Metabolic Signatures of the Exposome—Quantifying the Impact of Exposure to Environmental Chemicals on Human Health
Abstract
:1. Introduction
2. Exposomics Approach to Study Health and Disease
3. Analytical Methodologies
- Analytical techniques for comprehensive chemical profiling (exogeneous and endogenous compounds);
- Effect-directed analysis of the drivers of toxicity;
- Advanced bioinformatics methods, in order to integrate the highly complex data and to identify effect-based markers of exposure.
3.1. Analytical Methods
3.2. Data Analysis
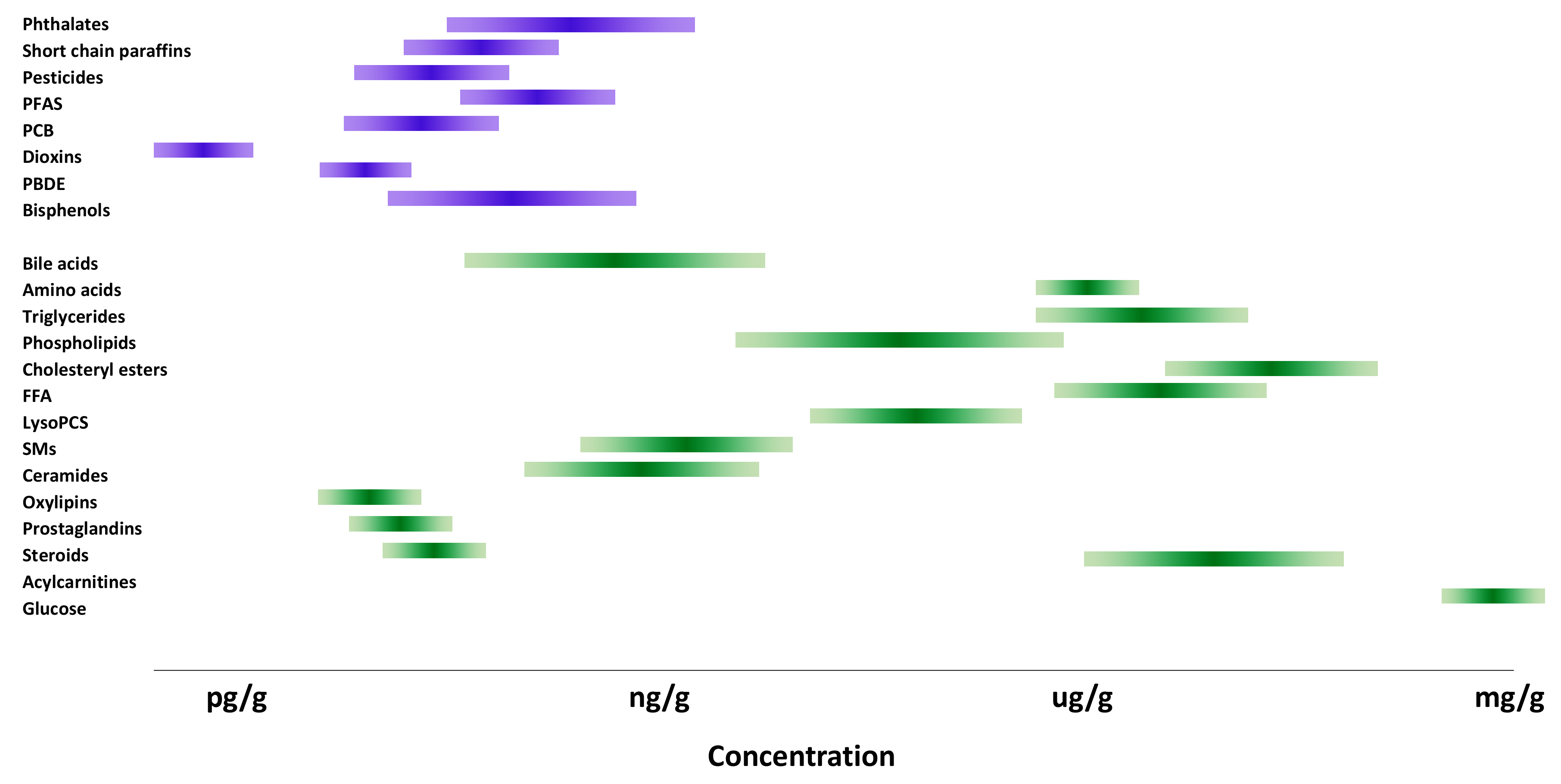
4. Metabolic Markers of Exposure to Environmental Chemicals
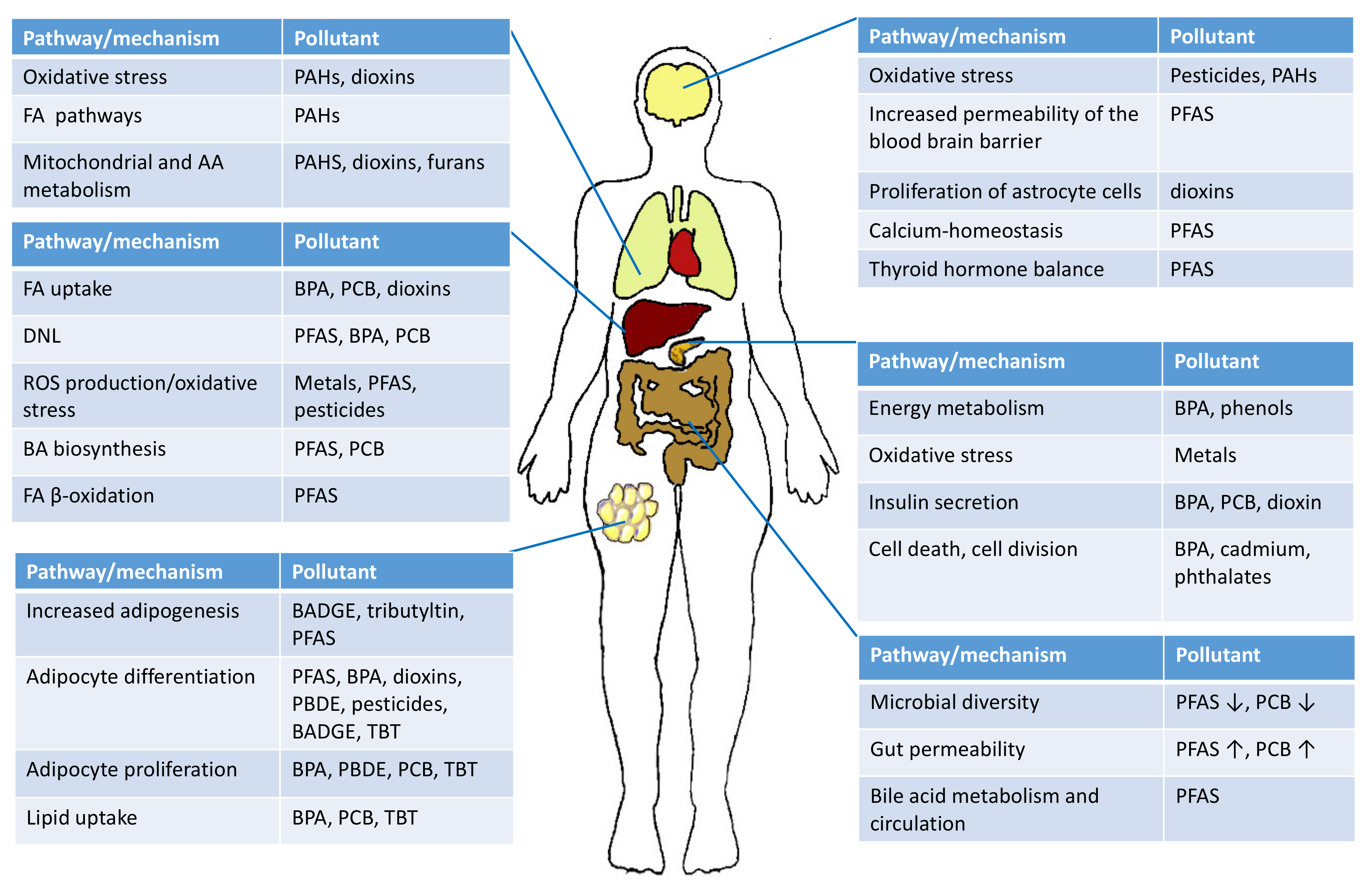
4.1. Lipid Metabolism in Liver and Adipose Tissue
4.2. Bile Acids
4.3. Amino Acid Metabolism
4.4. Energy Metabolism and Oxidative Stress
4.5. Impact of Environmental Exposure on Metabolome via Gut Microbiota
5. Health Impacts of Environmental Exposure
5.1. Exposures during Early Development
5.2. Type 2 Diabetes, NAFLD, Obesity, and Metabolic Syndrome
5.3. Type 1 Diabetes
5.4. Allergy and Obstructive Lung Disease
6. Conclusions
Funding
Conflicts of Interest
Abbreviations
| 12-oxo-LCA | 12-Oxolithocholic acid |
| 7-oxo-DCA | 7-Oxodeoxycholic acid |
| 7-oxo-HCA | 7-Oxohyocholic acid |
| ANN | Artificial neural networks |
| AT | Adipose tissue |
| BA | Bile acids |
| BAAT | Bile acid CoA: amino acid N-acetyltransferase |
| BACS | Bile acid-coenzyme A synthase |
| BADGE | Bisphenol A diglycidyl ether |
| BPA | Bisphenol A |
| BSE | Bile salt export pump |
| CA | Cholic acid |
| CDCA | Chenodeoxycholic acid |
| CE | Cholesteryl ester |
| Cer | Ceramide |
| CYP7A1 | Cytochrome P450 family 7 subfamily A member 1 |
| DCA | Deoxycholic acid |
| DDA | Data-dependent acquisition |
| DDE | Dichlorodiphenyldichloroethylene |
| DES | Diethylstilbestrol |
| DG | Diradylglycerol |
| DHCA | 3α,7α-Dihydroxycholestanoic acid |
| DIA | Data independent acquisition |
| EFSA | European Food Safety Authority |
| FT-ICR | Fourier transform ion cyclotron resonance |
| FXR | Farnesyl-X-receptor |
| FXR | Farnesoid X factor |
| GC | Gas chromatography |
| GCA | Glycocholic acid |
| GCDCA | Glycochenodeoxycholic acid |
