Proton NMR Enables the Absolute Quantification of Aqueous Metabolites and Lipid Classes in Unique Mouse Liver Samples
Abstract
:1. Introduction
2. Results
2.1. Comparison of Extraction Methods
2.2. Absolute Quantification of Lipidic and Aqueous Metabolites
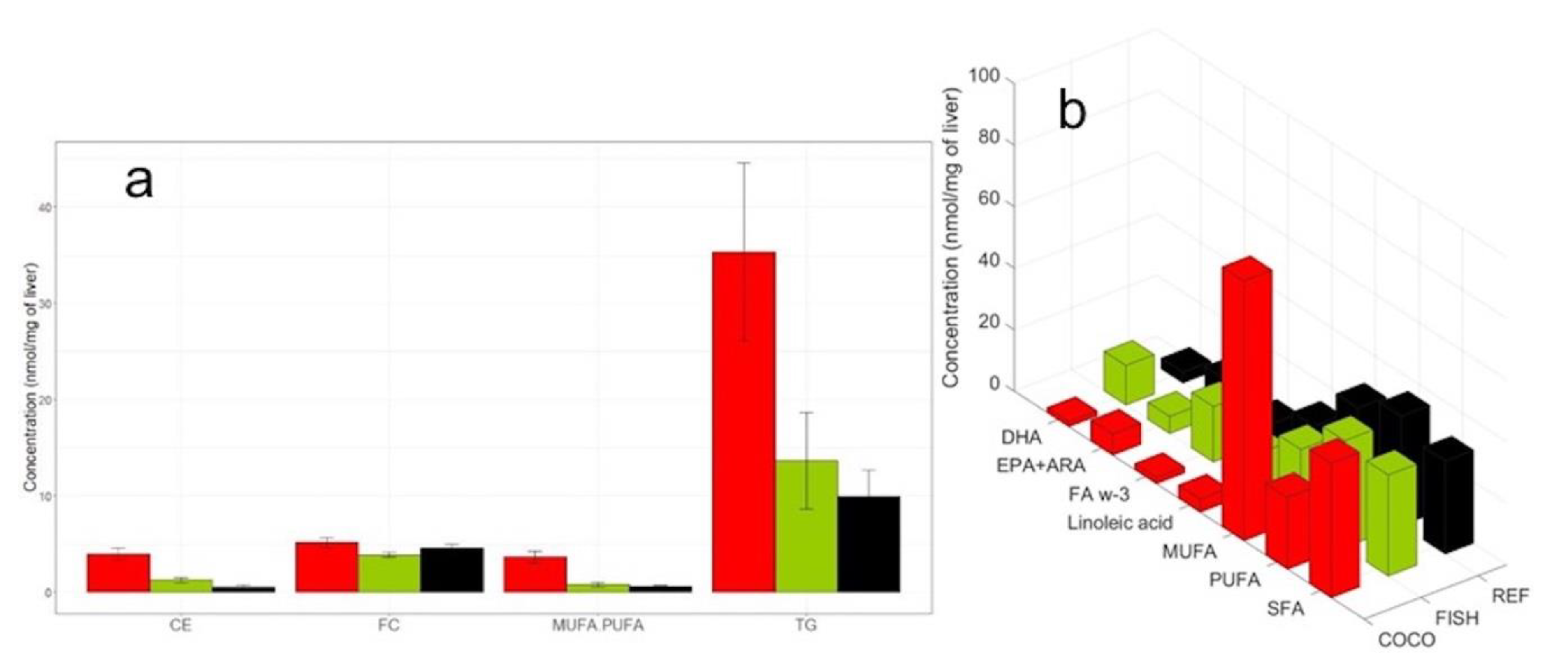
2.3. Analytical Validation with Liver Samples in a Dietary Intervention Study
2.3.1. Comparison with GC-FID Data
2.3.2. Comparison with LC-MS Data
2.3.3. Comparison with LipSpin Results
2.3.4. Biological Results
3. Discussion
3.1. Lipid Quantification: Comparison to Other Methods
3.2. Metabolic Differences between Livers of Mice Fed an Essential Fatty Acid-Deficient Diet or a Control Diet
3.3. Advantages and Limitations of 1H-NMR Spectroscopy for Metabolic Profiling in Liver
4. Materials and Methods
4.1. Animals
4.2. Extraction Procedure
4.3. GC Analysis of Neutral Lipids and Fatty Acids
4.4. HPLC-MS Analysis of Phospholipids
4.5. 1H-NMR Measurements
4.6. Data Processing and Multivariate Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.; Tacke, F.; Arrese, M.; Chander Sharma, B.; Mostafa, I.; Bugianesi, E.; Wai-Sun Wong, V.; Yilmaz, Y.; George, J.; Fan, J.; et al. Global Perspectives on Nonalcoholic Fatty Liver Disease and Nonalcoholic Steatohepatitis. Hepatology 2019, 69, 2672–2682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumas, M.-E.; Kinross, J.; Nicholson, J.K. Metabolic phenotyping and systems biology approaches to understanding metabolic syndrome and fatty liver disease. Gastroenterology 2014, 146, 46–62. [Google Scholar] [CrossRef] [PubMed]
- Mardinoglu, A.; Bjornson, E.; Zhang, C.; Klevstig, M.; Söderlund, S.; Ståhlman, M.; Adiels, M.; Hakkarainen, A.; Lundbom, N.; Kilicarslan, M.; et al. Personal model-assisted identification of NAD+ and glutathione metabolism as intervention target in NAFLD. Mol. Syst. Biol. 2017, 13, 916. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.-H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Moyec, L.; Triba, M.N.; Nahon, P.; Bouchemal, N.; Hantz, E.; Goossens, C.; Amathieu, R.; Savarin, P. Nuclear magnetic resonance metabolomics and human liver diseases: The principles and evidence associated with protein and carbohydrate metabolism. Biomed. Rep. 2017, 6, 387–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amathieu, R.; Triba, M.N.; Goossens, C.; Bouchemal, N.; Nahon, P.; Savarin, P.; Le Moyec, L. Nuclear magnetic resonance based metabolomics and liver diseases: Recent advances and future clinical applications. World J. Gastroenterol. 2016, 22, 417–426. [Google Scholar] [CrossRef]
- Jiang, L.; Si, Z.H.; Li, M.H.; Zhao, H.; Fu, Y.H.; Xing, Y.X.; Hong, W.; Ruan, L.Y.; Li, P.M.; Wang, J.S. 1H NMR-based metabolomics study of liver damage induced by ginkgolic acid (15:1) in mice. J. Pharm. Biomed. Anal. 2017, 136, 44–54. [Google Scholar] [CrossRef]
