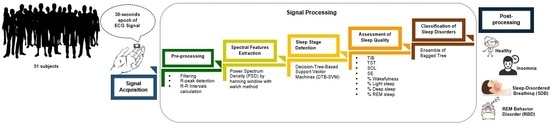
Automatic Sleep Disorders Classification Using Ensemble of Bagged Tree Based on Sleep Quality Features
Abstract
:1. Introduction
2. Related Works
- As mentioned above, different from most current ECG-based automatic sleep stage systems that applied a 5-minute epoch to observe the main frequency band of ECG signal, we perform a new pre-processing technique that suitable for 30-seconds epoch without detecting QRS. We take advantage that the proposed method more efficiently to be implemented in an embedded hardware device as a consideration of the complexity requirements and computational cost.
- A set of efficient ECG signal features (normalized LF and HF) is extracted by analyzing the HRV frequency band of Power Spectrum Density (PSD) using a Hanning window with the welch method, which is then used to identify the sleep stages.
- All sleep stage conditions are observed to patients and non-patients subjects. It is an essential factor for a robust sleep stage system.
- Since the proposed method present an effective and efficient in classifying sleep disorders of the elderly, we expect this method could be used as a general framework in modeling sleep disorders and become a fundamental model for future research. Moreover, we expect that our proposed method can contribute to the ICSD-3 study and aim as a new alternative for diagnosing the sleep disorders, besides using the questionnaire-based method, such as PSQI, BIQ, and RBDSQ.
3. Materials and Proposed Methods
3.1. Data Description
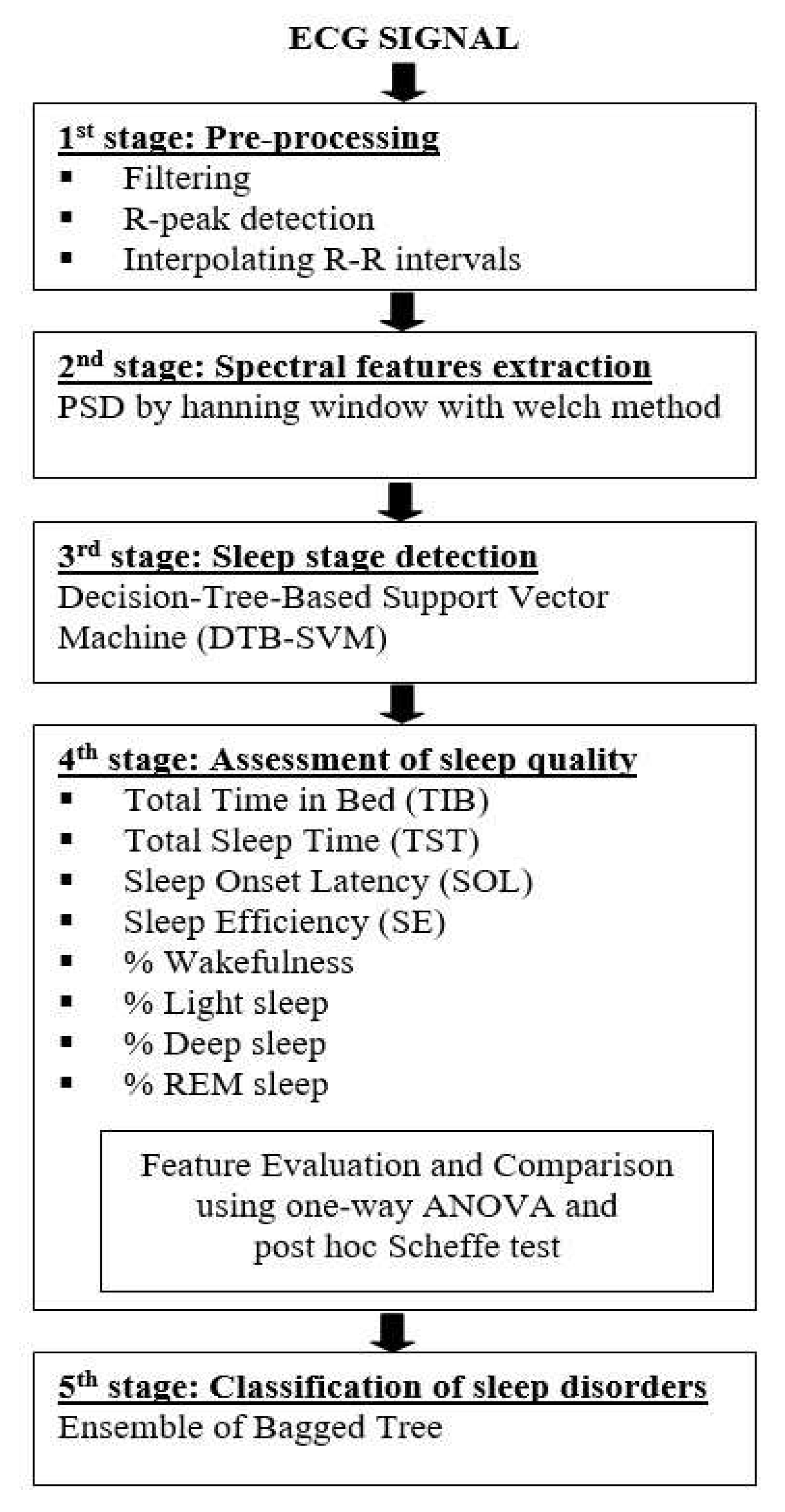
3.2. Pre-processing
- Removing the noise using a combination of the band-stop filter and moving average filter.