| GDCA | Glycodeoxycholic acid |
| GDHCA | Glycodehydrocholic acid |
| GHCA | Glycohyocholic acid |
| GHDCA | Glycohyodeoxycholic acid |
| GLCA | Glycolithocholic acid |
| GUDCA | Glycoursodeoxycholic acid |
| HCA | Hyocholic acid |
| HCB | Hexachlorobenzene |
| HDCA | Hyodeoxycholic acid |
| HLA | Human leukocyte antigen |
| HNF4a | Hepatocyte nuclear factor 4 alpha |
| HRMS | High-resolution mass spectrometry |
| IT–TOFMS | Ion trap–time-of-flight mass spectrometry |
| LCA | Lithocholic acid |
| LPC | Lysophosphatidylcholine |
| L-PFOS | Linear-perfluorooctane sulfonate |
| MITM | Meet-in-the-middle |
| MnBP | Mono-n-butyl phthalate |
| NAFLD | Non-alcoholic fatty liver disease |
| NO2 | Nitrogen dioxide |
| NTCP | Sodium taurocholate co-transporting polypeptide |
| O3 | Ozone |
| OSTα/β | Organic solute transporter α/β |
| PAH | Polyaromatic hydrocarbons |
| PAH | Polycyclic aromatic hydrocarbons |
| PBDE | Polybrominated diphenyl ether |
| PC | Phosphatidylcholine |
| PC ethers | Phosphatidylcholine ether |
| PCB | Polychlorinated biphenyls |
| PE | Phospatidylethanolamine |
| PFAS | Per- and polyfluoroalkyl substances |
| PFBA | Perfluorobutanoic acid |
| PFBS | Perfluorobutane sulfonate |
| PFDA | Perfluorodecanoic acid |
| PFDS | Perfluorodecane sulfonate |
| PFECHS | Potassium perfluoro-4-ethylcyclohexanesulfonate |
| PFHpA | Perfluoroheptanoic acid |
| PFHpS | Perfluoroheptane sulfonate |
| PFHxS | Perfluorohexane sulfonate |
| PFNA | Perfluorononanoic acid |
| PFNS | Perfluorononane sulfonate |
| PFOA | Perfluorooctanoic acid |
| PFOSA | Perfluorooctane sulfonamide |
| PFPeA | Perfluoropentanoic acid |
| PFPeS | Perfluoro pentane sulfonate |
| PFTDA | Perfluorotetradecanoic acid |
| PFTrDA | Perfluorotridecanoic acid |
| PFUnDA | Perfluoroundecanoic acid |
| PI | Phosphatidylinositol |
| PM | Particulate matter |
| POP | Persistent organic pollutants |
| ROS | Reactive oxygen species |
| SM | Sphingomyelin |
| T1D | Type 1 diabetes |
| T2D | Type 2 diabetes |
| TBT | Tributyltin |
| TCA | Taurocholic acid |
| TCA | Tricarboxylic acid cycle |
| TCDCA | Taurochenodeoxycholic acid |
| TDCA | Taurodeoxycholic acid |
| TDHCA | Taurohyodeoxycholic acid |