- Dagla, I.; Benaki, D.; Baira, E.; Lemonakis, N.; Poudyal, H.; Brown, L.; Tsarbopoulos, A.; Skaltsounis, A.L.; Mikros, E.; Gikas, E. Alteration in the liver metabolome of rats with metabolic syndrome after treatment with Hydroxytyrosol. A Mass Spectrometry and Nuclear Magnetic Resonance-based metabolomics study. Talanta 2018, 178, 246–257. [Google Scholar] [CrossRef]
- Bonvallot, N.; Canlet, C.; Blas-Y-Estrada, F.; Gautier, R.; Tremblay-Franco, M.; Chevolleau, S.; Cordier, S.; Cravedi, J.P. Metabolome disruption of pregnant rats and their offspring resulting from repeated exposure to a pesticide mixture representative of environmental contamination in Brittany. PLoS ONE 2018, 13, e0198448. [Google Scholar] [CrossRef]
- Chen, M.; Zheng, H.; Xu, M.; Zhao, L.; Zhang, Q.; Song, J.; Zhao, Z.; Lu, S.; Weng, Q.; Wu, X.; et al. Changes in hepatic metabolic profile during the evolution of STZ-induced diabetic rats via an 1H NMR-based metabonomic investigation. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, S.; Sengupta, A.; Sharma, S.; Sonawat, H.M. Metabolic fingerprints of serum, brain, and liver are distinct for mice with cerebral and noncerebral malaria: A 1H NMR spectroscopy-based metabonomic study. J. Proteome Res. 2012, 11, 4992–5004. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Rodado, V.; Nicoli, E.R.; Probert, F.; Smith, D.A.; Morris, L.; Wassif, C.A.; Platt, F.M.; Grootveld, M. 1H NMR-Linked Metabolomics Analysis of Liver from a Mouse Model of NP-C1 Disease. J. Proteome Res. 2016, 15, 3511–3527. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Cai, A.; Shu, Q.; Niu, Y.; Xu, P.; Li, C.; Lin, L.; Gao, H. Tissue-Specific Metabolomics Analysis Identifies the Liver as a Major Organ of Metabolic Disorders in Amyloid Precursor Protein/Presenilin 1 Mice of Alzheimer’s Disease. J. Proteome Res. 2019, 18, 1218–1227. [Google Scholar] [CrossRef] [PubMed]
- Fernando, H.; Bhopale, K.K.; Kondraganti, S.; Kaphalia, B.S.; Ansari, G.A.S. Lipidomic Changes in Rat Liver after Long-Term Exposure to Ethanol. Toxicol. Appl. Pharmacol. 2011, 255, 127–137. [Google Scholar] [CrossRef] [Green Version]
- Fernando, H.; Bhopale, K.K.; Kondraganti, S.S.; Kaphalia, B.S.; Ansari, G.A.S. Alcohol-Induced Hepatic Steatosis: A Comparative Study to Identify Possible Indicator(s) of Alcoholic Fatty Liver Disease. J. Drug Alcohol Res. 2018, 7. [Google Scholar] [CrossRef] [Green Version]
- Fernando, H.; Kondraganti, S.; Bhopale, K.K.; Volk, D.E.; Neerathilingam, M.; Kaphalia, B.S.; Luxon, B.A.; Boor, P.J.; Ansari, G.A.S. 1H and 31P NMR Lipidome of Ethanol-Induced Fatty Liver. Alcohol. Clin. Exp. Res. 2010, 34, 1937–1947. [Google Scholar] [CrossRef] [Green Version]