- Applying the simple R-peak detector using a 70% threshold from the maximum amplitude of the ECG signal to detect the R-peaks location and use it as a threshold.
- Interpolating the R-R intervals in the time domain using a cubic spline and re-sampled it at 2.5 Hz. A time-series signal should be re-sampled using frequency sampling at least two-times of the considered maximal frequency. It aims to estimate the HRV spectral (maximum HF band power is 0.4 Hz) for satisfying the Nyquist-Shannon sampling theorem.
3.3. Spectral Features Extraction
3.4. Sleep Stage Detection
3.5. Assessment of Sleep Quality
- TIB is the total investigation time or the total in-bed duration (in minutes). TIB has a clinical significance for diagnosing sufficient sleep.
- TST is the total sleep duration or total non-wake conditions (in minutes). TST has a relation for diagnosing the effects of medications, sleep deprivation, and medical condition.
- SOL is the duration time from the wake condition until getting the first non-wake condition (in minutes). SOL represents sleep time habits.
- SE is the ratio of total sleep duration (TST) and the total in-bed duration (TIB) (in percentage). In normal sleep conditions, it should at least 85% of TIB. SE represents how well the subject slept.
- The percentage of wakefulness stage is used to measures awake condition.
- The percentage of light sleep stage is associated with the transition between being awake and asleep. The increasing percentages of light sleep indicate the patient has a sleep disorder. Typically, the percentage of light sleep is around 55% of the total sleep duration for normal sleep conditions.
- The percentage of deep sleep stage is associated with the rebound sleep and side effect of medications. SDB disorders are indicated by increasing the percentages of deep sleep [46]. The normal percentage of deep sleep is around 20% of total sleep for normal sleep conditions.
- The percentage of the REM sleep stage is sensitive to the effect of medications and sleep deprivation. Nevertheless, the REM sleep stage remains approximately 25% of the total sleep in normal sleep conditions. The increasing percentages of the REM sleep indicate a recovery of sleep deprivation.
3.6. Classification of Sleep Disorders
4. Results and Discussion
4.1. Experimental Result
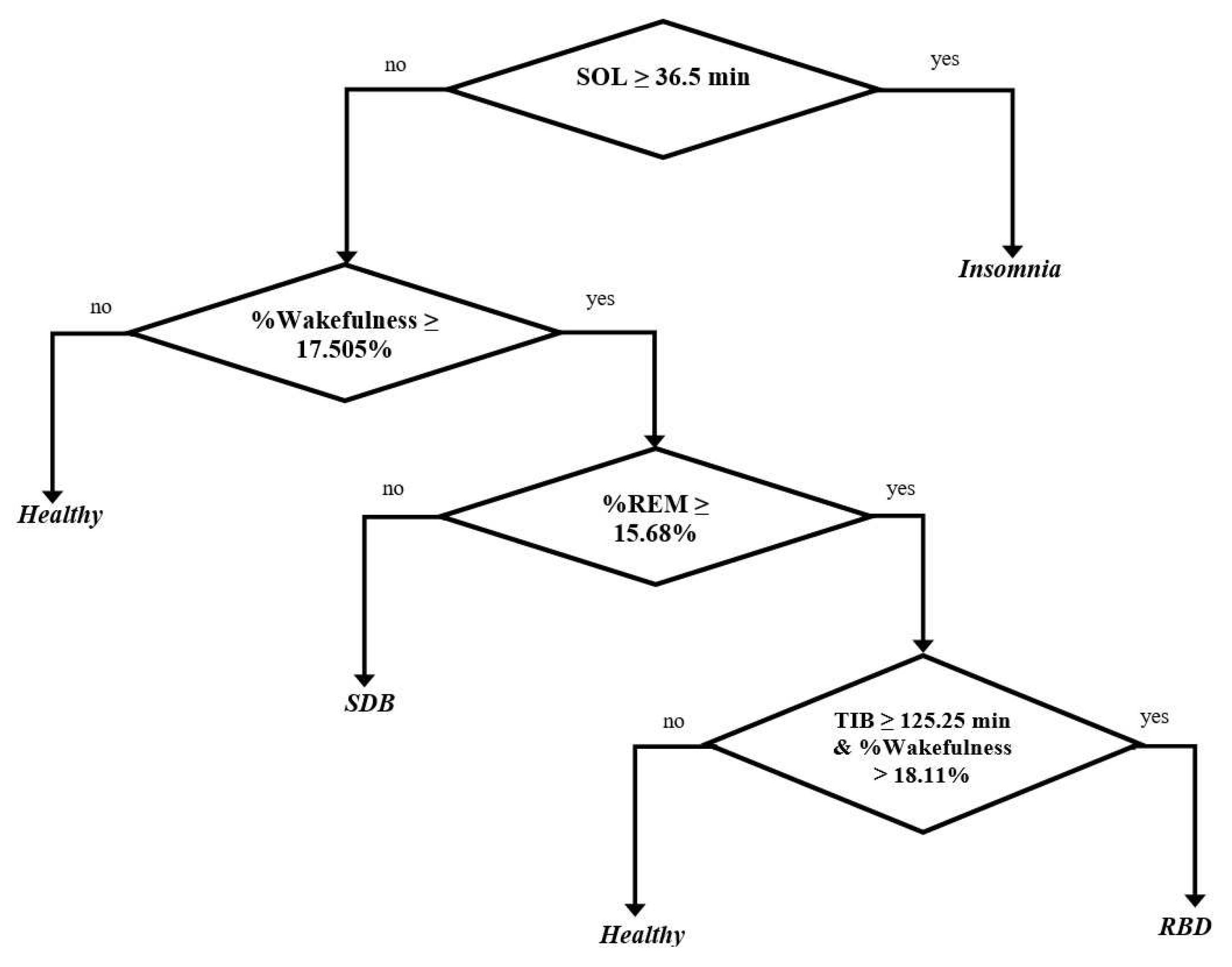
- The longest duration of SOL for SDB patients was below 36.5 min It corresponds to a clinical study that estimated the average longest duration of SDB patients was around 9.5 min [58], and we obtained below 36.5 min.
- The percentage of wakefulness was above 17.505%.
- The longest percentage of REM was below 15.68%. It corresponds to a clinical study that estimated the prevalence percentage of REM in the SDB patients (such as OSA patients) was around 13.5% [59], and we obtained below 15.68%.
- The longest duration of SOL for RBD patients was below 36.5 min. It corresponds to a related clinical study that estimated the average duration of SOL in 8 RBD patients was around 11.1 min [60], and we obtained below 36.5 min.
- The percentage of wakefulness was above 17.505%.
- The longest percentage of REM was above 15.68%. It corresponds to a related clinical study that estimated the characteristics percentage of REM in 94 RBD patients was around 22.4% [42], and we obtained above 15.68%.