| TG | Triradylglycerol |
| THCA | Taurodeoxycholic acid |
| THDCA | Taurohyodeoxycholic acid |
| TLCA | Taurolithocholic acid |
| TRAP | Traffic-related air pollutants |
| TUDCA | Tauroursodeoxycholic acid |
| TαβMCA | α,β-Tauromuricholic acid |
| TωMCA | ω-Tauromuricholic acid |
| UDCA | Ursodeoxycholic acid |
| UGT | UDP-glucosyltransferases |
| UHPLC | Ultra-performance liquid chromatography |
| β-HCH | β-hexachlorocyclohexane |
| βMCA | β-Muricholic acid |
| ωαMCA | ω-Tauromuricholic acid |
References
- Rappaport, S.M. Implications of the exposome for exposure science. J. Expo. Sci. Environ. Epidemiol. 2011, 21, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Rattray, N.J.W.; Deziel, N.C.; Wallach, J.D.; Khan, S.A.; Vasiliou, V.; Ioannidis, J.P.A.; Johnson, C.H. Beyond genomics: Understanding exposotypes through metabolomics. Hum. Genom. 2018, 12, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wild, C.P. Complementing the Genome with an “Exposome”: The Outstanding Challenge of Environmental Exposure Measurement in Molecular Epidemiology. Cancer Epidemiol. Biomark. Prev. 2005, 14, 1847–1850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rappaport, S.M.; Barupal, D.K.; Wishart, D.; Vineis, P.; Scalbert, A. The blood exposome and its role in discovering causes of disease. Environ. Health Perspect 2014, 122, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Wheelock, C.E.; Rappaport, S.M. The role of gene-environment interactions in lung disease: The urgent need for the exposome. Eur. Respir. J. 2020, 55. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, R.; Schymanski, E.L.; Barabasi, A.L.; Miller, G.W. The exposome and health: Where chemistry meets biology. Science 2020, 367, 392–396. [Google Scholar] [CrossRef]
- Miller, G.W.; Jones, D.P. The nature of nurture: Refining the definition of the exposome. Toxicol. Sci. 2014, 137, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Arora, M.; Chaleckis, R.; Isobe, T.; Jain, M.; Meister, I.; Melen, E.; Perzanowski, M.; Torta, F.; Wenk, M.R.; et al. Tackling the Complexity of the Exposome: Considerations from the Gunma University Initiative for Advanced Research (GIAR) Exposome Symposium. Metabolites 2019, 9, 106. [Google Scholar] [CrossRef] [Green Version]
