- Melillo, P.; Pacifici, E.; Orrico, A.; Iadanza, E.; Pecchia, L. Heart rate variability for automatic assessment of congestive heart failure severity. In Proceedings of the XIII Mediterranean Conference on Medical and Biological Engineering and Computing 2013, Seville, Spain, 25–28 September 2013; Volume 41, pp. 1342–1345. [Google Scholar] [CrossRef]
- Nirschl, J.J.; Janowczyk, A.; Peyster, E.G.; Frank, R.; Margulies, K.B.; Feldman, M.D.; Madabhushi, A. A deep-learning classifier identifies patients with clinical heart failure using whole-slide images of H&E tissue. PLoS ONE 2018, 13, e0192726. [Google Scholar]
- Rao, S.; Li, Y.; Ramakrishnan, R.; Hassaine, A.; Canoy, D.; Cleland, J.; Lukasiewicz, T.; Salimi-Khorshidi, G.; Rahimi, K. An explainable Transformer-based deep learning model for the prediction of incident heart failure. IEEE J. Biomed. Health Inform. 2022, 26, 3362–3372. [Google Scholar] [CrossRef]
- Gjoreski, M.; Gradišek, A.; Budna, B.; Gams, M.; Poglajen, G. Machine learning and end-to-end deep learning for the detection of chronic heart failure from heart sounds. IEEE Access 2020, 8, 20313–20324. [Google Scholar] [CrossRef]
- Pana, M.A.; Busnatu, S.S.; Serbanoiu, L.I.; Vasilescu, E.; Popescu, N.; Andrei, C.; Sinescu, C.J. Reducing the Heart Failure Burden in Romania by Predicting Congestive Heart Failure Using Artificial Intelligence: Proof of Concept. Appl. Sci. 2021, 11, 11728. [Google Scholar] [CrossRef]
- D’Addio, G.; Donisi, L.; Cesarelli, G.; Amitrano, F.; Coccia, A.; La Rovere, M.T.; Ricciardi, C. Extracting Features from Poincaré Plots to Distinguish Congestive Heart Failure Patients According to NYHA Classes. Bioengineering 2021, 8, 138. [Google Scholar] [CrossRef]
- Kwon, J.m.; Kim, K.H.; Jeon, K.H.; Kim, H.M.; Kim, M.J.; Lim, S.M.; Song, P.S.; Park, J.; Choi, R.K.; Oh, B.H. Development and validation of deep-learning algorithm for electrocardiography-based heart failure identification. Korean Circ. J. 2019, 49, 629–639. [Google Scholar] [CrossRef]
- Porumb, M.; Iadanza, E.; Massaro, S.; Pecchia, L. A convolutional neural network approach to detect congestive heart failure. Biomed. Signal Process. Control. 2020, 55, 101597. [Google Scholar] [CrossRef]
- Li, D.; Tao, Y.; Zhao, J.; Wu, H. Classification of congestive heart failure from ECG segments with a multi-scale residual network. Symmetry 2020, 12, 2019. [Google Scholar] [CrossRef]
- Chicco, D.; Jurman, G. Machine learning can predict survival of patients with heart failure from serum creatinine and ejection fraction alone. BMC Med. Inform. Decis. Mak. 2020, 20, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- UCI. Heart Disease Data Set. Available online: https://www.kaggle.com/datasets/redwankarimsony/heart-disease-data (accessed on 1 November 2022).