- The duration of TIB and the percentage of wakefulness were generated simultaneously. Thus, the RBD patient is the subject that has the duration of TIB above 125.25 min and the percentage of wakefulness above 18.11%. It corresponds to a clinical study that estimated the average duration of TIB in 4 RBD patients is around 452.75 min [41], and we obtained above 125.25 min. In addition, [57] evaluated that healthy subjects have an average percentage of wakefulness around 10.55%, and we obtained below 17.505% and 18.11%.
4.2. Implementation Planning
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AASM | American Academy of Sleep Medicine |
| AHI | Apnea–Hypopnea Index |
| ANOVA | Analysis of Variance |
| ANS | Autonomic Nervous System |
| BIQ | Brief Insomnia Questionnaire |
| CAP | Cyclic Alternating Pattern |
| CART | Classification and Regression Trees |
| DT | Decision Tree |
| DTB-SVM | Decision-Tree-Based Support Vector Machine |
| ECG | Electrocardiography |
| EEG | Electroencephalogram |
| ELS | Ensemble Learning Systems |
| EMG | Electromyogram |
| EOG | Electrooculogram |
| FFT | Fast Fourier Transform |
| FN | False-Negative |
| FP | False-Positive |
| HF | High Frequency |
| HRV | Heart Rate Variability |
| ICSD | International Classification of Sleep disorders |
| LF | Low Frequency |
| NN | Neural Network |
| NREM | Non Rapid Eye Movement |
| OSA | Obstructive Sleep Apnea |
| PCA | Principal Component Analysis |
| PLMS | Periodic Leg Movement of Sleep |
| PLS | Partial Least Squares |
| PSD | Power Spectrum Density |
| PSG | Polysomnography |
| PSQI | Pittsburgh Sleep Quality Index |
| R&K | Rechtschaffen & Kales |
| RBD | REM Behavior Disorder |
| RBDSQ | REM Sleep Behavior Disorder Screening Questionnaire |
| REM | Rapid Eye Movement |
| RLS | Restless Leg Syndrome |
| RMSSD | Standard Derivation of NN Intervals |
| RSWA | REM Sleep Without Atonia |
| SDB | sleep-Disordered Breathing |
| SDNN | Standard Derivation of NN Intervals |
| SE | Sleep Efficiency |
| SOL | Sleep Onset Latency |
| SVM | Support Vector Machine |
| TIB | Total Time in Bed |
| TN | True Negative |
| TP | True Positive |
| TSP | Total Spectral Power |
| TST | Total Sleep Time |
| VLF | Very Low Frequency |
References
- Suzuki, K.; Miyamoto, M.; Hirata, K. Sleep disorders in the elderly: Diagnosis and management. J. Gen. Fam. Med. 2017, 18, 61–71. [Google Scholar] [CrossRef]
- Susan, K.R.; Sonia, A.I. Sleep disorders in the elderly. Indian J. Med Res. 2010, 131, 302–310. [Google Scholar]
- Ariel, B.N.; Sonia, A.I. Sleep disorders in the older adult- a mini review. Gerontology 2010, 56, 181–189. [Google Scholar]
- Hiroki, K.; Takashi, K.; Keiichi, F. Sleep-disordered breathing in the elderly: Is it distinct from that in the younger or middle-aged populations? J. Thorac. Dis. 2018, 10, 1102–1107. [Google Scholar]
- Mahlknecht, P.; Seppi, K.; Frauscher, B.; Kiechl, S.; Willeit, J.; Stockner, H.; Djamshidian, A.; Nocker, M.; Rastner, V.; Defrancesco, M.; et al. Probable RBD and association with neurodegenerative disease markers: A population-based study. Mov. Disord. 2015, 30, 1417–1421. [Google Scholar] [CrossRef]
- Kushida, C.A.; Kushida, C.A.; Littner, M.R.; Morgenthaler, T.; Alessi, C.A.; Bailey, D.; Coleman, J., Jr.; Friedman, L.; Hirshkowitz, M.; Kapen, S.; et al. Practice parameters for the indications for polysomnography and related procedures: An update 2005. Sleep 2005, 28, 499–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schumann, A.Y.; Bartsch, R.P.; Penzel, T.; Ivanov, P.C.; Kantelhardt, J.W. Aging effects on cardiac and respiratory dynamic in healthy subjects across sleep stages. Sleep 2010, 33, 943–955. [Google Scholar] [CrossRef] [PubMed]
