Explainable AI to Predict Male Fertility Using Extreme Gradient Boosting Algorithm with SMOTE
Abstract
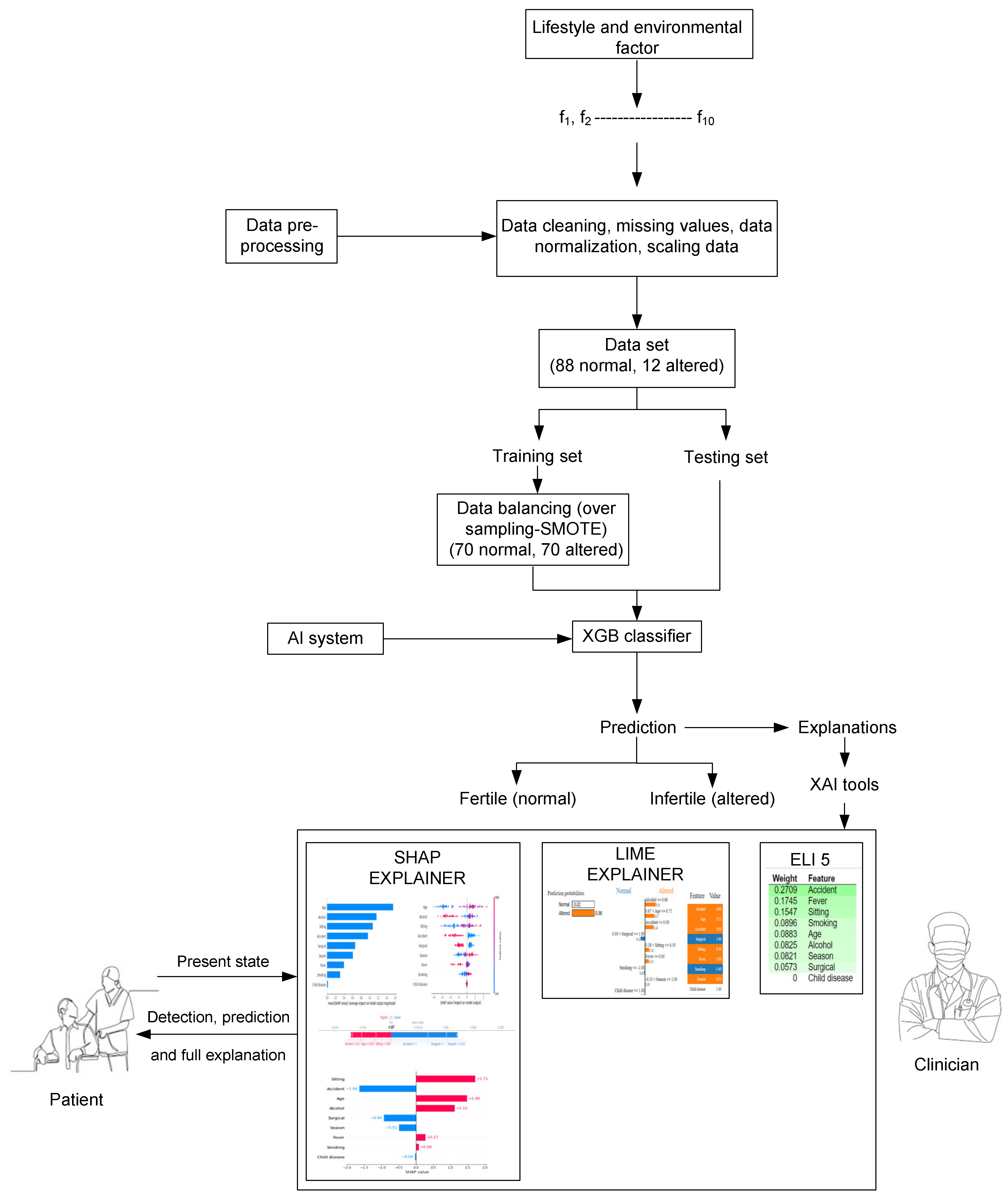
:1. Introduction
- To detect male fertility, a conventional XGB-SMOTE-based generalized AI system is proposed;
- Hold-out and five-fold cross-validation schemes are utilized for system testing;
- Benchmarking of the interpretability of the proposed system is performed via implemented XAI tools;
- To assess the performance of the proposed system, a comparative analysis is performed with existing AI systems.
