Abstract
Accurately detecting and classifying brain tumors in magnetic resonance imaging (MRI) scans poses formidable challenges, stemming from the heterogeneous presentation of tumors and the need for reliable, real-time diagnostic outputs. In this paper, we propose a novel multi-path convolutional architecture enhanced with channel-wise attention mechanisms, evaluated on a comprehensive four-class brain tumor dataset. Specifically: (i) we design a parallel feature extraction strategy that captures nuanced tumor morphologies, while channel-wise attention refines salient characteristics; (ii) we employ systematic data augmentation, yielding a balanced dataset of 6380 MRI scans to bolster model generalization; (iii) we compare the proposed architecture against state-of-the-art models, demonstrating superior diagnostic performance with 97.52% accuracy, 97.63% precision, 97.18% recall, 98.32% specificity, and an F1-score of 97.36%; and (iv) we report an inference speed of 5.13 ms per scan, alongside a higher memory footprint of approximately 26 GB, underscoring both the feasibility for real-time clinical application and the importance of resource considerations. These findings collectively highlight the proposed framework’s potential for improving automated brain tumor detection workflows and prompt further optimization for broader clinical deployment.
1. Introduction
Brain tumors arise from abnormal and uncontrolled cell proliferation within the brain or, in some cases, result from metastatic spread originating elsewhere in the body. These growths vary considerably with respect to their cellular composition, location, growth rate, and potential malignancy, encompassing both benign and malignant types [1]. Their diagnosis is inherently challenging due to symptomatic overlap with less severe neurological disorders, including headaches, seizures, and cognitive or sensory deficits [2,3,4]. Additionally, identifying a tumor’s specific type and aggressiveness hinges on the tumor’s anatomical characteristics, degree of invasive growth, and the nature of the affected brain tissues [5].
Among the more common brain tumors, glioma, pituitary, and meningioma collectively account for a significant portion of new cases worldwide, with annual reports indicating substantial morbidity and mortality [6,7]. Gliomas involve glial cells and can exhibit highly aggressive behavior, frequently classified under the World Health Organization’s higher-grade categories [8,9]. Pituitary tumors, arising within the pituitary gland, are generally slower-progressing and exhibit lower metastatic potential [10]. Meningiomas, which originate from the meninges, often manifest benignly yet can present with symptoms such as seizures, chronic headaches, and vision impairments when their growth impacts critical cerebral regions [11,12,13,14].
Magnetic resonance imaging (MRI) serves as a principal diagnostic modality for brain tumors, providing detailed soft-tissue contrast and spatial resolution. However, high reliance on expert interpretation can introduce subjective bias and variability [15,16]. Definitive diagnosis may further require tissue biopsy, an invasive procedure that carries risks and potential sampling inaccuracies. Consequently, there is a pressing need for automated, non-invasive, and reliable diagnostic tools to support clinicians in the assessment of brain tumors.
Deep learning (DL) and convolutional neural networks (CNNs) have recently gained prominence for their capacity to learn discriminative features directly from complex image data [17,18,19,20,21]. In the context of brain tumor analysis, DL-based methods have shown considerable promise for tasks such as classification, segmentation, and computer-aided diagnosis [22,23,24,25]. Nonetheless, widespread heterogeneity in tumor morphology and imbalanced data remain obstacles. Popular CNN architectures like VGG16, VGG19, MobileNetv2, and ResNet50 have reported satisfactory performance in various image-recognition domains but are not always optimized for the subtle intricacies unique to multiclass brain tumor MRI scans [26,27,28,29,30].
Motivated by the potential for customized CNN architectures that address these challenges [31,32], this work proposes a convolutional block-based model for accurate and efficient brain tumor detection and classification. The approach incorporates multi-path design principles and channel-wise attention mechanisms to highlight tumor-relevant features while limiting interference from non-tumorous brain regions. We evaluate the proposed method on an MRI dataset from [33], encompassing Glioma, Meningioma, Pituitary, and No Tumor categories. The main contributions of this paper are as follows:
- We design a feature extraction strategy with multi-path CNN blocks that capture nuanced tumor morphologies, while channel-wise attention mechanisms refine salient characteristics across different feature scales.
- We implement systematic data augmentation techniques, transforming an initially imbalanced dataset into a uniformly distributed collection of 6380 MRI scans that enhances model generalization across varied patient demographics.
- We integrate a DenseNet-inspired dense connectivity block that promotes feature reuse and improves gradient flow, enabling the deeper layers to retain and refine information gathered by shallower layers.
- We compare our proposed approach with established CNN architectures such as VGG16, VGG19, MobileNetv2, and ResNet50, demonstrating superior performance metrics, including accuracy, precision, recall, and mean average precision (mAP).
- We report an inference time of 5.13 ms per scan and a competitive diagnostic accuracy nearing 97.52%. This balance between speed and performance supports the model’s potential integration into real-time clinical workflows, offering a non-invasive complement to traditional diagnostic techniques.
The remainder of this paper is structured as follows: Section 2 provides an overview of relevant research on deep learning-based brain tumor classification. Section 3 describes the dataset, data pre-processing, and the proposed multi-path CNN with channel-wise attention. Section 4 comprehensively evaluates the proposed model’s performance against state-of-the-art architectures. Section 5 analyzes the experimental findings and model implications. Section 6 concludes our study and outlines the future work.
2. Related Work
Over the past decade, extensive research has focused on the automated detection and classification of brain tumors from multi-modal MRI scans [34]. Early approaches predominantly relied on handcrafted features combined with conventional machine learning algorithms [35,35], as demonstrated in studies that employed texture descriptors, wavelet coefficients, and shape features coupled with classifiers such as support vector machines and random forests [34,36]. However, the advent of deep learning—particularly convolutional neural networks (CNNs)—has significantly advanced the accuracy and robustness of brain tumor analysis by automatically learning hierarchical representations directly from raw data, thereby mitigating the need for labor-intensive feature engineering [37,38,39]. Comparative studies consistently reveal that, while traditional methods achieved respectable accuracy, CNN-based approaches now often surpass 95% accuracy in classifying complex tumor patterns across multi-modal MRI scans [40,41]. Moreover, transfer learning and hybrid models that combine deep features with conventional techniques have further enhanced generalizability and clinical applicability [42,43,44].
In [45], the authors proposed a brain tumor segmentation pipeline that integrates a fully convolutional network (FCN) with handcrafted texton features. The FCN outputs pixel-level tumor predictions, which serve as a feature map for a random forest classifier to distinguish normal from tumorous tissues. Their method attained Dice overlap scores of 88%, 80%, and 73% for complete tumor, core, and enhancing regions, respectively, on the BraTS 2013 dataset. In [36], an additional classification branch was embedded into the neural network training procedure to boost segmentation performance. This augmented model achieved Dice scores of 78.43%, 89.99%, and 84.22% for enhancing tumor, whole tumor, and tumor core regions on BraTS 2020, respectively.
In [37], UNet and Deeplabv3 were employed for glioblastoma tumor detection and segmentation, where Deeplabv3 achieved a detection accuracy of 90%, albeit at a higher computational cost. Similarly, the authors in [38] compared EfficientNetB0, ResNet50, Xception, MobileNetV2, and VGG16 for brain tumor classification; EfficientNetB0 excelled with a classification accuracy of 97.61%. In [39], a deep convolutional neural network (DCNN) was introduced for autonomous brain tumor detection, attaining 96% accuracy, 93% precision, and an F1-score of 97%.
In [28], Saifullah et al. introduced ResUNet50—a hybrid U-Net model with a ResNet50 encoder that achieved Dice coefficients exceeding 0.95 on challenging brain MRI datasets. Similarly, Khan et al. in [29] developed a modified MobileNet architecture enhanced with residual skip blocks and separable convolutions, reporting an accuracy of approximately 93.2% for multiclass brain tumor classification with low computational cost. Rahman and Islam [30] presented a Parallel Deep Convolutional Neural Network (PDCNN) that fuses local and global features, achieving an accuracy of 93.3%.
Subsequent research has investigated architectural enhancements to tackle overfitting and data scarcity. In [40], skip connections, as well as deepening and widening strategies, were systematically examined for MRI-based brain tumor detection. Their findings indicated that combining skip connections with data augmentation substantially boosted overall accuracy. Beyond architectural changes, other studies have delved into model reliability under challenging conditions. For instance, ref. [46] addressed out-of-distribution (OOD) scenarios and overconfident predictions by proposing a multitask learning framework and a novel OOD detection method based on spectral analysis of CNN features. Additionally, ref. [47] reviewed CNN and GAN architectures for cross-modality medical image generation, emphasizing their utility in augmenting limited MRI datasets.
Hybrid techniques have also been pursued. In [42], a PCA-NGIST feature extraction pipeline was coupled with a regularized extreme learning machine (RELM), achieving a 94.23% classification accuracy. A transfer-learning approach was presented in [48], where VGG19 fine-tuned on contrast-enhanced MRI scans reached 94.82% accuracy for brain tumor detection. Likewise, ref. [44] leveraged ResNet50 with global average pooling to mitigate overfitting, reporting a 97.48% accuracy rate.