- Kantelhardt, J.W.; Havlin, S.; Ivanov, P.C. Modeling transient correlations in heartbeat dynamics during sleep. Europhys. Lett. 2003, 62, 147–153. [Google Scholar] [CrossRef]
- Penzel, T.; Kantelhardt, J.W.; Bartsch, R.P.; Riedl, M.; Kraemer, J.F.; Wessel, N.; Garcia, C.; Glos, M.; Fietze, I.; Schöbel1, C. Modulations of heart rate, ECG, and cardio-respiratory coupling observed in polysomnography. Front. Physiol. 2016, 7, 460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebrahimi, F.; Setarehdan, S.K.; Ayala-Moyeda, J.; Nazeran, H. Automatic sleep staging using empirical mode decomposition, discrete wavelet transform, time-domain, and non linear dynamics features of heart rate variability signals. Comput. Methods Programs Biomed. 2013, 112, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Tompkins, W.J. A real-time QRS detection algorithm. IEEE Trans. Biomed. Eng. 1985, 32, 230–236. [Google Scholar] [CrossRef]
- Lin, C.C.; Chang, H.Y.; Huang, Y.H.; Yeh, C.Y. A novel wavelet-based algorithm for detection of QRS complex. Appl. Sci. 2019, 9, 2142. [Google Scholar] [CrossRef] [Green Version]
- Versace, F.; Mozzato, M.; Tona, G.D.M.; Cavallero, C.; Stegagno, L. Heart rate variability during sleep as a function of the sleep cycle. Biol. Psychol. 2003, 63, 149–162. [Google Scholar] [CrossRef]
- Bonnet, M.H.; Arand, D.L. Heart rate variability: Sleep stage, tome of night, and arousal influences. Electroencephalogr. Clin. Neurophysiol. 1997, 102, 390–396. [Google Scholar] [CrossRef]
- Long, X.; Fonseca, P.; Haakma, R.; Aarts, R.M.; Foussier, J. Spectral boundary adaption on heart rate variability for sleep and wake classification. Int. J. Artif. Intell. Tools 2014, 23, 1460002-1–1460002-20. [Google Scholar] [CrossRef]
- Aboalayon, K.A.I.; Faezipour, M.; Almuhammadi, W.S.; Moslehpour, S. Sleep stage classification using EEG signal analysis: A comprehensive survey and new investigation. Entropy 2016, 18, 272. [Google Scholar] [CrossRef]
- Werteni, H.; Yacoub, S.; Ellouze, N. An automatic sleep-wake classifier using ECG signals. IJCSI Int. J. Comput. Sci. Issues 2014, 11, 84–93. [Google Scholar]
- Khemiri, S.; Alouri, K.; Nacaeur, M.S. Automatic detection of slow wave sleep and REM-sleep stages using polysomnographic ECG signals. In Proceedings of the 8th International Multi-Conference on Systems, Signals and Devices, Sousse, Tunisia, 22–25 March 2011; pp. 1–4. [Google Scholar]
- Singh, J.; Sharma, R.K.; Gupta, A.K. A method of REM-NREM sleeps distinction using ECG signal for unobtrusive personal monitoring. Comput. Biol. Med. 2016, 78, 138–143. [Google Scholar] [CrossRef]
- Moriyama, Y.; Ogasawara, H.; Yuda, E.; Hayano, J. Assessment of sleep quality by electrocardiogram: Usefulness for risk stratification among hemodialysis patients with end-stage renal disease. In Proceedings of the IEEE 7th Global Conference on Consumer Electronics (GCCE), Nara, Japan, 9–12 October 2018; pp. 242–243. [Google Scholar]
- Bsoul, M.; Minn, H.; Nourani, M.; Gupta, G.; Tamil, L. Real-time sleep quality assessment using single-lead ECG and multi-stage SVM classifier. In Proceedings of the Annual International Conference of IEEE Engineering in Medicine and Biology Society, Buenos Aires, Argentina, 31 August–4 September 2010; pp. 1178–1181. [Google Scholar]