- Ishaq, A.; Sadiq, S.; Umer, M.; Ullah, S.; Mirjalili, S.; Rupapara, V.; Nappi, M. Improving the prediction of heart failure patients’ survival using SMOTE and effective data mining techniques. IEEE Access 2021, 9, 39707–39716. [Google Scholar] [CrossRef]
- Ghosh, P.; Azam, S.; Jonkman, M.; Karim, A.; Shamrat, F.J.M.; Ignatious, E.; Shultana, S.; Beeravolu, A.R.; De Boer, F. Efficient prediction of cardiovascular disease using machine learning algorithms with relief and LASSO feature selection techniques. IEEE Access 2021, 9, 19304–19326. [Google Scholar] [CrossRef]
- Guo, A.; Pasque, M.; Loh, F.; Mann, D.L.; Payne, P.R. Heart failure diagnosis, readmission, and mortality prediction using machine learning and artificial intelligence models. Curr. Epidemiol. Rep. 2020, 7, 212–219. [Google Scholar] [CrossRef]
- Alotaibi, F.S. Implementation of machine learning model to predict heart failure disease. Int. J. Adv. Comput. Sci. Appl. 2019, 10, 261–268. [Google Scholar] [CrossRef] [Green Version]
- Aljanabi, M.; Qutqut, M.H.; Hijjawi, M. Machine learning classification techniques for heart disease prediction: A review. Int. J. Eng. Technol. 2018, 7, 5373–5379. [Google Scholar]
- Kannan, R.; Vasanthi, V. Machine learning algorithms with ROC curve for predicting and diagnosing the heart disease. In Soft Computing and Medical Bioinformatics; Springer: Singapore, 2019; pp. 63–72. [Google Scholar]
- Sudoh, T.; Kangawa, K.; Minamino, N.; Matsuo, H. A new natriuretic peptide in porcine brain. Nature 1988, 332, 78–81. [Google Scholar] [CrossRef]
- Lee, Y.K.; Choi, D.O.; Kim, G.Y. Development of a Rapid Diagnostic Kit for Congestive Heart Failure Using Recombinant NT-proBNP Antigen. Medicina 2021, 57, 751. [Google Scholar] [CrossRef]
- Kasahara, S.; Sakata, Y.; Nochioka, K.; Miura, M.; Abe, R.; Sato, M.; Aoyanagi, H.; Fujihashi, T.; Yamanaka, S.; Shiroto, T.; et al. Conversion formula from B-type natriuretic peptide to N-terminal proBNP values in patients with cardiovascular diseases. Int. J. Cardiol. 2019, 280, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Nabeshima, Y.; Sakanishi, Y.; Otani, K.; Higa, Y.; Honda, M.; Otsuji, Y.; Takeuchi, M. Estimation of B-type natriuretic peptide values from n-terminal proBNP levels. J. UOEH 2020, 42, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cameron, S.J.; Green, G.B.; White, C.N.; Laterza, O.F.; Clarke, W.; Kim, H.; Sokoll, L.J. Assessment of BNP and NT-proBNP in emergency department patients presenting with suspected acute coronary syndromes. Clin. Biochem. 2006, 39, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Feng, Q.P.; Wu, P.; Lupu, R.A.; Wilke, R.A.; Wells, Q.S.; Denny, J.C.; Wei, W.Q. Learning from Longitudinal Data in Electronic Health Record and Genetic Data to Improve Cardiovascular Event Prediction. Sci. Rep. 2019, 9, 717. [Google Scholar] [CrossRef] [Green Version]
- Rongali, S.; Rose, A.J.; McManus, D.D.; Bajracharya, A.S.; Kapoor, A.; Granillo, E.; Yu, H. Learning latent space representations to predict patient outcomes: Model development and validation. J. Med Internet Res. 2020, 22, e16374. [Google Scholar] [CrossRef]
- Golas, S.B.; Shibahara, T.; Agboola, S.; Otaki, H.; Sato, J.; Nakae, T.; Hisamitsu, T.; Kojima, G.; Felsted, J.; Kakarmath, S.; et al. A machine learning model to predict the risk of 30-day readmissions in patients with heart failure: A retrospective analysis of electronic medical records data. BMC Med. Inform. Decis. Mak. 2018, 18, 44. [Google Scholar] [CrossRef] [Green Version]
- Choi, E.; Schuetz, A.; Stewart, W.F.; Sun, J. Using recurrent neural network models for early detection of heart failure onset. J. Am. Med Inform. Assoc. 2017, 24, 361–370. [Google Scholar] [CrossRef] [Green Version]
- Gron, A. Hands-On Machine Learning with Scikit-Learn and TensorFlow: Concepts, Tools, and Techniques to Build Intelligent Systems, 1st ed.; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2017. [Google Scholar]