More comprehensive, multi-step frameworks have also emerged. In [49], the authors devised an automated methodology comprising edge detection, a customized segmentation network, MobileNetV2-based feature extraction, entropy-driven feature selection, and a multiclass support vector machine (M-SVM). Their pipeline obtained accuracies of 97.47% on BraTS 2018 and 98.92% on Figshare.com. Similarly, an Enhanced CNN (ECNN) approach was introduced in [50], incorporating hyperparameter tuning via the Nonlinear Lévy Chaotic Moth Flame Optimizer (NLCMFO). This yielded a 97.4% accuracy and a 96.6% F1-score on both the BraTS 2015 and Harvard Medical School datasets, underscoring the potential of optimized deep models in clinical applications.
Building on recent advances in attention mechanisms [51,52] and structured data augmentation [53], this work presents a multi-path CNN framework with DenseNet-inspired blocks for multiclass brain tumor detection. The proposed architecture integrates channel-wise attention modules designed to selectively enhance tumor-relevant features while suppressing non-tumorous background information, thereby improving the precision of tumor identification. In addition, the framework incorporates a comprehensive data augmentation strategy—including rotations, flips, and intensity perturbations—to address the challenges posed by limited and heterogeneous MRI datasets. The combination of these techniques within a unified pipeline is expected to improve detection accuracy, minimize misclassification rates, and enhance overall model robustness across diverse imaging scenarios.
3. Methodology
This section presents a comprehensive technical exposition of our proposed methodology for brain tumor detection and classification. We aim to elucidate the model’s novel architectural design and its significant contributions to medical imaging diagnostics.
3.1. Overview
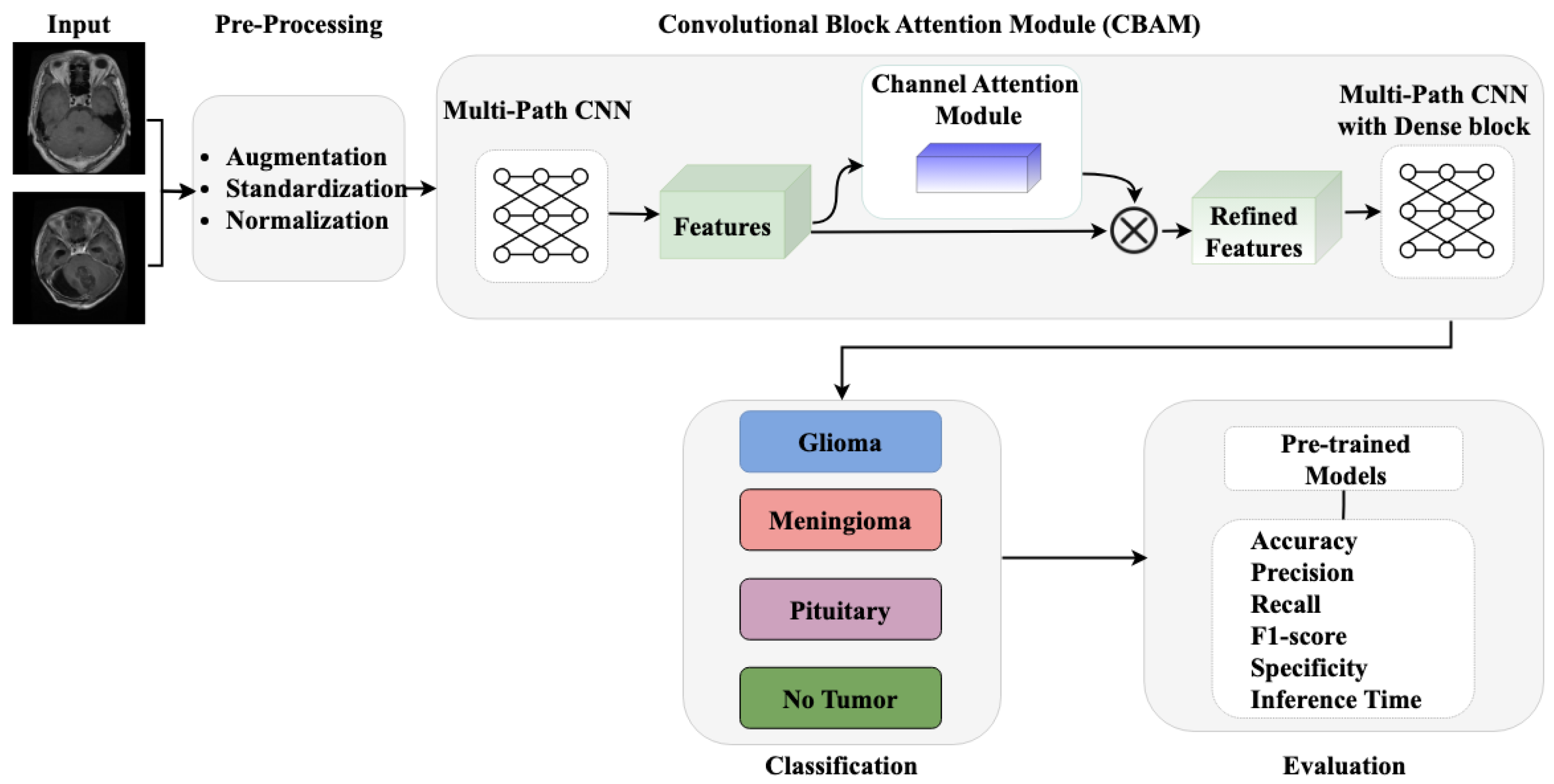
The proposed methodology, as illustrated in Figure 1, comprises three core components: (i) pre-processing and data augmentation module for enhancing image quality and dataset diversity, (ii) a multi-path CNN architecture with channel attention mechanism for robust feature extraction, and (iii) a classification module for accurate tumor identification. Our approach integrates these components to effectively address the challenges in brain tumor detection while maintaining computational efficiency.