- Adnane, M.; Jiang, Z.; Yan, Z. Sleep-wake stages classification and sleep efficiency estimation using single-lead electrocardiogram. Expert Syst. Appl. 2012, 39, 1401–1413. [Google Scholar] [CrossRef]
- Balakrishman, G.; Burli, D.; Behbehani, K.; Burk, J.; Lucas, E. Comparison of a sleep quality index between normal and obstructive sleep apnea patients. In Proceedings of the Annual conference of IEEE Engineering in Medicine and Biology Society, Shanghai, China, 17–18 January 2006; pp. 1154–1157. [Google Scholar]
- Widasari, E.R.; Tanno, K.; Tamura, H. Automatic sleep quality assessment for obstructive sleep apnea patients based on HRV spectrum analysis. In Proceedings of the IEEE International Conference on Systems, Man, and Cybernetics (SMC), Bari, Italy, 6–9 October 2019. [Google Scholar]
- Mosquera-Lopez, C.; Leitschuh, J.; Condon, J.; Hagen, C.C.; Rajhbeharrysingh, U.; Hanks, C.; Jacobs, P.G. Design and evaluation of a non-contact bed-mounted sensing device for automated in-home detection of obstructive sleep apnea: A pilot study. Biosensors 2019, 9, 90. [Google Scholar] [CrossRef] [Green Version]
- Buysse, D.J.; Reynolds, C.F.; Monk, T.H.; Berman, S.R.; Kupfer, D.J. The pittsburgh sleep quality index: A new instrument for psychiatric practice and research. Psychiatry Res. 1989, 28, 193–213. [Google Scholar] [CrossRef]
- Kessler, R.C.; Coulouvrat, C.; Hajak, G.; Lakoma, M.D.; Roth, T.; Sampson, N.; Shahly, V.; Shillington, A.; Stephenson, J.J.; Walsh, J.K.; et al. Reliability and validity of the brief insomnia questionnaire in the america insomnia survey. Sleep 2010, 33, 1539–1549. [Google Scholar] [CrossRef] [PubMed]
- Stiasny-Koister, K.; Mayer, G.; Schäfer, S.; Möller, J.C.; Heinzel-Gutenbrunner, M.; Oertel, W.H. REM sleep behavior disorder screening questionnaire (RBDSQ)—A new diagnostic instrument. Mov. Disord. 2007, 22, 2386–2393. [Google Scholar] [CrossRef] [PubMed]
- Halsband, C.; Zapf, A.; Sixel-Döring, F.; Trenkwalder, C.; Mollenhauer, B. The REM sleep behavior disorder screening questionnaire is not valid in de novo parkinson’s disease. Mov. Disord. Clin. Pract. 2018, 5, 171–176. [Google Scholar] [CrossRef]
- Shahrbabaki, S.S.; Dissanayaka, C.; Patti, C.R.; Cvetkovic, D. Automatic detection of sleep arousal events from polysomnographic biosignals. In Proceedings of the IEEE Biomedical Circuits and Systems Conference (BioCAS), Atlanta, GA, USA, 22–24 December 2015. [Google Scholar]
- Terzano, M.G.; Parrino, L.; Sherieri, A.; Chervin, R.; Chokroverty, S.; Guilleminault, C.; Hirshkowitz, M.; Mahowald, M.; Moldofsky, H.; Rosa, A.; et al. Atlas, rules, and recording techniques for the scoring of cyclic alternating pattern (CAP) in human sleep. Sleep Med. 2001, 2, 537–553. [Google Scholar] [CrossRef]
- Rechtschaffen, A.K. A manual of standardized terminology, techniques, and scoring system for sleep stages of human subjects. In Public Health Service; Government Printing Office: Washington, DC, USA, 1968. [Google Scholar]
- Iber, C.; Ancoli-Israel, S.; Chesson, A. The AASM manual for the scoring of sleep and associated events: Rules, terminology and technical specifications. In Case Book of Sleep Medicine; American Academy of Sleep Medicine: Winchester, IL, USA, 2007. [Google Scholar]
- Heart rate variability: Standards of measurement, physiological interpretation and clinical use. Task Force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology. Circulation 1996, 93, 1043–1065.