- Chollet, F. Deep Learning with Python; Simon and Schuster: New York, NY, USA, 2021. [Google Scholar]
- Dahouda, M.K.; Joe, I. A Deep-Learned Embedding Technique for Categorical Features Encoding. IEEE Access 2021, 9, 114381–114391. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Guidi, G.; Pollonini, L.; Dacso, C.C.; Iadanza, E. A multi-layer monitoring system for clinical management of Congestive Heart Failure. BMC Med. Inform. Decis. Mak. 2015, 15, S5. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.; Tran, T.; Phung, D.; Venkatesh, S. Predicting healthcare trajectories from medical records: A deep learning approach. J. Biomed. Inform. 2017, 69, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.H.; Liu, A.; Fuh, S.C.; Lian, Y.; Guo, L.; Yang, Y.; Marelli, A.; Li, Y. Recurrent disease progression networks for modelling risk trajectory of heart failure. PLoS ONE 2021, 16, e0245177. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.; Beheshti, R.; Khan, Y.M.; Langabeer, J.R.; Foraker, R.E. Predicting cardiovascular health trajectories in time-series electronic health records with LSTM models. BMC Med. Inform. Decis. Mak. 2021, 21, 5. [Google Scholar] [CrossRef]





| Continuous | Ordinal | Boolean |
|---|---|---|
| Age | NYHA class | Ischemic heart disease |
| Systolic arterial pressure | ACE-blockers dose level | Hypertension |
| Diastolic arterial pressure | Sartans dose level | Valvulopathy |
| Weight | Beta-blockers dose level | Cardiomyopathy |
| Height | Diuretics dose level | Toxic heart disease |
| Cardiac frequency | compliance | Diabetes |
| Ejection fraction | COPD | |
| ACE-blockers Dose | Kidney failure | |
| Sartans dose | Dyslipidemia | |
| Betablockers dose | Cerebrovascular pathologies | |
| Diuretics dose | Thyropathy | |
| BNP (or proBNP) | Hepatopathy | |
| Oxigen saturation | Sinus rhythm | |
| Atrial fibrillation | ||
| Brachial block | ||
| Pacemaker ICD | ||
| Pacemaker ICDCRT | ||
| Digitalic | ||
| Antialdosterone | ||
| Antiplatelet agents | ||
| Anticoagulants | ||
| Nitrates | ||
| Statins | ||
| Amiodarone | ||
| Ivabradine | ||
| Surgical therapy |
| Model | Accuracy | AUROC | AUPRC | Parameters | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| mean | std | variance | mean | std | variance | mean | std | variance | ||
| 1 | 61.71 | 4.65 | 51.58 | 79.15 | 3.45 | 11.93 | 59.67 | 6.13 | 37.63 | 0 padding Weight set 1 |
| 2 | 60.89 | 4.25 | 18.02 | 77.99 | 3.49 | 12.20 | 58.71 | 6.12 | 37.40 | 0 padding Weight set 1 |
| 3 | 61.56 | 4.36 | 18.98 | 78.64 | 2.50 | 6.25 | 57.84 | 4.46 | 19.86 | Custom padding Weight set 1 |
| 4 | 62.74 | 1.55 | 2.39 | 79.24 | 1.68 | 2.82 | 61.64 | 3.71 | 13.74 | 0 padding Weight set 1 |
| Model 1 | Predicted Labels | Model 2 | Predicted Labels | ||||||
| Mild | Moderate | Severe | Mild | Moderate | Severe | ||||
| True | Mild | 11 | 12 | 0 | True | Mild | 11 | 11 | 1 |
| labels | Moderate | 3 | 53 | 12 | labels | Moderate | 5 | 45 | 18 |
| Severe | 0 | 4 | 5 | Severe | 0 | 2 | 7 | ||
| Model 3 | Predicted Labels | Model 4 | Predicted labels | ||||||
| Mild | Moderate | Severe | Mild | Moderate | Severe | ||||
| True | Mild | 8 | 13 | 2 | True | Mild | 15 | 8 | 0 |
| labels | Moderate | 3 | 34 | 31 | labels | Moderate | 13 | 55 | 0 |
| Severe | 0 | 1 | 8 | Severe | 0 | 9 | 0 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goretti, F.; Oronti, B.; Milli, M.; Iadanza, E. Deep Learning for Predicting Congestive Heart Failure. Electronics 2022, 11, 3996. https://doi.org/10.3390/electronics11233996
Goretti F, Oronti B, Milli M, Iadanza E. Deep Learning for Predicting Congestive Heart Failure. Electronics. 2022; 11(23):3996. https://doi.org/10.3390/electronics11233996
Chicago/Turabian StyleGoretti, Francesco, Busola Oronti, Massimo Milli, and Ernesto Iadanza. 2022. "Deep Learning for Predicting Congestive Heart Failure" Electronics 11, no. 23: 3996. https://doi.org/10.3390/electronics11233996