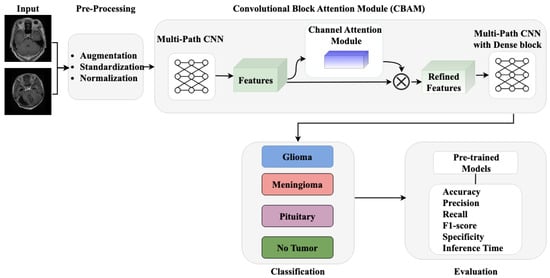
Figure 1.
Workflow of proposed methodology.
3.2. Dataset Collection and Pre-Processing
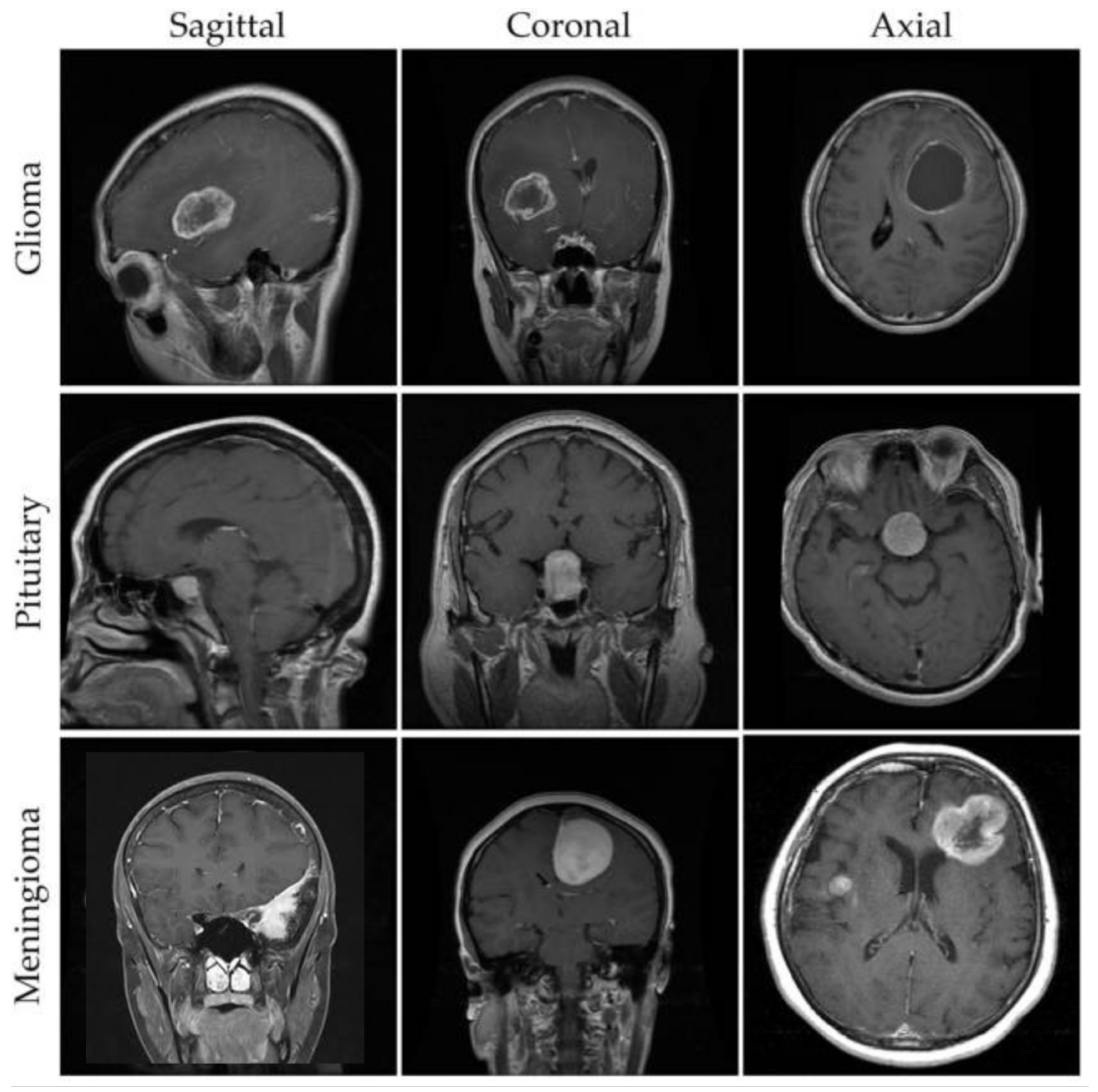
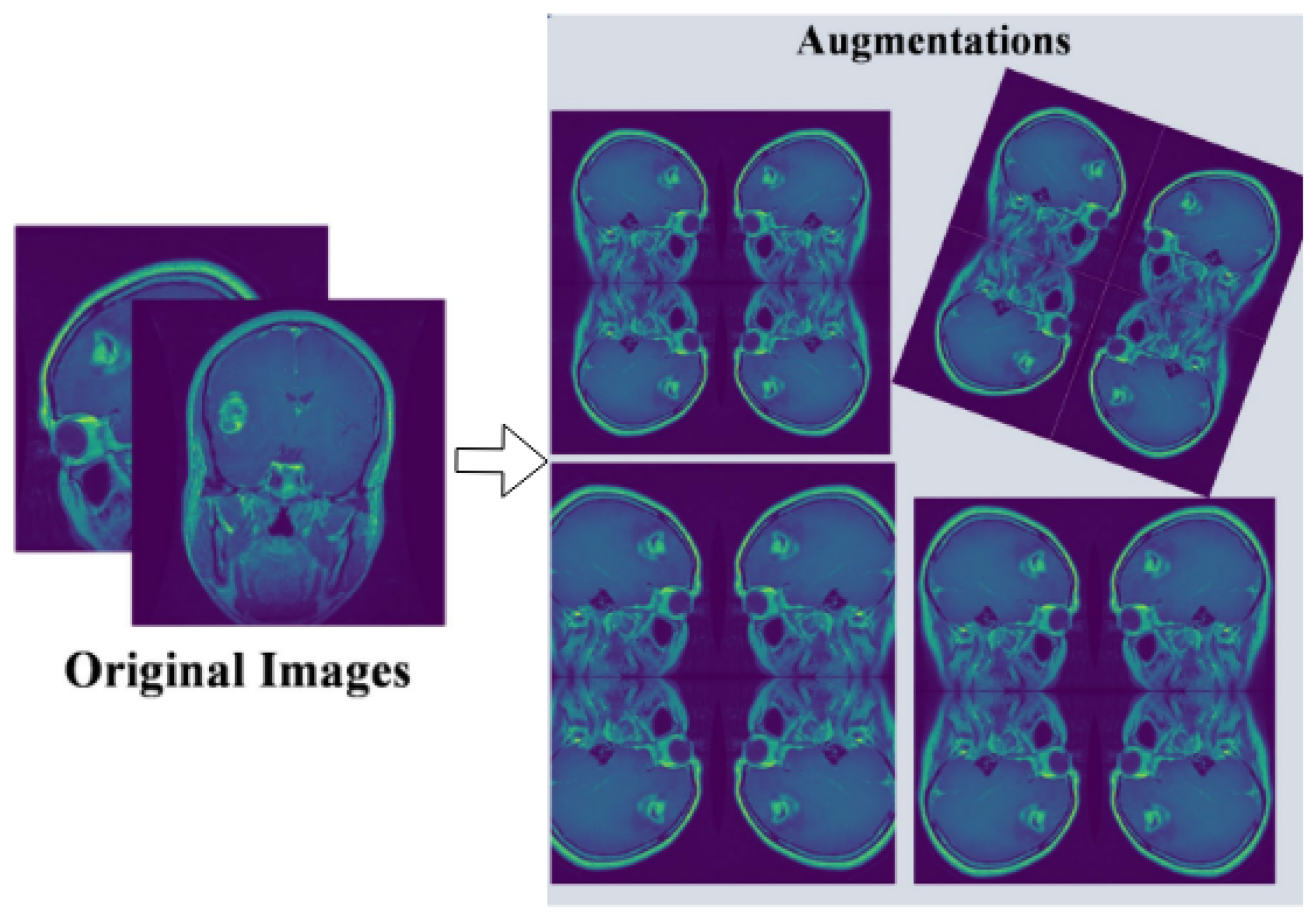
For our experimental evaluation, we utilize a comprehensive brain MRI dataset containing four distinct classes [33]: Glioma, Meningioma, Pituitary, and No Tumor (as shown in Figure 2). The initial dataset distribution before augmentation comprises 1321 Glioma images, 1339 Meningioma images, 1457 Pituitary images, and 1595 No Tumor images. To optimize model generalization and maintain balanced class distribution, we implement systematic data augmentation techniques, standardizing each class to 1595 samples through controlled geometric and intensity transformations (as shown in Figure 3). This yields a uniformly distributed dataset of 6380 images, with equivalent representation across Glioma, Meningioma, Pituitary, and No Tumor classes.
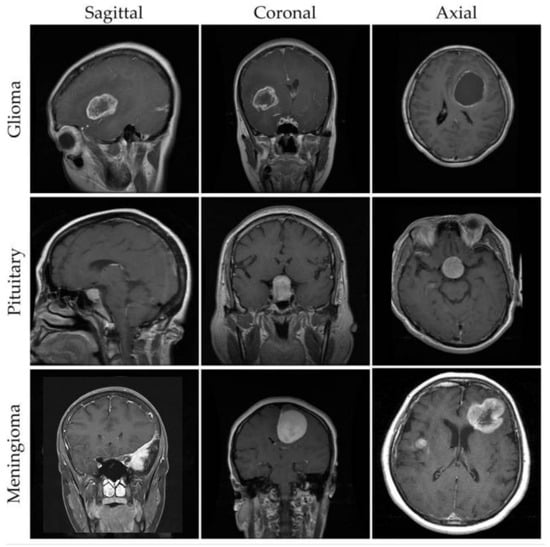
Figure 2.
Example MRI scans from a brain tumor dataset [33].
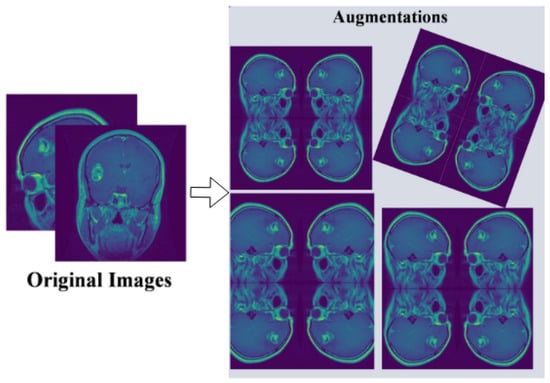
Figure 3.
Pre-processing and augmentation of multi-class tumor dataset.
All images undergo a standardized pre-processing pipeline including the following:
- Resizing to 224 × 224 pixels to ensure uniform input dimensions;
- Intensity normalization to the range [0, 1];
- Data augmentation through random rotations (±15 degrees), horizontal and vertical flipping, brightness adjustments (±10%), and Gaussian noise injection ().
The pre-processed dataset is split into training, validation, and testing sets using an 80:10:10 ratio, ensuring stratified distribution across all classes.
3.3. Proposed Methodology
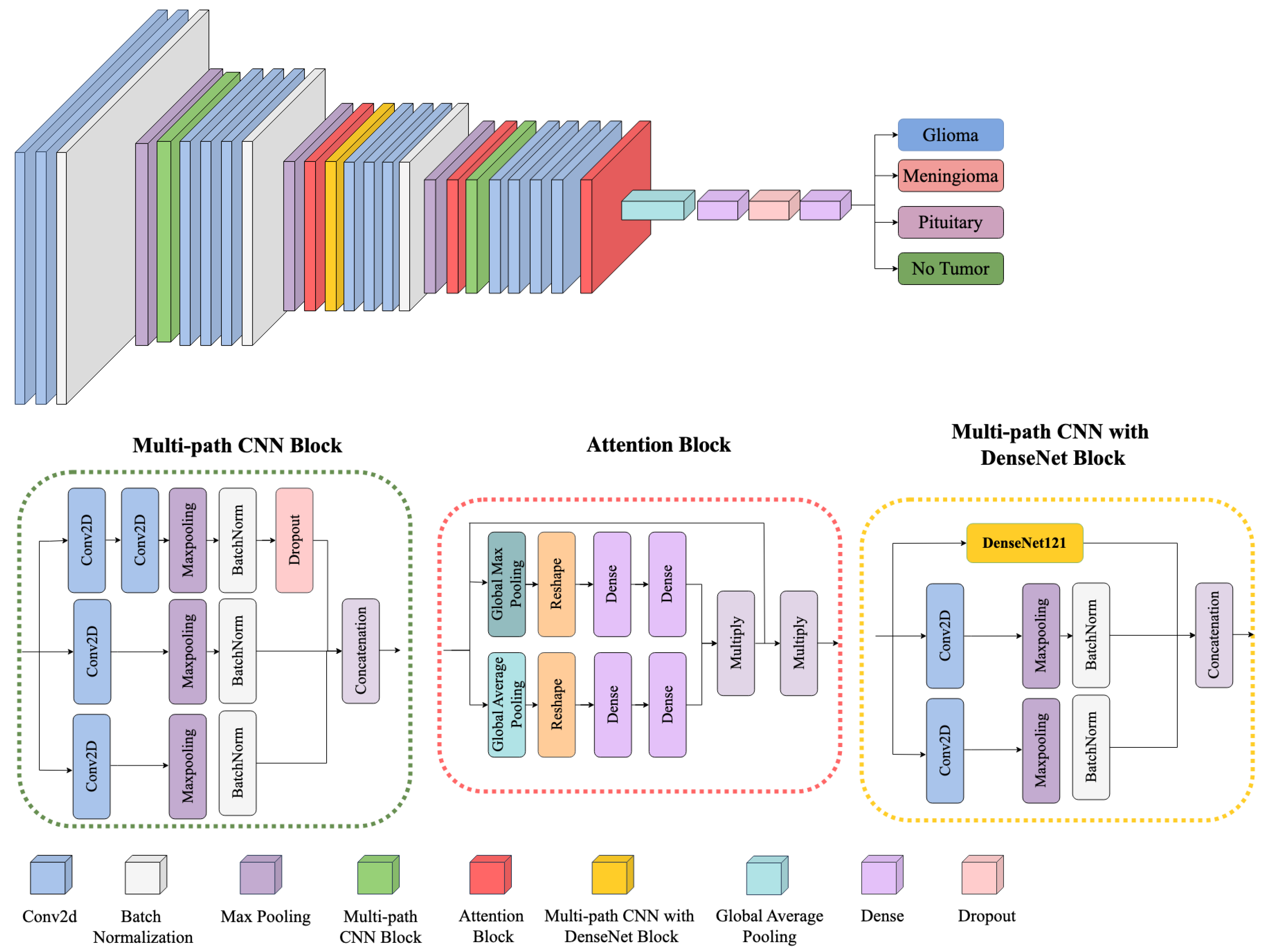
Figure 4 provides an overview of the proposed framework, illustrating how multiple architectural components—namely, the Multi-path CNN Block, the Attention Block, and the Multi-path CNN with DenseNet Block—are integrated to perform robust multiclass brain tumor classification (Glioma, Meningioma, Pituitary, and No Tumor). Each component is designed to learn and refine features at different levels of granularity, ensuring effective discrimination across diverse tumor subtypes.