- Fred, S.; Ginsberg, J.P. An overview of Heart rate variability metrics and norms. Front. Public Health 2017, 5, 1–17. [Google Scholar]
- Bartsch, R.P.; Schumann, A.Y.; Kantelhardt, J.W.; Penzel, T.; Ivanova, P.C. Phase transitions in physiologic coupling. Proc. Natl. Acad. Sci. USA 2012, 109, 10181–10186. [Google Scholar] [CrossRef] [Green Version]
- Schmitt, D.T.; Ivanov, P.C. Fractal scale-invariant and nonlinear properties of cardiac dynamics remain stable with advanced age: A new mechanistic picture of cardiac control in healthy elderly. Am. J. Physiol. Integr. Comp. Physiol. 2012, 293, 1923–1937. [Google Scholar] [CrossRef] [Green Version]
- Widasari, E.R.; Tanno, K.; Tamura, H. A new investigation of automatic sleep stage detection using decision-tree-based support vector machine and spectral features extraction. IEEJ Trans. Electron. Inf. Syst. 2019, 139, 820–827. [Google Scholar] [CrossRef]
- Sonka, K.; Sos, P.; Susta, M. Past and present in drug treatment of sleep disorders. Neuroendocrinol. Lett. 2014, 35, 186–197. [Google Scholar] [PubMed]
- Yang, J.S.C.; Nicholas, C.L.; Nixon, G.M.; Davey, M.J.; Anderson, V.; Walker, A.M.; Trinder, J.A.; Horne, R.S. Determining sleep quality in children with sleep disordered breathing: EEG spectral analysis compared with Conventional polysomnography. Sleep 2010, 33, 1165–1172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabater, L.; Gaig, C.; Gelpi, E.; Bataller, L.; Lewerenz, J.; Torres-Vega, E.; Contreras, A.; Giometto, B.; Compta, Y.; Embid, C.; et al. A novel NREM and REM parasomnia with sleep breathing disorder associated with antibodies against IgLON5: A case series, pathological features, and characterization of the antigen. Lancet Neurol. 2014, 13, 575–586. [Google Scholar] [CrossRef] [Green Version]
- Lee, G.L.; Choi, J.W.; Lee, Y.J.; Jeong, D.U. Depressed REM sleep behavior disorder patients are less likely to recall enacted dreams than non-depressed ones. Psychiatry Investig. 2016, 13, 227–231. [Google Scholar] [CrossRef] [Green Version]
- Shivastava, D.; Jung, S.; Saadat, M.; Sirohi, R.; Crewson, K. How to interpret the results of a sleep study. J. Community Hosp. Intern. Med. Perspect. 2014, 4, 1–4. [Google Scholar]
- Jung, D.W.; Hwang, S.H.; Yoon, H.N.; Lee, Y.J.G.; Jeong, D.U.; Park, K.S. Nocturnal awakening and sleep efficiency estimation using unobtrusively measured ballistocardiogram. IEEE Trans. Biomed. Eng. 2014, 61, 131–138. [Google Scholar] [CrossRef]
- Pressman, M.R.; Stoneham, M.A. Primer of polysomnogram interpretation. In Bookshelf; Devanathan, S., Ed.; Butterworth Heinemann: Boston, UK, 2002; p. 2030. [Google Scholar]
- Ratnavadivel, R.; Chau, N.; Stadler, D.; Yeo, A.; McEvoy, R.D.; Catcheside, P.G. Marked reduction in obstructive sleep apnea severity in slow wave sleep. J. Clin. Sleep Med. 2009, 5, 519–524. [Google Scholar] [CrossRef] [Green Version]
- Gilchrist, M.; Samuels, P. One-Way Analysis of Variance (ANOVA). In Statistics Teaching Resources; Loughborough University Mathematics Learning Support Centre and Coventry, University Mathematics Support Centre: Maynooth, Ireland, 2014. [Google Scholar]
- Williams, L.J.; Abdi, H. Post-Hoc Comparisons. In Encyclopedia of Research Design; Sage: Thousand Oaks, CA, USA, 2010. [Google Scholar]
- Zhou, Z.H. Ensemble Methods: Foundations and Algorithms. In Bookshelf; Chapman & Hall/CRC: Horsham Township, PA, USA, 2012. [Google Scholar]