2. The Model Development
2.1. Synthetic Minority Oversampling Technique (SMOTE)
2.2. XGB Algorithm
2.3. XAI
3. Experimental Setting
3.1. Dataset
3.2. Feature Importance
3.3. Performance Evalutaion
4. Numerical Results and Analysis
4.1. Analysis for Dataset
4.2. Performance Evalutaion
4.3. Explainability of Male Fertility Prediction
- Global explainability
- Local explainability
4.4. Comparision with Existing Systems
5. Conclusions and Future Work
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bongaarts, J. A method for the estimation of fecundability. Demography 1975, 12, 645–660. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Singh, A.K. Trends of male factor infertility, an important cause of infertility: A review of literature. J. Hum. Reprod. Sci. 2015, 8, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Mulgund, A.; Hamada, A.; Chyatte, M.R. A unique view on male infertility around the globe. Reprod. Biol. Endocrinol. 2015, 13, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durairajanayagam, D. Lifestyle causes of male infertility. Arab. J. Urol. 2018, 16, 10–20. [Google Scholar] [CrossRef] [Green Version]
- Yap, M.; Johnston, R.L.; Foley, H.; MacDonald, S.; Kondrashova, O.; Tran, K.A.; Nones, K.; Koufariotis, L.T.; Bean, C.; Pearson, J.V.; et al. Verifying explain-ability of a deep learning tissue classifier trained on RNA-seq data. Sci. Rep. 2021, 11, 2641. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.; Naik, N.; Somani, B.K.; Hameed, B.M.Z. Artificial intelligence (AI) in urology-Current use and future directions: An iTRUE study. Turk. J. Urol. 2020, 46, S27–S39. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Pan, W.; Jin, L.; Li, Y.; Geng, Y.; Gao, C.; Chen, G.; Wang, H.; Ma, D.; Liao, S. Artificial intelligence in reproductive medicine. Reproduction 2019, 158, R139–R154. [Google Scholar] [CrossRef]
- Azodi, C.B.; Tang, J.; Shiu, S.-H. Opening the Black Box: Interpretable Machine Learning for Geneticists. Trends Genet. 2020, 36, 442–455. [Google Scholar] [CrossRef]
- Moncada-Torres, A.; van Maaren, M.C.; Hendriks, M.P.; Siesling, S.; Geleijnse, G. Explainable machine learning can outperform Cox regression predictions and provide insights in breast cancer survival. Sci. Rep. 2021, 11, 6968. [Google Scholar] [CrossRef]
- Gadaleta, M.; Radin, J.M.; Baca-Motes, K.; Ramos, E.; Kheterpal, V.; Topol, E.J.; Steinhubl, S.R.; Quer, G. Passive detection of COVID-19 with wearable sensors and explainable machine learning algorithms. NPJ Digit. Med. 2021, 4, 166. [Google Scholar] [CrossRef]
- El-Sappagh, S.; Alonso, J.M.; Islam, S.M.R.; Sultan, A.M.; Kwak, K.S. A multilayer multimodal detection and prediction model based on explainable artificial intelligence for Alzheimer’s disease. Sci. Rep. 2021, 11, 2660. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Liu, X. The challenges of explainable AI in biomedical data science. BMC Bioinform. 2022, 22, 1–3. [Google Scholar] [CrossRef]
- Loh, H.W.; Ooi, C.P.; Seoni, S.; Barua, P.D.; Molinari, F.; Acharya, U.R. Application of Explainable Artificial Intelligence for Healthcare: A Systematic Review of the Last Decade (2011–2022). Comput. Methods Programs Biomed. 2022, 226, 107161. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Afolabi, D.O.; Ren, J.; Zhen, A. Predicting Seminal Quality via Imbalanced Learning with Evolutionary Safe-Level Synthetic Minority Over-Sampling Technique. Cogn. Comput. 2019, 13, 833–844. [Google Scholar] [CrossRef]
- Yibre, A.M.; Koçer, B. Semen quality predictive model using Feed Forwarded Neural Network trained by Learn-ing-Based Artificial Algae Algorithm. Eng. Sci. Technol. Int. J. 2021, 24, 310–318. [Google Scholar]