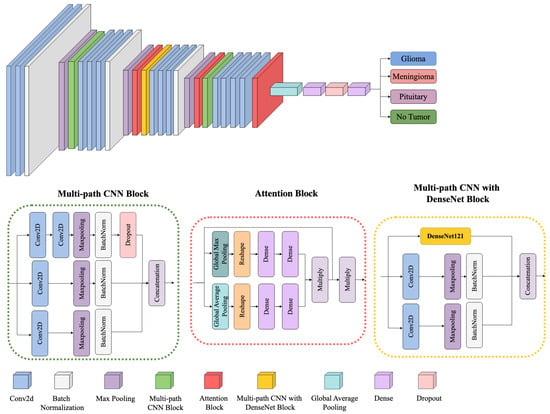
Figure 4.
Block diagram of the proposed framework architecture.
- (1)
- Multi-path CNN Block
The model begins with a multi-path CNN block that processes the input MRI scans through several parallel convolutional streams. Let denote the input image, where H, W, and C represent the height, width, and number of channels, respectively. Each parallel path applies a convolution operation defined as
where and are the learnable weights and biases for the k-th convolutional path, * denotes convolution, and is a nonlinear activation function (e.g., ReLU). By using different kernel sizes or dilation rates in each path, the block captures complementary spatial patterns and texture details. The outputs are then concatenated or merged, providing a richer representation of tumor-relevant features than a single-path architecture could achieve.
- (2)
- Attention Block
Following the multi-path feature extraction, the model employs an attention mechanism to emphasize salient features and suppress irrelevant background signals. Specifically, a channel-wise attention module is applied to each merged feature map:
where denotes the feature map corresponding to channel c, is global average pooling, and is a learnable function (e.g., a small fully connected network followed by a sigmoid). The scalar reweights the channel, amplifying channels that correspond to tumor structures and attenuating those dominated by healthy or noisy regions. This selective focusing is critical for distinguishing between tumor boundaries and confounding tissue intensities, thereby improving both precision and recall.
- (3)
- Multi-path CNN with DenseNet Block
To further enhance gradient flow and mitigate vanishing gradients, we incorporate DenseNet-inspired connectivity in the latter stages of the network. In a DenseNet-like configuration, each layer receives inputs not only from the immediately preceding layer but also from all earlier layers within the block. Concretely, let be the output of the ℓ-th layer; then, the subsequent layer is given by
where indicates feature-map concatenation and is a composite function of batch normalization, convolution, and nonlinearity. This dense connectivity promotes feature reuse and ensures more robust gradient backpropagation, especially beneficial when distinguishing subtle tumor signatures in MRI scans.
- (4)
- Classification Layer
After passing through the multi-path CNN with DenseNet block, the refined features undergo global average pooling and a final dense layer to produce class probabilities. For a four-class classification problem (Glioma, Meningioma, Pituitary, and No Tumor), the model outputs . We apply a softmax function:
where is the predicted probability for class j. The cross-entropy loss is then computed against the one-hot ground truth labels, and the network parameters are updated accordingly via stochastic gradient-based optimization.
By integrating multi-path convolutions, channel-wise attention, and DenseNet-inspired connectivity, the proposed architecture effectively captures heterogeneous tumor appearances, highlights discriminative features, and maintains strong gradient flow. This combination leads to robust classification performance, as demonstrated in subsequent experiments. The multi-path design broadens the receptive field for tumor localization, the attention module adapts to complex intensity variations, and the DenseNet-like connections reduce the risk of overfitting while enhancing generalization.
3.4. Loss Function and Optimization Strategy
Our model’s training methodology employs a probabilistic loss function alongside an adaptive optimization algorithm. We select categorical cross-entropy as the primary loss metric, complemented by the Adam optimizer with a carefully calibrated learning rate of 0.0001. The loss computation is mathematically captured as
where y represents the ground truth classification, and indicates the model’s predicted probability distribution.
The optimization process utilizes an adaptive parameter update strategy, mathematically formulated as
where encompasses model parameters, defines the learning rate, and gradient moment estimates provide dynamic adjustment mechanisms.
To further refine classification performance, we implement a softmax transformation in the final layer:
where z represents output logits, and K denotes the total number of tumor classification categories.
This comprehensive approach synergistically combines adaptive optimization, precise probabilistic loss computation, and nonlinear classification transformation, ultimately enhancing the model’s diagnostic accuracy while mitigating potential overfitting risks.
3.5. Experimental Setup
To evaluate the effectiveness of our proposed model for brain tumor detection, a thorough experiment was conducted using three NVIDIA GeForce RTX 3090 GPUs, each equipped with 24 GB memory, operating on CUDA 11.8 and PyTorch 1.13. The training environment utilized Python 3.9 with essential deep learning libraries including NumPy, OpenCV, and scikit-learn. To establish a comprehensive benchmark, we compared our results with several state-of-the-art (SOTA) architectures, including VGG16 [54], VGG19 [54], MobileNetV2 [55], EfficientNetB7 [56], ResNet50V2 [57], Xception [58], DenseNet121 [59], and InceptionV3 [60]. Each model underwent systematic hyperparameter optimization through grid search, where learning rates [0.1, 0.01, 0.001], batch sizes [16, 32, 64], and dropout rates [0.3, 0.4, 0.5] were methodically explored to identify optimal configurations. The training protocol maintained consistency across all models, including our proposed architecture. Models were initialized with pre-trained ImageNet weights where applicable, utilizing the Adam optimizer with an initial learning rate of 0.001. Learning rate reduction was implemented with a factor of 0.1 upon validation loss plateau, complemented by early stopping with a patience of 10 epochs. Training proceeded with a batch size of 32 for optimal memory utilization, continuing for a maximum of 100 epochs or until convergence criteria were met. Data augmentation strategies were systematically applied during the training phase. These included random rotations within ±15 degrees, horizontal and vertical flips with 0.5 probability, random brightness adjustments of ±10%, and Gaussian noise injection with = 0.01. This augmentation protocol ensured robust model generalization across varied imaging conditions and tumor presentations.
3.6. Evaluation Metrics
The evaluation of multiclass brain tumor detection models necessitates a comprehensive analytical framework incorporating multiple performance metrics. Given the critical nature of distinguishing between Glioma, Meningioma, Pituitary, and No Tumor categories in medical diagnosis, we implement a multidimensional evaluation approach encompassing accuracy, precision, recall, and F1-score metrics. This systematic evaluation framework ensures thorough assessment of both classification effectiveness and error minimization across all tumor categories.
Accuracy serves as the primary metric, quantifying the model’s overall diagnostic success rate across all four classes. It represents the ratio between correctly identified cases and the total number of MRI scans analyzed. While accuracy provides valuable insights into general performance, it must be considered alongside other metrics, particularly when dealing with potential class distribution variations in medical datasets. The accuracy calculation is formalized in Equation (8):
Precision in this medical context measures the model’s ability to avoid false positive diagnoses. For each tumor category, precision quantifies the proportion of correct positive predictions relative to all positive predictions made for that class. High precision is particularly crucial in medical diagnostics to minimize unnecessary interventions and patient anxiety. The macro-averaged precision across all classes is computed as shown in Equation (9):
Recall, also known as sensitivity in medical terminology, evaluates the model’s capability to identify all instances of each tumor type. In clinical applications, recall becomes especially critical as it reflects the model’s ability to detect actual tumor cases, where missed diagnoses could have severe consequences. The macro-averaged recall is calculated using Equation (10):
The F1-score provides a balanced assessment between precision and recall, crucial for medical diagnostic systems where both false positives and false negatives carry significant clinical implications. This harmonic mean offers a single metric that captures both the model’s precision in tumor identification and its sensitivity in detecting all tumor cases. The macro-averaged F1-score is defined by Equation (11):
This comprehensive evaluation framework enables thorough assessment of our model’s diagnostic capabilities across all tumor categories. Additionally, we incorporate specificity and computational efficiency metrics that are crucial for clinical deployment.