- Dietterich, T.G. Ensemble Methods in Machine Learning. In Proceedings of the International Workshop on Multiple Classifier Systems, London, UK, 21–23 June 2000; pp. 1–5. [Google Scholar]
- Mousavi, R.; Eftekhari, M. A new ensemble learning methodology based on hybridization of classifier ensemble selection approaches. Appl. Soft Comput. J. 2015, 37, 652–666. [Google Scholar] [CrossRef]
- Mishra, P.K.; Yadav, A.; Pazoki, M. A Novel Fault Classification Scheme for Series Capacitor Compensated Transmission Line Based on Bagged Tree Ensemble Classifier. IEEE Access 2018, 6, 27373–27382. [Google Scholar] [CrossRef]
- Boudreau, P.; Yeh, W.H.; Dumont, G.A.; Boivin, D.B. Circadian variation of heart rate variability across sleep stages. Sleep 2013, 36, 1919–1928. [Google Scholar] [CrossRef] [Green Version]
- McCarter, S.J.; St Louis, E.K.; Boeve, B.F. REM Sleep Behavior Disorder and REM Sleep Without Atonia as an Early Manifestation of Degenerative Neurological Disease. Curr. Neurol. Neurosci. Rep. 2012, 12, 182–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sateia, M.J.; Buysse, D.J.; Krystal, A.D.; Neubauer, D.N.; Heald, J.L. Clinical practice guideline for the pharmacologic treatment of chronic insomnia in adults: Am american academy of sleep medicine clinical practice guideline. J. Clin. Sleep Med. 2017, 13, 307–349. [Google Scholar] [CrossRef] [PubMed]
- Schutte-Rodin, S.; Broch, L.; Buysse, D.; Dorsey, C.; Sateia, M. Clinical guideline for the evaluation and management of chronic insomnia in adults. J. Clin. Sleep Med. 2008, 5, 487–504. [Google Scholar] [CrossRef]
- Hertenstein, E.; Gabryelska, A.; Spiegelhalder, K.; Nissen, C.; Johann, A.F.; Umarova, R.; Riemann, D.; Baglioni, C.; Feige, B. References data for polysomnography-measured and subjective sleep in healthy adults. J. Clin. Sleep Med. 2018, 14, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Foster, S.N.; Hansen, S.L.; Scalzitti, N.J.; Matsangas, P.; Moore, B.A.; Mysliwiec, V. Residual excessive daytime sleepiness in patients with obstructive sleep apnea treated with positive airway pressure therapy. Sleep Breath. 2019, 6, 1–6. [Google Scholar]
- Alzoubaidi, M.; Mokhlesi, B. Obstructive sleep apnea during REM sleep: Clinical relevance and therapeutic implications. Curr. Opin. Pulm. Med. 2016, 22, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.H.; Koshal, V.B.; Schmidht, H.S. Use of pramipexole in REM sleep behavior disorder: Results from a case series. Sleep Med. 2006, 7, 418–423. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [Green Version]
- Espiritu, H.; Metsis, V. Automated detection of sleep disorder-related events from polysomnographic data. In Proceedings of the International Conference on Healthcare Informatics, Dallas, TX, USA, 21–23 October 2015; pp. 562–569. [Google Scholar]
- David, L.G.; Chaibi, S.; Ruby, P.; Aguera, P.E.; Eichenlaub, J.B.; Samet, M.; Kachouri, A.; Jerbi, K. Automatic detection of sleep disorders: Multi-class automatic classification algorithms based on Support Vector Machines. In Proceedings of the International Conference on Time Series and Forecasting, Granada, Sapin, 19–21 September 2018; pp. 1270–1280. [Google Scholar]




| Subject | Sleep Stage | |||
|---|---|---|---|---|
| Wakefulness | Light Sleep | Deep Sleep | REM Sleep | |
| Healthy | 400 | 2080 | 1185 | 1067 |
| (8.45%) | (43.95%) | (25.05%) | (22.55%) | |
| Insomnia | 1114 | 1227 | 437 | 411 |
| (34.93%) | (38.48%) | (13.71%) | (12.88%) | |
| SDB | 196 | 473 | 231 | 65 |