- Dash, S.R.; Ray, R. Predicting Seminal Quality and its Dependence on Life Style Factors through Ensemble Learning. Int. J. E-Health Med. Commun. 2020, 11, 78–95. [Google Scholar] [CrossRef]
- Ahmed, M.T.; Imtiaz, M.N. Prediction of Seminal Quality Based on Naïve Bayes Approach. PUST 2020, 4. [Google Scholar]
- Engy, E.L.; Ali, E.L.; Sally, E.G. An optimized artificial neural network approach based on sperm whale optimization algorithm for predicting fertility quality. Stud. Inform. Control 2018, 27, 349–358. [Google Scholar]
- Candemir, C. Estimating the Semen Quality from Life Style Using Fuzzy Radial Basis Functions. Int. J. Mach. Learn. Comput. 2018, 8, 44–48. [Google Scholar] [CrossRef] [Green Version]
- Soltanzadeh, S.; Zarandi, M.H.F.; Astanjin, M.B. A Hybrid Fuzzy Clustering Approach for Fertile and Unfertile Analysis. In Proceedings of the 2016 Annual Conference of the North American Fuzzy Information Processing Society (NAFIPS), El Paso, TX, USA, 13 February 2016; pp. 1–6. [Google Scholar]
- Simfukwe, M.; Kunda, D.; Chembe, C. Comparing naive bayes method and artificial neural network for semen quality categorization. Int. J. Innov. Sci. Eng. Technol. 2015, 2, 689–694. [Google Scholar]
- Mendoza-Palechor, F.E.; Ariza-Colpas, P.P.; Sepulveda-Ojeda, J.A.; De-la-Hoz-Manotas, A.; Piñeres Melo, M. Fertility analysis method based on supervised and unsupervised data mining techniques. Int. J. Appl. Eng. Res. 2016, 11, 10374–10379. [Google Scholar]
- Rhemimet, A.; Raghay, S.; Bencharef, O. Comparative Analysis of Classification, Clustering and Regression Techniques to Explore Men’s Fertility. In Proceedings of the Mediterranean Conference on Information & Communication Technologies, Paphos, Cyprus, 4–6 September 2016; Springer: Cham, Switzerland, 2016; pp. 455–462. [Google Scholar]
- Bidgoli, A.A.; Komleh, H.E.; Mousavirad, S.J. Seminal Quality Prediction Using Optimized Artificial Neural Network with Genetic Algorithm. In Proceedings of the 9th International Conference on Electrical and Electronics Engineering (ELECO), Bursa, Turkey, 26–28 November 2015; pp. 695–699. [Google Scholar]
- Sahoo, A.J.; Kumar, Y. Seminal quality prediction using data mining methods. Technol. Health Care 2014, 22, 531–545. [Google Scholar] [CrossRef] [PubMed]
- Girela, J.L.; Gil, D.; Johnsson, M.; Gomez-Torres, M.J.; De Juan, J. Semen parameters can be predicted from environmental factors and lifestyle using artificial intelligence methods. Biol. Reprod. 2013, 88, 99–100. [Google Scholar] [CrossRef] [PubMed]
- Gil, D.; Girela, J.L.; De Juan, J.; Gomez-Torres, M.J.; Johnsson, M. Predicting seminal quality with artificial intelligence methods. Expert Syst. Appl. 2012, 39, 12564–12573. [Google Scholar] [CrossRef]
- Wang, H.; Xu, Q.; Zhou, L. Seminal Quality Prediction Using Clustering-Based Decision Forests. Algorithms 2014, 7, 405–417. [Google Scholar] [CrossRef]
- Roy, D.G.; Alvi, P.A. Detection of Male Fertility Using AI-Driven Tools. In International Conference on Recent Trends in Image Processing and Pattern Recognition; Springer: Cham, Switzerland, 2022; pp. 14–25. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Ijaz, M.F.; Alfian, G.; Syafrudin, M.; Rhee, J. Hybrid Prediction Model for Type 2 Diabetes and Hypertension Using DBSCAN-Based Outlier Detection, Synthetic Minority Over Sampling Technique (SMOTE), and Random Forest. Appl. Sci. 2018, 8, 1325. [Google Scholar] [CrossRef] [Green Version]
- Javale, D.P.; Desai, S.S. Machine learning ensemble approach for healthcare data analytics. Indones. J. Electr. Eng. Comput. Sci. 2022, 28, 926–933. [Google Scholar]