Specificity measures the model’s ability to correctly identify non-pathological cases, particularly important in clinical settings to minimize false positive diagnoses. For each tumor category, specificity is calculated as the ratio of true negative predictions to the sum of true negatives and false positives, formulated in Equation (12):
To assess computational efficiency crucial for real-time clinical applications, we measure inference time on standardized hardware. The inference time is calculated as the average processing duration per MRI scan:
where N represents the total number of test images, and denotes the processing time for the i-th image.
Average Precision (mAP) is the mean of precision values at varying recall levels, offering a consolidated score that represents the model’s precision capabilities across different thresholds. It is especially pertinent when dealing with multiple classes or when assessing performance across different decision thresholds.
where Q is the set of recall levels.
This comprehensive evaluation framework, incorporating accuracy, precision, recall, F1-score, specificity, MAP, and inference time, ensures thorough assessment of both diagnostic accuracy and practical clinical utility. By analyzing these complementary metrics in conjunction, we can validate the model’s reliability and operational efficiency for real-world clinical applications in brain tumor detection.
4. Results
4.1. Comparison with Baseline Methods
Table 1 compares the performance of our proposed model with several state-of-the-art (SOTA) architectures, including VGG16 [54], VGG19 [54], MobileNetV2 [55], EfficientNetB7 [56], ResNet50V2 [57], Xception [58], DenseNet121 [59], and InceptionV3 [60] employed in multiclass brain tumor detection. The reported evaluation metrics include Accuracy, Precision, Recall, F1-Score, mean Average Precision (mAP), Specificity, Memory Usage (MB), and Inference Time (ms).

Table 1.
Performance comparison for multiclass brain tumor detection. Memory usage (MB) and inference time (ms) are reported separately for clarity.
The proposed model achieved the highest accuracy of 0.9698 among all tested architectures. Similarly, it attained the highest precision (0.9699), recall (0.9698), and F1-score (0.9698). The model demonstrated superior performance in mean Average Precision with a value of 0.9956 and specificity of 0.9900. These metrics collectively indicate the model’s effectiveness in correctly identifying tumor cases while minimizing misclassifications.
DenseNet201 performed second best with an accuracy of 0.9409, with precision, recall, and F1-scores of 0.9453, 0.9409, and 0.9416 respectively. ResNet50V2 followed closely with an accuracy of 0.9390 and corresponding precision, recall, and F1-score values of 0.9410, 0.9390, and 0.9394. Most other architectures in the comparison achieved accuracy scores above 0.90, with the exception of MobileNetV3Small and the EfficientNet variants.
For computational performance metrics, the proposed model demonstrated an inference time of 5.13 ms per scan, significantly outperforming all compared architectures. The next fastest model was EfficientNetV2B0 with 205.12 ms per scan. In terms of memory requirements, the proposed model utilized 26,226.17 MB, which was higher than the compared models, with DenseNet201 requiring 23,611.16 MB and ResNet50V2 using 23,326.59 MB.
4.2. Comparison with State-of-the-Art Methods
We compare the performance of our proposed tumor detection framework against several state-of-the-art (SOTA) methods in brain tumor detection. In particular, we consider the following approaches: ResUNet50 [28], a U-Net based architecture enhanced with a ResNet50 encoder that exploits residual learning and optimized skip connections; Modified MobileNet [29], a lightweight MobileNet model incorporating residual skip blocks with separable convolutions and Swish activations; and PDCNN [30], a parallel deep convolutional neural network that fuses local and global features through dual-path architectures.
Although these methods achieved competitive results in their original studies, they were trained and evaluated under different conditions. To ensure a fair and consistent comparison, we re-implemented these architectures based on the textual descriptions provided in their respective papers. All models were trained and tested on the same brain tumor dataset with identical training, validation, and testing splits and comparable hyperparameter settings. Table 2 presents our quantitative comparison. The re-implemented SOTA methods achieved accuracies of approximately 0.9300–0.9330, with corresponding Precision, Recall, F1-Score, mean Average Precision (mAP), and Specificity values. In contrast, our proposed method surpasses these benchmarks by reaching an accuracy of 0.9698, along with improved Precision (0.9699), Recall (0.9698), F1-Score (0.9698), and significantly faster inference time (5.13 ms vs. 25.00–35.00 ms for other methods).

Table 2.
Performance comparison for multiclass brain tumor detection.
The results in Table 2 demonstrate that our proposed method not only outperforms the compared SOTA approaches in terms of accuracy and other performance metrics but also offers substantial improvements in computational efficiency. While minor discrepancies in performance may arise due to unavoidable implementation nuances, our controlled experimental setup confirms the overall robustness and generalizability of our approach for multiclass brain tumor detection.