- European Chemicals Agency. Guidance for the Identification and Naming of the Substances under REACH and CLP; European Chemicals Agency: Helsinki, Finland, 2017. [Google Scholar] [CrossRef]
- Pollock, T.; Weaver, R.E.; Ghasemi, R.; deCatanzaro, D. A mixture of five endocrine-disrupting chemicals modulates concentrations of bisphenol A and estradiol in mice. Chemosphere 2018, 193, 321–328. [Google Scholar] [CrossRef]
- Appenzeller, B.M.R.; Chadeau-Hyam, M.; Aguilar, L. Skin exposome science in practice: Current evidence on hair biomonitoring and future perspectives. J. Eur. Acad. Derm. Venereol. 2020, 34 (Suppl. 4), 26–30. [Google Scholar] [CrossRef]
- Leung, M.H.Y.; Tong, X.; Bastien, P.; Guinot, F.; Tenenhaus, A.; Appenzeller, B.M.R.; Betts, R.J.; Mezzache, S.; Li, J.; Bourokba, N.; et al. Changes of the human skin microbiota upon chronic exposure to polycyclic aromatic hydrocarbon pollutants. Microbiome 2020, 8, 100. [Google Scholar] [CrossRef] [PubMed]
- Pumarega, J.; Gasull, M.; Lee, D.H.; Lopez, T.; Porta, M. Number of Persistent Organic Pollutants Detected at High Concentrations in Blood Samples of the United States Population. PLoS ONE 2016, 11, e0160432. [Google Scholar] [CrossRef] [PubMed]
- Rappaport, S.M. Redefining environmental exposure for disease etiology. Npj. Syst. Biol. Appl. 2018, 4, 30. [Google Scholar] [CrossRef] [PubMed]
- Andra, S.S.; Austin, C.; Patel, D.; Dolios, G.; Awawda, M.; Arora, M. Trends in the application of high-resolution mass spectrometry for human biomonitoring: An analytical primer to studying the environmental chemical space of the human exposome. Environ. Int. 2017, 100, 32–61. [Google Scholar] [CrossRef] [Green Version]
- Athersuch, T. Metabolome analyses in exposome studies: Profiling methods for a vast chemical space. Arch. Biochem. Biophys. 2016, 589, 177–186. [Google Scholar] [CrossRef]
- Athersuch, T.J.; Keun, H.C. Metabolic profiling in human exposome studies. Mutagenesis 2015, 30, 755–762. [Google Scholar] [CrossRef] [Green Version]