- Cabaton, N.J.; Poupin, N.; Canlet, C.; Tremblay-Franco, M.; Audebert, M.; Cravedi, J.P.; Riu, A.; Jourdan, F.; Zalko, D. An Untargeted Metabolomics Approach to Investigate the Metabolic Modulations of HepG2 Cells Exposed to Low Doses of Bisphenol A and 17β-Estradiol. Front. Endocrinol. 2018, 9, 571. [Google Scholar] [CrossRef]
- Wei, F.; Lamichhane, S.; Orešič, M.; Hyötyläinen, T. Lipidomes in health and disease: Analytical strategies and considerations. TrAC Trends Anal. Chem. 2019, 120, 115664. [Google Scholar] [CrossRef]
- Khoury, S.; Canlet, C.; Lacroix, M.Z.; Berdeaux, O.; Jouhet, J.; Bertrand-Michel, J. Quantification of Lipids: Model, Reality, and Compromise. Biomolecules 2018, 8, 174. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Granados, B.; Morales, J.M.; Rodrigo, J.M.; Del Olmo, J.; Serra, M.A.; Ferrández, A.; Celda, B.; Monleón, D. Metabolic profile of chronic liver disease by NMR spectroscopy of human biopsies. Int. J. Mol. Med. 2011, 27, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Cobbold, J.F.L.; Anstee, Q.M.; Goldin, R.D.; Williams, H.R.T.; Matthews, H.C.; North, B.V.; Absalom, N.; Thomas, H.C.; Thursz, M.R.; Cox, R.D.; et al. Phenotyping murine models of non-alcoholic fatty liver disease through metabolic profiling of intact liver tissue. Clin. Sci. Lond. Engl. 2009, 116, 403–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teilhet, C.; Morvan, D.; Joubert-Zakeyh, J.; Biesse, A.S.; Pereira, B.; Massoulier, S.; Dechelotte, P.; Pezet, D.; Buc, E.; Lamblin, G.; et al. Specificities of Human Hepatocellular Carcinoma Developed on Non-Alcoholic Fatty Liver Disease in Absence of Cirrhosis Revealed by Tissue Extracts 1H-NMR Spectroscopy. Metabolites 2017, 7, 49. [Google Scholar] [CrossRef] [PubMed]
- Vinaixa, M.; Rodríguez, M.A.; Rull, A.; Beltrán, R.; Bladé, C.; Brezmes, J.; Cañellas, N.; Joven, J.; Correig, X. Metabolomic assessment of the effect of dietary cholesterol in the progressive development of fatty liver disease. J. Proteome Res. 2010, 9, 2527–2538. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Wu, H.; Tjeerdema, R.S.; Viant, M.R. Evaluation of metabolite extraction strategies from tissue samples using NMR metabolomics. Metabolomics 2007, 3, 55–67. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar]
- Beckonert, O.; Keun, H.C.; Ebbels, T.M.D.; Bundy, J.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007, 2, 2692–2703. [Google Scholar] [CrossRef]
- Ulrich, E.L.; Akutsu, H.; Doreleijers, J.F.; Harano, Y.; Ioannidis, Y.E.; Lin, J.; Livny, M.; Mading, S.; Maziuk, D.; Miller, Z.; et al. BioMagResBank. Nucleic Acids Res. 2008, 36, D402–D408. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef]