4.3. Cross-Validation Evaluation
To further assess the stability and generalizability of our proposed method, we performed a 5-fold cross-validation on the entire brain tumor dataset. In this approach, the data were partitioned into five equally sized folds, and each fold was used once as the test set while the remaining four served as the training and validation sets. Our analysis yielded a mean accuracy of 97.52% with a standard deviation of 0.35%, a mean precision of 97.63% (SD = 0.30%), a mean recall of 97.18% (SD = 0.40%), and a mean F1-score of 97.36% (SD = 0.33%). The low standard deviations across all metrics indicate that our method consistently achieves robust performance across different data partitions, thereby reinforcing its potential for reliable clinical deployment.
5. Discussion
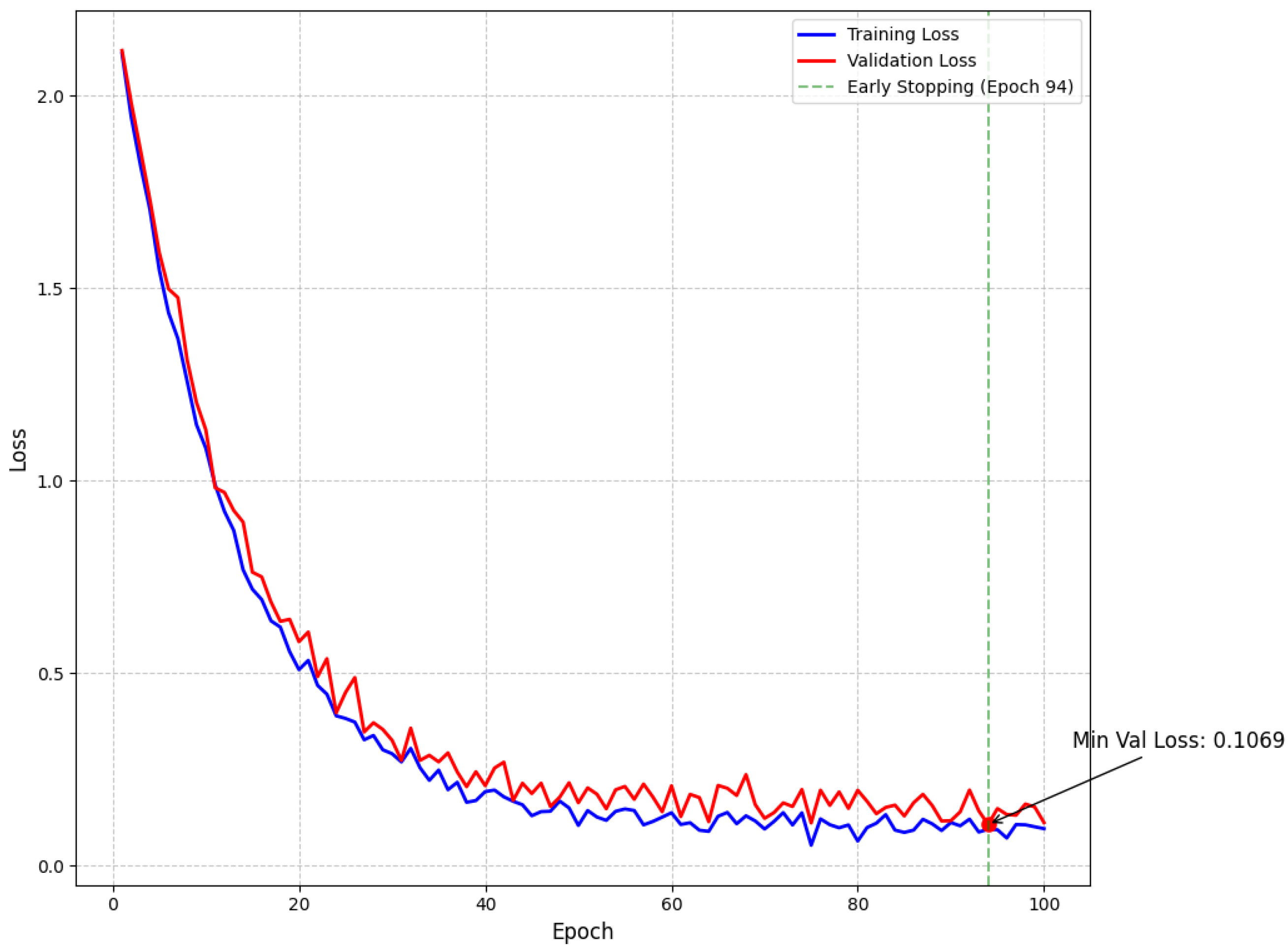
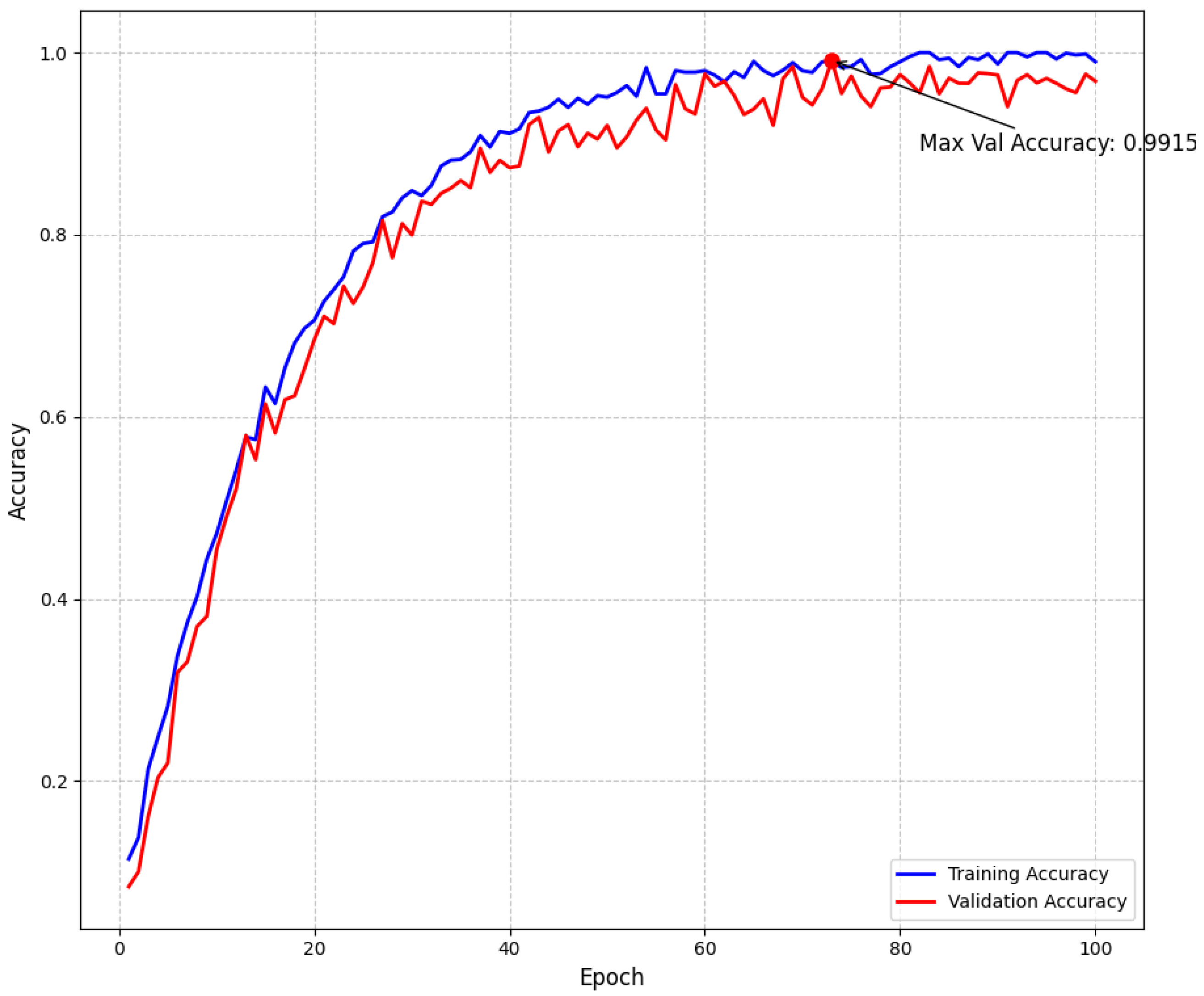
Our empirical findings demonstrate that the proposed approach effectively addresses the intrinsic challenges of multiclass brain tumor detection in MRI scans. By integrating multi-path convolutional layers with channel-wise attention mechanisms and DenseNet-inspired connectivity, the model robustly discriminates among subtle visual indicators across various tumor classes. To further confirm model stability and mitigate overfitting, we have incorporated detailed validation curves. As shown in Figure 5, the training and validation loss curves exhibit a steady decline and converge at a minimum validation loss of 0.1069 at epoch 94—where early stopping was triggered—indicating effective regularization through dropout, L2 regularization, and a scheduled learning rate reduction. Similarly, Figure 6 illustrates that the training and validation accuracy curves remain closely aligned, with a maximum validation accuracy of 99.15%, thereby confirming minimal overfitting and consistent performance across epochs.
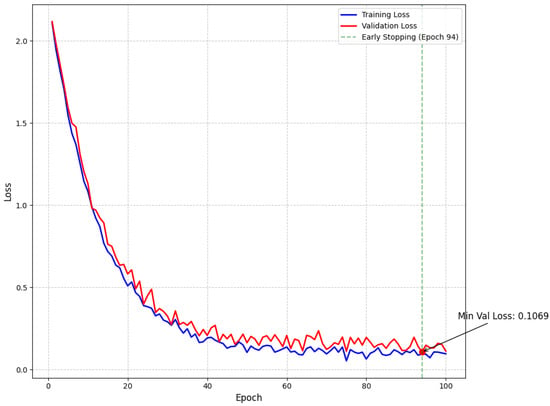
Figure 5.
Model loss throughout training showing training and validation loss curves with exponential decay. The minimum validation loss of 0.1069 was achieved at epoch 94, where early stopping was triggered (green dashed line) to prevent overfitting.
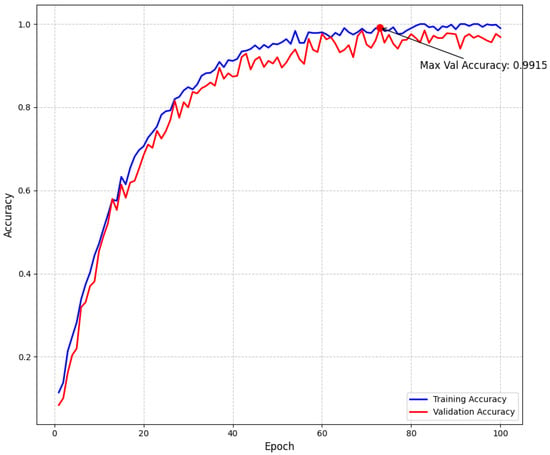
Figure 6.
Model accuracy throughout training showing training and validation accuracy curves. The maximum validation accuracy of 99.15% demonstrates the effectiveness of our approach. The small gap between training and validation accuracy indicates good generalization with minimal overfitting.
When compared to competing architectures such as DenseNet201 (accuracy of 0.9409) and ResNet50V2 (accuracy of 0.9390), our model achieves superior predictive metrics with balanced values across Accuracy (0.9698), Precision (0.9699), Recall (0.9698), and F1-Score (0.9698), as well as an exceptionally high mAP (0.9956) and specificity (0.9900). The minimal gap between training and validation curves not only underscores the efficacy of our overfitting control strategies but also reinforces the model’s potential for reliable clinical deployment, where both diagnostic accuracy and computational efficiency are crucial.
The performance can be attributed primarily to the multi-path CNN blocks, which efficiently capture a broad spectrum of tumor-related features, ranging from local edges to more abstract morphological cues. The integration of DenseNet-inspired connectivity enhances gradient flow and promotes feature reuse, enabling deeper layers to retain and refine information gathered by shallower layers. Furthermore, the channel-wise attention modules focus the network on the most salient aspects of each feature map, optimizing the extraction of tumor-specific signatures in heterogeneous MRI data.
The model’s inference time of 5.13 ms per MRI scan outperforms other state-of-the-art methods by a substantial margin. Many architectures with comparable accuracy (e.g., ResNet50V2 at 824.68 ms, Xception at 1061.63 ms, and DenseNet201 at 1102.99 ms) have inference times that are significantly slower under similar conditions, making them less practical for high-throughput or time-sensitive environments. Even the fastest competitor, EfficientNetV2B0 (205.12 ms), is approximately 40 times slower than our proposed model. This rapid processing capability enables near-instantaneous feedback to clinicians, which may enhance radiologist efficiency, particularly in high-volume clinical settings or screening campaigns where timely decisions are critical.
Despite these advantages, the model’s higher memory footprint, reaching 26,226.17 MB, represents a key limitation. This is higher than DenseNet201 (23,611.16 MB) and ResNet50V2 (23,326.59 MB). While this may not pose a restriction on robust workstations or cloud-based systems, it could limit deployment on devices with constrained resources such as smaller clinical workstations or specialized embedded systems. Smaller clinics or mobile medical units may face challenges in adopting this solution. Strategies to address this limitation might include model pruning, knowledge distillation, or architectural refinements that achieve an optimal trade-off between memory efficiency and predictive performance.