- Cohen, S. Artificial Intelligence and Deep Learning in Pathology; Elsevier Health Sciences: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Freund, Y.; Schapire, R.E. A Decision-Theoretic Generalization of On-Line Learning and an Application to Boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [Google Scholar] [CrossRef] [Green Version]
- Ogunleye, A.; Wang, Q.G. XGBoost model for chronic kidney disease diagnosis. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 17, 2131–2140. [Google Scholar] [CrossRef]
- Yang, G.; Ye, Q.; Xia, J. Unbox the black-box for the medical explainable AI via multi-modal and multi-centre data fusion: A mini-review, two showcases and beyond. Inf. Fusion 2022, 77, 29–52. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Wang, M.; Zhang, H.; Elahe, F.; Jin, M. An Explainable AI Approach for the Rapid Diagnosis of COVID-19 Using Ensemble Learning Algorithms. Front. Public Health 2022, 10, 874455. [Google Scholar] [CrossRef] [PubMed]
- Kerasidou, A. Ethics of artificial intelligence in global health: Explainability, algorithmic bias and trust. J. Oral Biol. Craniofacial Res. 2021, 11, 612–614. [Google Scholar] [CrossRef] [PubMed]
- Petch, J.; Di, S.; Nelson, W. Opening the Black Box: The Promise and Limitations of Explainable Machine Learning in Cardiology. Can. J. Cardiol. 2021, 38, 204–213. [Google Scholar] [CrossRef]
- Novakovsky, G.; Dexter, N.; Libbrecht, M.W.; Wasserman, W.W.; Mostafavi, S. Obtaining genetics insights from deep learning via explainable artificial intelligence. Nat. Rev. Genet. 2022, 1–13. [Google Scholar] [CrossRef]
- Guo, H.; Zhuang, X.; Chen, P.; Alajlan, N.; Rabczuk, T. Stochastic deep collocation method based on neural architecture search and transfer learning for heterogeneous porous media. Eng. Comput. 2022, 38, 5173–5198. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.Y. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017, 30, 1–9. [Google Scholar]
- Dorogush, A.V.; Ershov, V.; Gulin, A. CatBoost: Gradient boosting with categorical features support. arXiv 2018, arXiv:1810.11363. [Google Scholar]
- Santos, M.S.; Soares, J.P.; Abreu, P.H.; Araujo, H.; Santos, J. Cross-Validation for Imbalanced Datasets: Avoiding Overoptimistic and Overfitting Approaches. IEEE Comput. Intell. Mag. 2018, 13, 59–76. [Google Scholar] [CrossRef]






| Features. No | Feature’s Name | Values Range | Normalized |
|---|---|---|---|
| Season | winter, spring, summer, and fall | (−1, −0.33, 033, 1) | |
| Age | 18–36 | (0, 1) | |
| Childhood Disease | yes or no | (0, 1) | |
| Accident /Trauma | yes or no | (0, 1) | |
| Surgical Interventional | yes or no | (0, 1) | |
| High Fever | less than 3 months ago, more than 3 months ago, no | (−1, 0, 1) | |
| Alcohol Intake | several times a day, every day, several times in a week, and hardly ever or never | (0, 1) | |
| Smoking Habit | never, occasional, and daily | (−1, 0, 1) | |
| Sitting Hours/day | 1–16 | (0, 1) | |
| Target Class | normal, altered | (1, 0) |
| Algorithm | Performance (in %) | ||||
|---|---|---|---|---|---|
| ACC | SEN | SPEC | F1-Score | AUC | |
| XGB | 90.00 | 86.12 | 84.93 | 90.06 | 91.49 |
| XGB-SMOTE | 94.05 | 91.79 | 90.02 | 95.97 | 97.00 |
| Algorithms | Performance (in %) | ||||
|---|---|---|---|---|---|
| ACC | SEN | SPEC | F1-Score | AUC | |
| SVM | 84.28 | 87.04 | 82.91 | 82.75 | 81.90 |
| SVM-SMOTE | 85.71 | 87.47 | 86.96 | 86.63 | 83.89 |
| ADA | 88.57 | 83.45 | 87.45 | 86.12 | 89.60 |
| ADA-SMOTE | 90.8 | 88.31 | 86.98 | 89.26 | 88.43 |
| Algorithms | Performance (in %) | ||||
|---|---|---|---|---|---|
| ACC | SEN | SPEC | F1-Score | AUC | |
| RF | 91.17 | 96.00 | 100.00 | 98.00 | 79.67 |
