An Image Recognition-Based Approach to Actin Cytoskeleton Quantification
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Preparation
2.1.1. Cell Culture
2.1.2. Actin Cytoskeleton Treatment
2.1.3. Actin Cytoskeleton Staining
2.2. Fluorescence Microscope
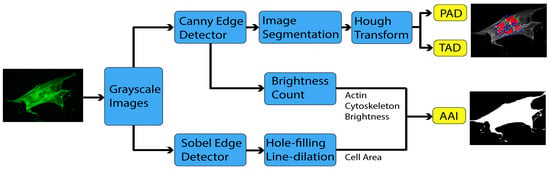
2.3. Image Processing
2.3.1. Sobel Edge Detector
2.3.2. Canny Edge Detector
2.3.3. Hough Transform
2.4. Actin Cytoskeleton Quantification
2.4.1. Actin Alignment Deviation
2.4.2. Actin Intensity
2.5. Statistical Analysis
3. Results and Discussion
3.1. Approach Validation
3.2. Actin Cytoskeleton Quantification
3.2.1. Actin Deviation
3.2.2. Actin Intensity
3.3. Quantification Result Comparison
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Elosegui-Artola, A.; Jorge-Peñas, A.; Moreno-Arotzena, O.; Oregi, A.; Lasa, M.; García-Aznar, J.M.; De Juan-Pardo, E.M.; Aldabe, R. Image analysis for the quantitative comparison of stress fibers and focal adhesions. PLoS ONE 2014, 9, e107393. [Google Scholar] [CrossRef] [PubMed]
- Lichtenstein, N.; Geiger, B.; Kam, Z. Quantitative analysis of cytoskeletal organization by digital fluorescent microscopy. Cytom. Part A 2003, 54, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Mollaeian, K.; Liu, Y.; Ren, J. Investigation of Nanoscale Poroelasticity of Eukaryotic Cells Using Atomic Force Microscopy. In Proceedings of the ASME 2017 Dynamic Systems and Control Conference American Society of Mechanical Engineers, Tysons, VI, USA, 11–13 October 2017; p. V001T08A005. [Google Scholar]
- Alioscha-Perez, M.; Benadiba, C.; Goossens, K.; Kasas, S.; Dietler, G.; Willaert, R.; Sahli, H. A robust actin filaments image analysis framework. PLoS Comput. Biol. 2016, 12, e1005063. [Google Scholar] [CrossRef] [PubMed]
- Weichsel, J.; Urban, E.; Small, J.V.; Schwarz, U.S. Reconstructing the orientation distribution of actin filaments in the lamellipodium of migrating keratocytes from electron microscopy tomography data. Cytom. Part A 2012, 81, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Mollaeian, K.; Liu, Y.; Bi, S.; Ren, J. Atomic force microscopy study revealed velocity-dependence and nonlinearity of nanoscale poroelasticity of eukaryotic cells. J. Mech. Behav. Biomed. Mater. 2018, 78, 65–73. [Google Scholar] [CrossRef]
- Tamiello, C.; Buskermolen, A.B.; Baaijens, F.P.; Broers, J.L.; Bouten, C.V. Heading in the right direction: Understanding cellular orientation responses to complex biophysical environments. Cell. Mol. Bioeng. 2016, 9, 12–37. [Google Scholar] [CrossRef]
- Pullarkat, P.A.; Fernández, P.A.; Ott, A. Rheological properties of the eukaryotic cell cytoskeleton. Phys. Rep. 2007, 449, 29–53. [Google Scholar] [CrossRef]
- Gupta, M.; Sarangi, B.R.; Deschamps, J.; Nematbakhsh, Y.; Callan-Jones, A.; Margadant, F.; Mège, R.M.; Lim, C.T.; Voituriez, R.; Ladoux, B. Adaptive rheology and ordering of cell cytoskeleton govern matrix rigidity sensing. Nat. Commun. 2015, 6, 7525. [Google Scholar] [CrossRef]
- Wakatsuki, T.; Schwab, B.; Thompson, N.C.; Elson, E.L. Effects of cytochalasin D and latrunculin B on mechanical properties of cells. J. Cell Sci. 2001, 114, 1025–1036. [Google Scholar] [PubMed]
- Sims, J.R.; Karp, S.; Ingber, D.E. Altering the cellular mechanical force balance results in integrated changes in cell, cytoskeletal and nuclear shape. J. Cell Sci. 1992, 103, 1215–1222. [Google Scholar]
- Mollaeian, K.; Liu, Y.; Bi, S.; Wang, Y.; Ren, J.; Lu, M. Nonlinear Cellular Mechanical Behavior Adaptation to Substrate Mechanics Identified by Atomic Force Microscope. Int. J. Mol. Sci. 2018, 19, 3461. [Google Scholar] [CrossRef]
- Kimori, Y.; Hikino, K.; Nishimura, M.; Mano, S. Quantifying morphological features of actin cytoskeletal filaments in plant cells based on mathematical morphology. J. Theor. Biol. 2016, 389, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Kunttu, I.; Lepistö, L.; Rauhamaa, J.; Visa, A. Multiscale Fourier descriptors for defect image retrieval. Pattern Recognit. Lett. 2006, 27, 123–132. [Google Scholar] [CrossRef]
- Liu, Y.; Mollaeian, K.; Ren, J. Finite element modeling of living cells for AFM indentation-based biomechanical characterization. Micron 2018. [Google Scholar] [CrossRef]
- Patel, M.N.; Tandel, P. A Survey on Feature Extraction Techniques for Shape based Object Recognition. Int. J. Comput. Appl. 2016, 0975–8887. [Google Scholar]
- Castañón, C.A.; Fraga, J.S.; Fernandez, S.; Gruber, A.; Costa, L.d.F. Biological shape characterization for automatic image recognition and diagnosis of protozoan parasites of the genus Eimeria. Pattern Recognit. 2007, 40, 1899–1910. [Google Scholar] [CrossRef]