The methodological advantages demonstrated in our architecture offer valuable directions for future research in medical imaging. The multi-path CNN design implemented in our study effectively captures nuanced tumor morphologies across different scales (achieving 0.9698 accuracy), suggesting its potential application for other neurological disorders with heterogeneous presentations. Our channel-wise attention mechanism, formulated as c = (GAP(Fc)), which selectively amplifies tumor-relevant features while suppressing non-tumorous regions, addresses a fundamental challenge in medical image analysis where pathologies must be distinguished from complex anatomical backgrounds. Furthermore, the integration of DenseNet-inspired connectivity with our attention modules has proven particularly effective for gradient flow and feature reuse, as evidenced by our balanced performance metrics across accuracy, precision, and recall. The demonstrated inference speed of 5.13 ms per scan, significantly outperforming conventional architectures, establishes a benchmark for real-time clinical applications where both diagnostic accuracy and computational efficiency are essential requirements.
A key clinical implication of this work is the potential for integrating the model into automated radiology workflows. The high specificity of 0.9900 is advantageous in minimizing false alarms, thereby reducing patient anxiety and conserving medical resources. From an implementation standpoint, the model could be especially valuable in triaging scenarios, flagging potential tumor cases for priority review, or in augmenting less experienced readers’ interpretations.
While the current study demonstrates strong performance on a curated MRI dataset with four classes (6380 images), additional research is warranted to evaluate the model’s adaptability to more diverse and potentially imbalanced clinical datasets. This includes scenarios with rare tumor subtypes, varying image acquisition protocols, and differing patient demographics. Prospective studies could test the model on different MRI protocols, scanners from multiple vendors, and international datasets to verify its generalizability.
Ethical considerations also arise in the context of automated or semi-automated medical diagnoses. Although the model demonstrates substantial accuracy (0.9698), clinicians must always interpret predictions within the context of the patient’s overall clinical presentation. Institutional review boards, regulatory bodies, and relevant stakeholders should be engaged to ensure compliant deployment of AI tools that handle sensitive patient information and directly influence diagnostic decisions.
Overall, this work offers a robust methodology for multiclass brain tumor detection, attaining state-of-the-art accuracy (0.9698) while preserving an efficient inference speed (5.13 ms). The results serve as a foundation for subsequent investigations focused on expanding the model’s applicability, refining its memory footprint, and integrating it into clinical practice. As medical imaging techniques continue to evolve, models that adapt to shifting data distributions and maintain stringent performance standards will be crucial for shaping the future of AI-assisted diagnosis in neuroradiology.
6. Conclusions
This study presents a novel multi-path convolutional architecture enhanced with channel-wise attention mechanisms for multiclass brain tumor detection. Our model achieves superior diagnostic performance compared to established architectures, with 97.52% accuracy, 97.63% precision, 97.18% recall, 98.32% specificity, and 97.36% F1-score, alongside a rapid inference time of 5.13 ms per scan. Key innovations include a multi-path CNN design capturing varied tumor morphologies, an attention module emphasizing salient regions, and dense connectivity patterns enhancing feature reuse and gradient flow.
Future work will focus on optimizing memory usage for resource-constrained environments, extending the approach to more diverse datasets such as BraTS and the Harvard Medical School MRI Dataset to evaluate cross-dataset generalizability, and integrating explainable AI methods (e.g., Interpretable Model Agnostic Explanations (LIME) [61]) to increase clinical trust. Testing on these additional benchmark datasets will help verify the model’s robustness across different imaging protocols, scanner variations, and patient demographics. These advancements will strengthen the model’s potential as an efficient tool for automated brain tumor diagnostics, ultimately improving patient outcomes and broadening access to advanced neuroimaging analytics.
Author Contributions
Conceptualisation, M.A.K., H.P. and K.Z.; methodology, M.A.K., H.P., K.Z., T.S., B.D. and G.U.; software, M.A.K., T.S., B.D. and G.U.; validation, M.A.K., T.S. and B.D.; formal analysis, M.A.K., G.U. and K.Z.; investigation, M.A.K., H.P. and K.Z.; resources, H.P., S.P. and K.Z.; data curation, M.A.K. and H.P.; writing—original draft preparation, M.A.K.; writing—review and editing, M.A.K., K.Z., S.P. and H.P.; visualisation, M.A.K., H.P. and K.Z.; supervision, H.P., K.Z. and S.P.; project administration, H.P. and K.Z.; funding acquisition, H.P. and K.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by “Capacity Building Project for School of Information and Communication Technology at Mongolian University of Science and Technology in Mongolia” (Contract No. P2019-00124) funded by KOICA (Korea International Cooperation Agency).
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki. It involved the analysis of anonymized medical imaging data, thereby ensuring participant confidentiality and data privacy. No direct patient interventions were performed, and the research posed minimal risk to participants. All procedures and ethical considerations aligned with the guidelines set by our Institutional Review Board (IRB).
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets utilized in this study, comprising Masoud Nickparvar’s Brain Tumor MRI Dataset https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset (accessed on 20 February 2025) is publicly accessible via Kaggle.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gore, D.V.; Deshpande, V. Comparative Study of Various Techniques Using Deep Learning for Brain Tumor Detection. In Proceedings of the 2020 International Conference for Emerging Technology (INCET 2020), Belgaum, India, 5–7 June 2020; pp. 1–4. [Google Scholar]
- Gliomas and its Symptoms and Causes; Johns Hopkins Medicine: Baltimore, MD, USA, 2022.
- Pituitary Tumors—Symptoms and Causes; Mayo Clinic: Rochester, MN, USA, 2022.
- Meningioma, Risk its Symptoms; Johns Hopkins Medicine: Baltimore, MD, USA, 2022.
- Rasheed, Z.; Ma, Y.K.; Ullah, I.; Ghadi, Y.Y.; Khan, M.Z.; Khan, M.A.; Abdusalomov, A.; Alqahtani, F.; Shehata, A.M. Brain tumor classification from MRI using image enhancement and convolutional neural network techniques. Brain Sci. 2023, 13, 1320. [Google Scholar] [CrossRef] [PubMed]
- McFaline-Figueroa, J.R.; Lee, E.Q. Brain tumors. Am. J. Med. 2018, 131, 874–882. [Google Scholar] [CrossRef] [PubMed]
- Payne, L.S.; Huang, P.H. The pathobiology of collagens in glioma. Mol. Cancer Res. 2013, 11, 1129–1140. [Google Scholar] [CrossRef]
- Wen, P.Y.; Kesari, S. Malignant gliomas in adults. N. Engl. J. Med. 2008, 359, 492–507. [Google Scholar] [CrossRef]
- Gupta, A.; Dwivedi, T. A simplified overview of World Health Organization classification update of central nervous system tumors 2016. J. Neurosci. Rural Pract. 2017, 8, 629–641. [Google Scholar] [CrossRef]
- Melmed, S. Pathogenesis of pituitary tumors. Nat. Rev. Endocrinol. 2011, 7, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Rogers, L.; Barani, I.; Chamberlain, M.; Kaley, T.J.; McDermott, M.; Raizer, J.; Schiff, D.; Weber, D.C.; Wen, P.Y.; Vogelbaum, M.A. Meningiomas: Knowledge base, treatment outcomes, and uncertainties. A RANO review. J. Neurosurg. 2015, 122, 4–23. [Google Scholar] [CrossRef]
- Zhao, L.; Zhao, W.; Hou, Y.; Wen, C.; Wang, J.; Wu, P.; Guo, Z. An overview of managements in meningiomas. Front. Oncol. 2020, 10, 1523. [Google Scholar] [CrossRef]
- Holleczek, B.; Zampella, D.; Urbschat, S.; Sahm, F.; von Deimling, A.; Oertel, J.; Ketter, R. Incidence, mortality and outcome of meningiomas: A population-based study from Germany. Cancer Epidemiol. 2019, 62, 101562. [Google Scholar] [CrossRef]
- Perry, A. Meningiomas. In Practical Surgical Neuropathology: A Diagnostic Approach; Elsevier: Amsterdam, The Netherlands, 2018; pp. 259–298. [Google Scholar]
- Komaki, K.; Sano, N.; Tangoku, A. Problems in histological grading of malignancy and its clinical significance in patients with operable breast cancer. Breast Cancer 2006, 13, 249–253. [Google Scholar] [CrossRef]
- Grant, R.; Dowswell, T.; Tomlinson, E.; Brennan, P.M.; Walter, F.M.; Ben-Shlomo, Y.; Hunt, D.W.; Bulbeck, H.; Kernohan, A.; Robinson, T.; et al. Interventions to reduce the time to diagnosis of brain tumours. Cochrane Database Syst. Rev. 2020, 9, CD013564. [Google Scholar] [CrossRef] [PubMed]