- Reis, A.; Rudnitskaya, A.; Blackburn, G.J.; Mohd Fauzi, N.; Pitt, A.R.; Spickett, C.M. A comparison of five lipid extraction solvent systems for lipidomic studies of human LDL. J. Lipid Res. 2013, 54, 1812–1824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Southam, A.D.; Hines, A.; Viant, M.R. High-throughput tissue extraction protocol for NMR-and MS-based metabolomics. Anal. Biochem. 2008, 372, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Viant, M.R.; Ebbels, T.M.D.; Beger, R.D.; Ekman, D.R.; Epps, D.J.T.; Kamp, H.; Leonards, P.E.G.; Loizou, G.D.; MacRae, J.I.; van Ravenzwaay, B.; et al. Use cases, best practice and reporting standards for metabolomics in regulatory toxicology. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bharti, S.K.; Roy, R. Quantitative 1H NMR spectroscopy. TrAC Trends Anal. Chem. 2012, 35, 5–26. [Google Scholar] [CrossRef]
- Vidal, N.P.; Manzanos, M.J.; Goicoechea, E.; Guillén, M.D. Quality of farmed and wild sea bass lipids studied by (1) H NMR: Usefulness of this technique for differentiation on a qualitative and a quantitative basis. Food Chem. 2012, 135, 1583–1591. [Google Scholar] [CrossRef] [PubMed]
- Barrilero, R.; Gil, M.; Amigó, N.; Dias, C.B.; Wood, L.G.; Garg, M.L.; Ribalta, J.; Heras, M.; Vinaixa, M.; Correig, X. LipSpin: A New Bioinformatics Tool for Quantitative 1H NMR Lipid Profiling. Anal. Chem. 2018, 90, 2031–2040. [Google Scholar] [CrossRef]
- Ducheix, S.; Montagner, A.; Polizzi, A.; Lasserre, F.; Marmugi, A.; Bertrand-Michel, J.; Podechard, N.; Al Saati, T.; Chétiveaux, M.; Baron, S.; et al. Essential fatty acids deficiency promotes lipogenic gene expression and hepatic steatosis through the liver X receptor. J. Hepatol. 2013, 58, 984–992. [Google Scholar] [CrossRef]
- Jiang, C.; Yang, K.; Yang, L.; Miao, Z.; Wang, Y.; Zhu, H. A 1H NMR-Based Metabonomic Investigation of Time-Related Metabolic Trajectories of the Plasma, Urine and Liver Extracts of Hyperlipidemic Hamsters. PLoS ONE 2013, 8, e66786. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Vosegaard, T.; Guo, Z. Applications of nuclear magnetic resonance in lipid analyses: An emerging powerful tool for lipidomics studies. Prog. Lipid Res. 2017, 68, 37–56. [Google Scholar] [CrossRef]
- Botolin, D.; Wang, Y.; Christian, B.; Jump, D.B. Docosahexaneoic acid (22:6, n-3) regulates rat hepatocyte SREBP-1 nuclear abundance by Erk-and 26S proteasome-dependent pathways. J. Lipid Res. 2006, 47, 181–192. [Google Scholar] [CrossRef] [Green Version]