- Ellis, J.K.; Athersuch, T.J.; Thomas, L.D.; Teichert, F.; Pérez-Trujillo, M.; Svendsen, C.; Spurgeon, D.J.; Singh, R.; Järup, L.; Bundy, J.G.; et al. Metabolic profiling detects early effects of environmental and lifestyle exposure to cadmium in a human population. BMC Med. 2012, 10, 61. [Google Scholar] [CrossRef] [Green Version]
- Lai, Y.; Xue, J.; Liu, C.-W.; Gao, B.; Chi, L.; Tu, P.; Lu, K.; Ru, H. Serum Metabolomics Identifies Altered Bioenergetics, Signaling Cascades in Parallel with Exposome Markers in Crohn’s Disease. Molecules 2019, 24, 449. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Cirillo, P.; Hu, X.; Tran, V.; Krigbaum, N.; Yu, S.; Jones, D.P.; Cohn, B. Understanding mixed environmental exposures using metabolomics via a hierarchical community network model in a cohort of California women in 1960′s. Reprod. Toxicol. 2019. [Google Scholar] [CrossRef]
- Maitre, L.; Robinson, O.; Martinez, D.; Toledano, M.B.; Ibarluzea, J.; Marina, L.S.; Sunyer, J.; Villanueva, C.M.; Keun, H.C.; Vrijheid, M.; et al. Urine Metabolic Signatures of Multiple Environmental Pollutants in Pregnant Women: An Exposome Approach. Environ. Sci. Technol. 2018, 52, 13469–13480. [Google Scholar] [CrossRef] [Green Version]
- Mapesa, J.O.; Maxwell, A.L.; Ryan, E.P. An Exposome Perspective on Environmental Enteric Dysfunction. Environ. Health Perspect 2016, 124, 1121–1126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lind, L.; Zethelius, B.; Salihovic, S.; van Bavel, B.; Lind, P.M.J.D. Circulating levels of perfluoroalkyl substances and prevalent diabetes in the elderly. Diabetologia 2014, 57, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Salihovic, S.; Fall, T.; Ganna, A.; Broeckling, C.D.; Prenni, J.E.; Hyötyläinen, T.; Kärrman, A.; Lind, P.M.; Ingelsson, E.; Lind, L. Identification of metabolic profiles associated with human exposure to perfluoroalkyl substances. J. Expo. Sci. Environ. Epidemiol. 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salihovic, S.; Ganna, A.; Fall, T.; Broeckling, C.D.; Prenni, J.E.; van Bavel, B.; Lind, P.M.; Ingelsson, E.; Lind, L. The metabolic fingerprint of p,p′-DDE and HCB exposure in humans. Environ. Int. 2016, 88, 60–66. [Google Scholar] [CrossRef]