| RF-SMOTE | 92.45 | 95.00 | 83.00 | 91.00 | 86.79 |
| ET | 84.09 | 90.00 | 80.00 | 83.00 | 71.35 |
| ET-SMOTE | 85.87 | 88.00 | 83.00 | 84.00 | 76.85 |
| Dataset Fold | ACC | SEN | SPEC | F1-Score | AUC |
|---|---|---|---|---|---|
| F-1 | 92.91 | 0.89 | 0.91 | 0.93 | 1.0 |
| F-2 | 94.05 | 0.91 | 0.90 | 0.95 | 0.97 |
| F-3 | 93.78 | 0.98 | 0.96 | 0.89 | 1.0 |
| F-4 | 91.55 | 1.0 | 0.98 | 0.91 | 0.96 |
| F-5 | 93.81 | 0.98 | 1.0 | 0.93 | 1.0 |
| µ | 93.22 | 0.95 | 0.95 | 0.92 | 0.98 |
| σ | 0.92 | 0.04 | 0.03 | 0.02 | 0.01 |
| Weights | Features |
|---|---|
| 0.2861 | |
| 0.1758 | |
| 0.1697 | |
| 0.0929 | |
| 0.0846 | |
| 0.0706 | |
| 0.0587 | |
| 0.0475 | |
| 0.0143 |
| Authors [Ref] (Year) | Data Pre-Processing | AI Methods | Performance | |||
|---|---|---|---|---|---|---|
| ACC (in %) | SEN | SPEC | AUC | |||
| Gil et al. [27] (2012) | - | SVM, MLP, DT | 86, 86 and 84 (sperm concertation) 69, 69, 67 (sperm morphology) | 0.94, 0.97, 0.96 (Sperm concertation) 0.72, 0.73, 0.71 (Sperm morphology) | 0.4, 0.2, 0.13 (Sperm concentration) 0.25, 0.12, 0.12 (Sperm morphology) | - |
| Girela et al. [26] (2013) | - | ANN1, ANN2 | 97 (on training dataset) | 0.954, 0.892 | 0.5, 0.437 | - |
| Sahoo and Kumar [25] (2014) | Feature selection | DT, MLP, SVM, SVM-PSO, NB | 89, 92, 91, 94, 89 | - | - | 0.735, 0.728, 0.758, 0.932, 0.850 |
| Wang et al. [28] (2014) | - | CBDF | - | - | - | 0.80 |
| Bidgoli et al. [24] (2015) | - | Optimize MLP, NB, DT, SVM | 93.3, 73.10, 83.82, 80.88 | - | - | 0.933, 0.81, 0.858, 0.882 |
| Simfukwe et al. [21] (2015) | - | ANN, NB | 97 (on training dataset) | - | - | - |
| Soltanzadeh et al. [20] (2016) | Filtering | NB, NN, LR, Fuzzy C-means | - | - | - | 0.779, 0.7656, 0.3423, 0.73 |
| Rhemimet et al. [23] (2016) | - | DT, NB | 61.36, 88.63 | - | -- | - |
| Palechor et al. [22] (2016) | - | J48, SMO, NB, lazy IBK | 100, 100, 98.04, 100 (TP)0, 0, 1.5, 0 (FP) | - | - | - |
| Candemir et al. [19] (2018) | - | MLP, SVM, DT, FRBF | 69.0, 69.0, 67, 90 | 0.72, 0.73, 0.71, 0.92 | 0.25, 012, 0.12, 0.50 | - |
| Engy et al. [18] (2018) | - | ANN, ANN-GA, DT, SVM, ANN-SWA | 90, 95, 88, 95, 99.96 | 0.92, 0.97, 0.83 | 0.71, 0.70, 0.82, 0.72, 0.99 | - |
| Ma et al. [14] (2019) | ESLSMOTE | SVM, ADA, BPNN | 81.6, 95.1, 91.6 | - | - | - |
| Ahmed and Imtiaz [17] (2020) | - | NB | 87.75 | - | - | - |
| Dash and Ray [16] (2020) | - | soft voting, DT, NB, LR DT, DT bagged, RF, ET | 89, 78, 83, 88 88, 88, 84 (bagged) 78.80, 88.12, 89.07, 90.02 | - | - | 0.66 |
| Yibre and Kocer [15] (2021) | SMOTE | Feed forward neural network | 97.50 | 0.93 | 1 | 0.97 |
| Roy and Alvi [29] (2022) | - | KNN | 90 | - | - | - |
| Proposed | SMOTE | XGB | 93.22 | 0.95 | 0.95 | 0.98 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
GhoshRoy, D.; Alvi, P.A.; Santosh, K. Explainable AI to Predict Male Fertility Using Extreme Gradient Boosting Algorithm with SMOTE. Electronics 2023, 12, 15. https://doi.org/10.3390/electronics12010015
GhoshRoy D, Alvi PA, Santosh K. Explainable AI to Predict Male Fertility Using Extreme Gradient Boosting Algorithm with SMOTE. Electronics. 2023; 12(1):15. https://doi.org/10.3390/electronics12010015
Chicago/Turabian StyleGhoshRoy, Debasmita, Parvez Ahmad Alvi, and KC Santosh. 2023. "Explainable AI to Predict Male Fertility Using Extreme Gradient Boosting Algorithm with SMOTE" Electronics 12, no. 1: 15. https://doi.org/10.3390/electronics12010015
APA StyleGhoshRoy, D., Alvi, P. A., & Santosh, K. (2023). Explainable AI to Predict Male Fertility Using Extreme Gradient Boosting Algorithm with SMOTE. Electronics, 12(1), 15. https://doi.org/10.3390/electronics12010015