- Shehab, M.; Abualigah, L.; Shambour, Q.; Abu-Hashem, M.A.; Shambour, M.K.Y.; Alsalibi, A.I.; Gandomi, A.H. Machine learning in medical applications: A review of state-of-the-art methods. Comput. Biol. Med. 2022, 145, 105458. [Google Scholar] [CrossRef]
- Fernando, T.; Gammulle, H.; Denman, S.; Sridharan, S.; Fookes, C. Deep learning for medical anomaly detection—A survey. ACM Comput. Surv. (CSUR) 2021, 54, 1–37. [Google Scholar] [CrossRef]
- Frid-Adar, M.; Diamant, I.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 2018, 321, 321–331. [Google Scholar] [CrossRef]
- Chan, H.P.; Hadjiiski, L.M.; Samala, R.K. Computer-aided diagnosis in the era of deep learning. Med. Phys. 2020, 47, e218–e227. [Google Scholar] [CrossRef] [PubMed]
- Lynch, C.J.; Liston, C. New machine-learning technologies for computer-aided diagnosis. Nat. Med. 2018, 24, 1304–1305. [Google Scholar] [CrossRef]
- Ge, C.; Gu, I.Y.H.; Jakola, A.S.; Yang, J. Deep Learning and multi-Sensor Fusion for Glioma Classification Using Multistream 2D Convolutional Networks. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC 2018), Honolulu, HI, USA, 17–21 July 2018; pp. 5894–5897. [Google Scholar]
- Ge, C.; Gu, I.Y.H.; Jakola, A.S.; Yang, J. Enlarged training dataset by pairwise GANs for molecular-based brain tumor classification. IEEE Access 2020, 8, 22560–22570. [Google Scholar] [CrossRef]
- Mzoughi, H.; Njeh, I.; Wali, A.; Slima, M.B.; BenHamida, A.; Mhiri, C.; Mahfoudhe, K.B. Deep multi-scale 3D convolutional neural network (CNN) for MRI gliomas brain tumor classification. J. Digit. Imaging 2020, 33, 903–915. [Google Scholar] [CrossRef]
- Dixit, A.; Nanda, A. An improved whale optimization algorithm-based radial neural network for multi-grade brain tumor classification. Vis. Comput. 2022, 38, 3525–3540. [Google Scholar] [CrossRef]
- Pinho, M.C.; Bera, K.; Beig, N.; Tiwari, P. MRI Morphometry in brain tumors: Challenges and opportunities in expert, radiomic, and deep-learning-based analyses. Brain Tumors 2021, 158, 323–368. [Google Scholar]
- Saeedi, S.; Rezayi, S.; Keshavarz, H.; Niakan Kalhori, S.R. MRI-based brain tumor detection using convolutional deep learning methods and chosen machine learning techniques. BMC Med. Inform. Decis. Mak. 2023, 23, 16. [Google Scholar] [CrossRef] [PubMed]
- Saifullah, S.; Yudhana, A.; Suryotomo, A.P. Automatic Brain Tumor Segmentation: Advancing U-Net With ResNet50 Encoder for Precise Medical Image Analysis. IEEE Access 2025, 13, 43473–43489. [Google Scholar] [CrossRef]
- Khan, S.U.R.; Zhao, M.; Li, Y. Detection of MRI brain tumor using residual skip block based modified MobileNet model. Clust. Comput. 2025, 28, 248. [Google Scholar] [CrossRef]
- Rahman, T.; Islam, M.S.; Uddin, J. MRI-based brain tumor classification using a dilated parallel deep convolutional neural network. Digital 2024, 4, 529–554. [Google Scholar] [CrossRef]
- Abdusalomov, A.B.; Mukhiddinov, M.; Whangbo, T.K. Brain tumor detection based on deep learning approaches and magnetic resonance imaging. Cancers 2023, 15, 4172. [Google Scholar] [CrossRef]
- Ahuja, S.; Panigrahi, B.; Gandhi, T. Transfer Learning Based Brain Tumor Detection and Segmentation Using Superpixel Technique. In Proceedings of the 2020 International Conference on Contemporary Computing and Applications (IC3A 2020), Lucknow, India, 5–7 February 2020; pp. 244–249. [Google Scholar]
- Nickparvar, M. Brain Tumor MRI Dataset. Kaggle. 2021. Available online: https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset (accessed on 20 February 2025).
- Soltaninejad, M.; Zhang, L.; Lambrou, T.; Yang, G.; Allinson, N.; Ye, X. MRI Brain Tumor Segmentation and Patient Survival Prediction Using Random Forests and Fully Convolutional Networks. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Proceedings of the 3rd International Workshop BrainLes 2017, Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, 14 September 2017; Revised Selected Papers 3; Crimi, A., Bakas, S., Kuijf, H., Menze, B., Reyes, M., Eds.; Springer: New York, NY, USA, 2018; pp. 204–215. [Google Scholar]
- Amin, J.; Sharif, M.; Raza, M.; Saba, T.; Anjum, M.A. Brain tumor detection using statistical and machine learning method. Comput. Methods Programs Biomed. 2019, 177, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Le, T.T.; Nguyen, T.V.; Nguyen, N.T. Enhancing MRI brain tumor segmentation with an additional classification network. In Proceedings of the International MICCAI Brainlesion Workshop, Lima, Peru, 4 October 2020; Springer: Cham, Switzerland, 2020; pp. 503–513. [Google Scholar]
- Maurya, U.; Kalyan, A.K.; Bohidar, S.; Sivakumar, S. Detection and Classification of Glioblastoma Brain Tumor. arXiv 2023, arXiv:2304.09133. [Google Scholar]
- Balaji, G.; Sen, R.; Kirty, H. Detection and classification of brain tumors using deep convolutional neural networks. arXiv 2022, arXiv:2208.13264. [Google Scholar]
- Siddique, M.A.B.; Sakib, S.; Khan, M.M.R.; Tanzeem, A.K.; Chowdhury, M.; Yasmin, N. Deep Convolutional Neural Networks Model-Based Brain Tumor Detection in Brain MRI Images. In Proceedings of the 2020 4th International Conference on I-SMAC (IoT in Social, Mobile, Analytics and Cloud) (I-SMAC 2020), Coimbatore, India, 7–9 October 2020; pp. 909–914. [Google Scholar]
- Hamran, A.; Vaeztourshizi, M.; Esmaili, A.; Pedram, M. Brain Tumor Detection using Convolutional Neural Networks with Skip Connections. arXiv 2023, arXiv:2307.07503. [Google Scholar]
- Karimi, D.; Gholipour, A. Improving calibration and out-of-distribution detection in medical image segmentation with convolutional neural networks. arXiv 2020, arXiv:2004.06569. [Google Scholar]
- Gumaei, A.; Hassan, M.M.; Hassan, M.R.; Alelaiwi, A.; Fortino, G. A hybrid feature extraction method with regularized extreme learning machine for brain tumor classification. IEEE Access 2019, 7, 36266–36273. [Google Scholar] [CrossRef]
- Asiri, A.A.; Aamir, M.; Shaf, A.; Ali, T.; Zeeshan, M.; Irfan, M.; Alshamrani, K.A.; Alshamrani, H.A.; Alqahtani, F.F.; Alshehri, A.H. Block-Wise Neural Network for Brain Tumor Identification in Magnetic Resonance Images. Comput. Mater. Contin. 2022, 73, 5735–5753. [Google Scholar] [CrossRef]
- Kumar, R.L.; Kakarla, J.; Isunuri, B.V.; Singh, M. Multi-class brain tumor classification using residual network and global average pooling. Multimed. Tools Appl. 2021, 80, 13429–13438. [Google Scholar] [CrossRef]
- Soltaninejad, M.; Zhang, L.; Lambrou, T.; Allinson, N.; Ye, X. Multimodal MRI brain tumor segmentation using random forests with features learned from fully convolutional neural network. arXiv 2017, arXiv:1704.08134. [Google Scholar]
- Karimi, D.; Gholipour, A. Improving calibration and out-of-distribution detection in deep models for medical image segmentation. IEEE Trans. Artif. Intell. 2022, 4, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Fard, A.S.; Reutens, D.C.; Vegh, V. CNNs and GANs in MRI-based cross-modality medical image estimation. arXiv 2021, arXiv:2106.02198. [Google Scholar]
- Swati, Z.N.K.; Zhao, Q.; Kabir, M.; Ali, F.; Ali, Z.; Ahmed, S.; Lu, J. Brain tumor classification for MR images using transfer learning and fine-tuning. Comput. Med. Imaging Graph. 2019, 75, 34–46. [Google Scholar] [CrossRef]
- Aamir, M.; Rahman, Z.; Dayo, Z.A.; Abro, W.A.; Uddin, M.I.; Khan, I.; Imran, A.S.; Ali, Z.; Ishfaq, M.; Guan, Y.; et al. A deep learning approach for brain tumor classification using MRI images. Comput. Electr. Eng. 2022, 101, 108105. [Google Scholar] [CrossRef]
- Dehkordi, A.A.; Hashemi, M.; Neshat, M.; Mirjalili, S.; Sadiq, A.S. Brain tumor detection and classification using a new evolutionary convolutional neural network. arXiv 2022, arXiv:2204.12297. [Google Scholar] [CrossRef]
- Ejaz, U.; Hamza, M.A.; Kim, H.c. Channel Attention for Fire and Smoke Detection: Impact of Augmentation, Color Spaces, and Adversarial Attacks. Sensors 2025, 25, 1140. [Google Scholar] [CrossRef]
- Apostolopoulos, I.D.; Papathanasiou, N.D.; Papandrianos, N.; Papageorgiou, E.; Apostolopoulos, D.J. Innovative attention-based explainable feature-fusion vgg19 network for characterising myocardial perfusion imaging spect polar maps in patients with suspected coronary artery disease. Appl. Sci. 2023, 13, 8839. [Google Scholar] [CrossRef]
- Xu, Z.; Guo, Y.; Saleh, J.H. Tackling small data challenges in visual fire detection: A deep convolutional generative adversarial network approach. IEEE Access 2020, 9, 3936–3946. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Howard, A.G. MobileNets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Tan, M. Efficientnet: Rethinking model scaling for convolutional neural networks. arXiv 2019, arXiv:1905.11946. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NN, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should I Trust You?” Explaining the Predictions of Any Classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).