- Jump, D.B. Fatty acid regulation of hepatic lipid metabolism. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 115–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dentin, R.; Benhamed, F.; Pégorier, J.P.; Foufelle, F.; Viollet, B.; Vaulont, S.; Girard, J.; Postic, C. Polyunsaturated fatty acids suppress glycolytic and lipogenic genes through the inhibition of ChREBP nuclear protein translocation. J. Clin. Investig. 2005, 115, 2843–2854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, M.S.; Dorn, C.; Saugspier, M.; Hellerbrand, C.; Oefner, P.J.; Gronwald, W. Discrimination of steatosis and NASH in mice using nuclear magnetic resonance spectroscopy. Metabolomics 2011, 7, 237–246. [Google Scholar] [CrossRef]
- Yang, Y.; Li, C.; Nie, X.; Feng, X.; Chen, W.; Yue, Y.; Tang, H.; Deng, F. Metabonomic studies of human hepatocellular carcinoma using high-resolution magic-angle spinning 1H NMR spectroscopy in conjunction with multivariate data analysis. J. Proteome Res. 2007, 6, 2605–2614. [Google Scholar] [CrossRef]
- Schofield, Z.; Reed, M.A.; Newsome, P.N.; Adams, D.H.; Günther, U.L.; Lalor, P.F. Changes in human hepatic metabolism in steatosis and cirrhosis. World J. Gastroenterol. 2017, 23, 2685–2695. [Google Scholar] [CrossRef]
- Chen, I.S.; Shen, C.S.J.; Sheppard, A.J. Comparison of methylene chloride and chloroform for the extraction of fats from food products. J. Am. Oil Chem. Soc. 1981, 58, 599–601. [Google Scholar] [CrossRef]
- Barrans, A.; Collet, X.; Barbaras, R.; Jaspard, B.; Manent, J.; Vieu, C.; Chap, H.; Perret, B. Hepatic lipase induces the formation of pre-beta 1 high density lipoprotein (HDL) from triacylglycerol-rich HDL2. A study comparing liver perfusion to in vitro incubation with lipases. J. Biol. Chem. 1994, 269, 11572–11577. [Google Scholar]
- Lillington, J.M.; Trafford, D.J.; Makin, H.L. A rapid and simple method for the esterification of fatty acids and steroid carboxylic acids prior to gas-liquid chromatography. Clin. Chim. Acta Int. J. Clin. Chem. 1981, 111, 91–98. [Google Scholar] [CrossRef]
- Wold, S.; Antti, H.; Lindgren, F.; Öhman, J. Orthogonal signal correction of near-infrared spectra. Chemom. Intell. Lab. Syst. 1998, 44, 175–185. [Google Scholar] [CrossRef]
- McCombie, G.; Browning, L.M.; Titman, C.M.; Song, M.; Shockcor, J.; Jebb, S.A.; Griffin, J.L. omega-3 oil intake during weight loss in obese women results in remodelling of plasma triglyceride and fatty acids. Metab. Off. J. Metab. Soc. 2009, 5, 363–374. [Google Scholar] [CrossRef] [Green Version]
- Lapins, M.; Eklund, M.; Spjuth, O.; Prusis, P.; Wikberg, J.E. Proteochemometric modeling of HIV protease susceptibility. BMC Bioinform. 2008, 9, 181. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Lipid Species | External Standard (TSP) | Internal Standard (TMS) | ||||
|---|---|---|---|---|---|---|
| Pearson’s r | p-value a | Slope | Pearson’s r | p-value a | Slope | |