| (20.31%) | (49.02%) | (23.94%) | (6.73%) | |
| RBD | 1322 | 2084 | 1330 | 878 |
| (23.55%) | (37.12%) | (23.69%) | (16.64%) |
| Features | Insomnia | SDB | RBD |
|---|---|---|---|
| [39] | [40,41] | [41,42] | |
| Total Time in Bed (min) | Decrease | Decrease | Decrease |
| Total Sleep Time (min) | Decrease | Decrease | Decrease |
| Sleep Onset Latency (min) | Increase | Increase | Increase |
| Sleep Efficiency (%) | Decrease | Decrease | Decrease |
| Wakefulness (%) | Increase | Increase | Increase |
| Light sleep (%) | Increase | Increase | Increase |
| Deep sleep (%) | Decrease | Increase | Decrease |
| REM sleep (%) | Decrease | Decrease | Decrease |
| Automatic Scoring | |||||
|---|---|---|---|---|---|
| Wakefulness | Light Sleep | Deep Sleep | REM Sleep | ||
| wakefulness | 238 | 155 | 0 | 0 | |
| PSG | light sleep | 120 | 1873 | 0 | 0 |
| Scoring | deep sleep | 0 | 0 | 1185 | 0 |
| REM sleep | 0 | 0 | 169 | 894 |
| Automatic Scoring | |||||
|---|---|---|---|---|---|
| Wakefulness | Light Sleep | Deep Sleep | REM Sleep | ||
| wakefulness | 893 | 6 | 35 | 0 | |
| PSG | light sleep | 0 | 1145 | 0 | 0 |
| Scoring | deep sleep | 19 | 3 | 432 | 0 |
| REM sleep | 0 | 7 | 0 | 321 |
| Automatic Scoring | |||||
|---|---|---|---|---|---|
| Wakefulness | Light Sleep | Deep Sleep | REM Sleep | ||
| wakefulness | 1372 | 0 | 0 | 0 | |
| PSG | light sleep | 0 | 2329 | 0 | 0 |
| Scoring | deep sleep | 0 | 0 | 1418 | 20 |
| REM sleep | 51 | 1 | 15 | 898 |
| Automatic Scoring | |||||
|---|---|---|---|---|---|
| Wakefulness | Light Sleep | Deep Sleep | REM Sleep | ||
| PSG Scoring | wakefulness | 332 | 0 | 0 | 0 |
| light sleep | 0 | 620 | 0 | 0 | |
| deep sleep | 0 | 0 | 324 | 41 | |
| REM sleep | 0 | 0 | 106 | 31 |
| Healthy | Insomnia | SDB | RBD | p Value | |
|---|---|---|---|---|---|
| TIB | 0.000016 | ||||
| TST | 0.75 | ||||
| SOL | 0.000011 | ||||
| SE | 0.00083 | ||||
| % Wakefulness | 0.0000017 | ||||
| % Light sleep | 0.23 | ||||
| % Deep sleep | 0.19 | ||||
| % REM sleep | 0.00000046 |
| Automatic Scoring | |||||
|---|---|---|---|---|---|
| Healthy | Insomnia | SDB | RBD | ||
| Healthy | 13 | 0 | 0 | 3 | |
| PSG | Insomnia | 0 | 8 | 0 | 1 |
| Scoring | SDB | 0 | 0 | 3 | 1 |
| RBD | 2 | 0 | 0 | 20 | |
| Sensitivity (%) | Specificity (%) | Accuracy (%) | Cohen’s Kappa (%) | |
|---|---|---|---|---|
| Healthy | 81.25 | 93.94 | 89.80 | 0.71 |
| Insomnia | 88.89 | 100.00 | 97.78 | 0.71 |
| SDB | 75.00 | 100.00 | 97.78 | 0.60 |
| RBD | 90.91 | 82.76 | 86.27 | 0.73 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Widasari, E.R.; Tanno, K.; Tamura, H. Automatic Sleep Disorders Classification Using Ensemble of Bagged Tree Based on Sleep Quality Features. Electronics 2020, 9, 512. https://doi.org/10.3390/electronics9030512
Widasari ER, Tanno K, Tamura H. Automatic Sleep Disorders Classification Using Ensemble of Bagged Tree Based on Sleep Quality Features. Electronics. 2020; 9(3):512. https://doi.org/10.3390/electronics9030512
Chicago/Turabian StyleWidasari, Edita Rosana, Koichi Tanno, and Hiroki Tamura. 2020. "Automatic Sleep Disorders Classification Using Ensemble of Bagged Tree Based on Sleep Quality Features" Electronics 9, no. 3: 512. https://doi.org/10.3390/electronics9030512
APA StyleWidasari, E. R., Tanno, K., & Tamura, H. (2020). Automatic Sleep Disorders Classification Using Ensemble of Bagged Tree Based on Sleep Quality Features. Electronics, 9(3), 512. https://doi.org/10.3390/electronics9030512