- Takamatsu, A.; Takaba, E.; Takizawa, G. Environment-dependent morphology in plasmodium of true slime mold Physarum polycephalum and a network growth model. J. Theor. Biol. 2009, 256, 29–44. [Google Scholar] [CrossRef]
- Pantic, I.; Harhaji-Trajkovic, L.; Pantovic, A.; Milosevic, N.T.; Trajkovic, V. Changes in fractal dimension and lacunarity as early markers of UV-induced apoptosis. J. Theor. Biol. 2012, 303, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Verkhovsky, A.B.; Chaga, O.Y.; Schaub, S.; Svitkina, T.M.; Meister, J.J.; Borisy, G.G. Orientational order of the lamellipodial actin network as demonstrated in living motile cells. Mol. Biol. Cell 2003, 14, 4667–4675. [Google Scholar] [CrossRef]
- Higaki, T.; Kutsuna, N.; Sano, T.; Kondo, N.; Hasezawa, S. Quantification and cluster analysis of actin cytoskeletal structures in plant cells: Role of actin bundling in stomatal movement during diurnal cycles in Arabidopsis guard cells. Plant J. 2010, 61, 156–165. [Google Scholar] [CrossRef]
- Basu, S.; Liu, C.; Rohde, G.K. Localizing and extracting filament distributions from microscopy images. J. Microsc. 2015, 258, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Tondon, A.; Kaunas, R. The direction of stretch-induced cell and stress fiber orientation depends on collagen matrix stress. PLoS ONE 2014, 9, e89592. [Google Scholar] [CrossRef] [PubMed]
- Vijayarani, S.; Vinupriya, M. Performance analysis of canny and sobel edge detection algorithms in image mining. Int. J. Innov. Res. Comput. Commun. Eng. 2013, 1, 1760–1767. [Google Scholar]
- Ojala, T.; Pietikainen, M.; Maenpaa, T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 971–987. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Q.; Zhang, B. Image enhancement based on equal area dualistic sub-image histogram equalization method. IEEE Trans. Consum. Electron. 1999, 45, 68–75. [Google Scholar] [CrossRef]
- Feibush, E.A.; Levoy, M.; Cook, R.L. Synthetic texturing using digital filters. In ACM SIGGRAPH Computer Graphics; ACM: New York, NY, USA, 1980; Volume 14, pp. 294–301. [Google Scholar]
- Ballard, D.H. Generalizing the Hough transform to detect arbitrary shapes. In Readings in Computer Vision; Elsevier: Amsterdam, The Netherlands, 1987; pp. 714–725. [Google Scholar]
- Liu, Y.; Ren, J. Modeling and Control of Dynamic Cellular Mechanotransduction (I): Actin Cytoskeleton Quantification; ASME: New York, NY, USA, 2018. [Google Scholar]
- Cooper, J.A. Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 1987, 105, 1473–1478. [Google Scholar] [CrossRef]
- Maini, R.; Aggarwal, H. Study and comparison of various image edge detection techniques. Int. J. Image Process. 2009, 3, 1–11. [Google Scholar]
- Baxes, G.A. Digital Image Processing: Principles and Applications; Wiley: New York, NY, USA, 1994. [Google Scholar]
- Bear, J.E.; Svitkina, T.M.; Krause, M.; Schafer, D.A.; Loureiro, J.J.; Strasser, G.A.; Maly, I.V.; Chaga, O.Y.; Cooper, J.A.; Borisy, G.G.; et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 2002, 109, 509–521. [Google Scholar] [CrossRef]
- Wang, S.; Arellano-Santoyo, H.; Combs, P.A.; Shaevitz, J.W. Actin-like cytoskeleton filaments contribute to cell mechanics in bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 9182–9185. [Google Scholar] [CrossRef]
- Chen, T.; Teng, N.; Wu, X.; Wang, Y.; Tang, W.; Šamaj, J.; Baluška, F.; Lin, J. Disruption of actin filaments by latrunculin B affects cell wall construction in Picea meyeri pollen tube by disturbing vesicle trafficking. Plant Cell Physiol. 2007, 48, 19–30. [Google Scholar] [CrossRef]










© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Mollaeian, K.; Ren, J. An Image Recognition-Based Approach to Actin Cytoskeleton Quantification. Electronics 2018, 7, 443. https://doi.org/10.3390/electronics7120443
Liu Y, Mollaeian K, Ren J. An Image Recognition-Based Approach to Actin Cytoskeleton Quantification. Electronics. 2018; 7(12):443. https://doi.org/10.3390/electronics7120443
Chicago/Turabian StyleLiu, Yi, Keyvan Mollaeian, and Juan Ren. 2018. "An Image Recognition-Based Approach to Actin Cytoskeleton Quantification" Electronics 7, no. 12: 443. https://doi.org/10.3390/electronics7120443
APA StyleLiu, Y., Mollaeian, K., & Ren, J. (2018). An Image Recognition-Based Approach to Actin Cytoskeleton Quantification. Electronics, 7(12), 443. https://doi.org/10.3390/electronics7120443