| Total FA | 0.98 | 0.002 | 1.3 | 0.82 | 0.089 | 0.8 |
| Saturated FA | 0.98 | 0.002 | 1.2 | 0.97 | 0.006 | 1.0 |
| ω-3 FA | 0.98 | 0.002 | 1.0 | 0.94 | 0.019 | 1.8 |
| MUFA | 0.98 | 0.001 | 1.2 | 0.93 | 0.019 | 2.0 |
| PUFA | 0.92 | 0.03 | 0.8 | 0.95 | 0.015 | 1.7 |
| UFA | 0.94 | 0.017 | 0.8 | 0.95 | 0.014 | 1.7 |
| DHA | 0.99 | 0.0001 | 1.1 | 0.93 | 0.024 | 1.9 |
| Linoleic acid | 0.98 | 0.002 | 0.9 | 0.93 | 0.025 | 1.7 |
| TC | 0.99 | 0.0003 | 1.2 | 0.89 | 0.04 | 2.5 |
| FC | 0.95 | 0.011 | 1.0 | 0.89 | 0.04 | 1.7 |
| CE | 0.99 | 0.00007 | 0.8 | 0.98 | 0.004 | 1.4 |
| Triglycerides | 0.99 | 0.001 | 1.2 | 0.98 | 0.004 | 1.0 |
| PC | 0.95 | 0.012 | 1.2 | 0.96 | 0.009 | 1.2 |
| PE | 0.55 | 0.33 | 0.1 | 0.99 | 0.0001 | 0.2 |
| SM | 0.94 | 0.018 | 0.8 | 0.98 | 0.002 | 0.9 |
| Total PL | 0.91 | 0.03 | 0.9 | 0.95 | 0.015 | 1.0 |
| Lipid Species | External Standard (TSP) | Internal Standard (TMS) | ||||
|---|---|---|---|---|---|---|
| Pearson’s r | p-value a | Slope | Pearson’s r | p-value a | Slope | |
| Total FA | 0.99 | 5 × 10−5 | 1.0 | 0.98 | 2 × 10−3 | 1.2 |
| Saturated FA | 0.99 | 2 × 10−5 | 1.0 | 0.98 | 2 × 10−3 | 1.2 |
| ω-3 FA | 0.99 | 5 × 10−5 | 1.0 | 0.98 | 2 × 10−3 | 1.0 |
| MUFA | 0.99 | 3 × 10−5 | 1.1 | 0.99 | 4 × 10−4 | 1.0 |
| PUFA | 0.99 | 5 × 10−4 | 1.0 | 0.97 | 6 × 10−3 | 1.5 |
| UFA | 0.99 | 3 × 10−4 | 1.0 | 0.97 | 5 × 10−3 | 1.4 |
| DHA | 0.99 | 1 × 10−5 | 1.1 | 0.99 | 4 × 10−5 | 0.7 |
| Linoleic acid | 0.99 | 8 × 10−6 | 0.9 | 0.99 | 2 × 10−4 | 1.0 |
| TC | 0.98 | 2 × 10−3 | 1.1 | 0.96 | 9 × 10−3 | 1.6 |
| FC | 0.99 | 6 × 10−4 | 1.0 | 0.93 | 2 × 10−2 | 1.5 |
| CE | 0.99 | 8 × 10−4 | 0.9 | 0.98 | 2 × 10−3 | 0.9 |
| Triglycerides | 0.99 | 3 × 10−6 | 1.0 | 0.99 | 7 × 10−4 | 1.0 |
| PC | 0.99 | 5 × 10−5 | 1.2 | 0.99 | 2 × 10−4 | 1.1 |
| PE | 0.98 | 3 × 10−3 | 0.6 | 0.91 | 3 × 10−2 | 0.5 |
| SM | 0.99 | 8 × 10−4 | 0.9 | 0.96 | 8 × 10−3 | 0.8 |
| Total PL | 0.99 | 3 × 10−5 | 1.0 | 0.99 | 2 × 10−5 | 1.2 |
| Lipid Species | Pearson’s r | p-value a |
|---|---|---|
| Total FA | 0.93 | 8.8 × 10−8 |
| Saturated FA | 0.85 | 1.43 × 10−5 |
| ω-3 FA | 0.80 | 1.1 × 10−4 |
| MUFA | 0.96 | 4.5 × 10−10 |
| PUFA | 0.80 | 9.6 × 10−5 |
| ARA+EPA | 0.69 | 2 × 10−3 |
| DHA | 0.95 | 3.4 × 10−9 |
| Linoleic acid | 0.96 | 6.7 × 10−10 |
| MUFA/PUFA | 0.89 | 1.3 × 10−6 |
| Total cholesterol | 0.99 | 1.2 × 10−14 |
| Free cholesterol | 0.91 | 1.3 × 10−6 |
| Cholesterol ester | 0.98 | 6.6 × 10−12 |
| Triglycerides | 0.98 | 7.7 × 10−12 |
| Concentration Ratio | PE | PC + LPC | SM | Total PL, Except LPC | ||||
|---|---|---|---|---|---|---|---|---|
| LC-MS | NMR | LC-MS | NMR | LC-MS | NMR | LC-MS | NMR | |
| COCO/REF | 0.94 | 0.95 | 1.07 | 1.01 | 0.90 | 0.81 | 1.05 | 1.03 |
| FISH/REF | 1.24 | 1.17 | 1.13 | 1.06 | 1.00 | 1.10 | 1.12 | 1.08 |
| Lipid Species | Pearson’s r | p-value a |
|---|---|---|
| Saturated FA | 0.66 | 5.7 × 10−3 |
| ω-3 FA | 0.98 | 7.4 × 10−12 |
| MUFA | 0.97 | 3.0 × 10−10 |
| ARA+EPA | 0.001 | 0.99 |
| DHA | 0.31 | 0.24 |
| Linoleic acid | 0.95 | 2.8 × 10−8 |
| Free cholesterol | 0.54 | 0.031 |
| Esterified cholesterol | 0.80 | 1.8 × 10−4 |
| Triglycerides | 0.97 | 9.2 × 10−10 |
| Total phospholipids | 0.27 | 0.32 |
| PE | 0.58 | 0.017 |
| SM | 0.14 | 0.59 |
| PC+LPC | 0.37 | 0.15 |
| Metabolites a | FC b COCO | FC b FISH |
|---|---|---|
| FA (CH2)n | 0.71 * | 0.82 |
| EPA+ARA | 0.67 * | 0.80 |
| FA CH3 | 0.73 * | 0.80 |
| Linoleic acid | 0.28 * | 0,92 |
| MUFA | 1.99 * | 0.84 |
| PC+LPC+SM | 0.61 * | 0.94 |
| PL (Except LPC) | 0.65 * | 1.15 |
| TG | 1.63 * | 0.98 |
| 3-Hydroxybutyrate | 1.21 * | 1.10 |
| Alanine | 1.30 * | 0.97 |
| Choline | 1.02 | 1.68 * |
| Glucose | 0.88 * | 0.88 * |
| Glutamine | 1 | 1.18 * |
| Glutathione | 1.07 | 1.30 * |
| GPC | 1.39 * | 1.61 * |
| Inosine | 1.24 * | 1 |
| Lactate | 1.27 * | 1.14 * |
| Leucine | 1.23 * | 1.11 * |
| Phenylalanine | 1.32 * | 1.15 |
| Succinate | 1.50 * | 1.38 * |
| Threonine | 1.28 * | 1.03 |
| Tyrosine | 1.47 * | 1.28 * |
| Valine | ↑1.21 * | 1.07 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amiel, A.; Tremblay-Franco, M.; Gautier, R.; Ducheix, S.; Montagner, A.; Polizzi, A.; Debrauwer, L.; Guillou, H.; Bertrand-Michel, J.; Canlet, C. Proton NMR Enables the Absolute Quantification of Aqueous Metabolites and Lipid Classes in Unique Mouse Liver Samples. Metabolites 2020, 10, 9. https://doi.org/10.3390/metabo10010009
Amiel A, Tremblay-Franco M, Gautier R, Ducheix S, Montagner A, Polizzi A, Debrauwer L, Guillou H, Bertrand-Michel J, Canlet C. Proton NMR Enables the Absolute Quantification of Aqueous Metabolites and Lipid Classes in Unique Mouse Liver Samples. Metabolites. 2020; 10(1):9. https://doi.org/10.3390/metabo10010009
Chicago/Turabian StyleAmiel, Aurélien, Marie Tremblay-Franco, Roselyne Gautier, Simon Ducheix, Alexandra Montagner, Arnaud Polizzi, Laurent Debrauwer, Hervé Guillou, Justine Bertrand-Michel, and Cécile Canlet. 2020. "Proton NMR Enables the Absolute Quantification of Aqueous Metabolites and Lipid Classes in Unique Mouse Liver Samples" Metabolites 10, no. 1: 9. https://doi.org/10.3390/metabo10010009
APA StyleAmiel, A., Tremblay-Franco, M., Gautier, R., Ducheix, S., Montagner, A., Polizzi, A., Debrauwer, L., Guillou, H., Bertrand-Michel, J., & Canlet, C. (2020). Proton NMR Enables the Absolute Quantification of Aqueous Metabolites and Lipid Classes in Unique Mouse Liver Samples. Metabolites, 10(1), 9. https://doi.org/10.3390/metabo10010009