- Sun, Q.; Zong, G.; Valvi, D.; Nielsen, F.; Coull, B.; Grandjean, P. Plasma Concentrations of Perfluoroalkyl Substances and Risk of Type 2 Diabetes: A Prospective Investigation among U.S. Women. Environ. Health Perspect. 2018, 126, 037001. [Google Scholar] [CrossRef] [Green Version]
- Dzierlenga, M.W.; Yoon, M.; Wania, F.; Ward, P.L.; Armitage, J.M.; Wood, S.A.; Clewell, H.J.; Longnecker, M.P. Quantitative bias analysis of the association of type 2 diabetes mellitus with 2,2′,4,4′,5,5′-hexachlorobiphenyl (PCB-153). Environ. Int. 2019, 125, 291–299. [Google Scholar] [CrossRef]
- Lee, D.-H.; Porta, M.; Jacobs, D.R., Jr.; Vandenberg, L.N. Chlorinated Persistent Organic Pollutants, Obesity, and Type 2 Diabetes. Endocr. Rev. 2014, 35, 557–601. [Google Scholar] [CrossRef] [Green Version]
- Tornevi, A.; Sommar, J.; Rantakokko, P.; Åkesson, A.; Donat-Vargas, C.; Kiviranta, H.; Rolandsson, O.; Rylander, L.; Wennberg, M.; Bergdahl, I.A. Chlorinated persistent organic pollutants and type 2 diabetes—A population-based study with pre- and post- diagnostic plasma samples. Environ. Res. 2019, 174, 35–45. [Google Scholar] [CrossRef]
- Beggs, K.M.; McGreal, S.R.; McCarthy, A.; Gunewardena, S.; Lampe, J.N.; Lau, C.; Apte, U. The role of hepatocyte nuclear factor 4-alpha in perfluorooctanoic acid- and perfluorooctanesulfonic acid-induced hepatocellular dysfunction. Toxicol. Appl. Pharmacol. 2016, 304, 18–29. [Google Scholar] [CrossRef] [Green Version]
- Sarigiannis, D.A. Assessing the impact of hazardous waste on children’s health: The exposome paradigm. Environ. Res 2017, 158, 531–541. [Google Scholar] [CrossRef]
- Kawamoto, T.; Nitta, H.; Murata, K.; Toda, E.; Tsukamoto, N.; Hasegawa, M.; Yamagata, Z.; Kayama, F.; Kishi, R.; Ohya, Y.; et al. Rationale and study design of the Japan environment and children’s study (JECS). BMC Public Health 2014, 14, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinisalu, L.; Sen, P.; Salihovic, S.; Virtanen, S.M.; Hyoty, H.; Ilonen, J.; Toppari, J.; Veijola, R.; Oresic, M.; Knip, M.; et al. Early-life exposure to perfluorinated alkyl substances modulates lipid metabolism in progression to celiac disease. Environ. Res. 2020, 188, 109864. [Google Scholar] [CrossRef] [PubMed]
- McGlinchey, A.; Sinioja, T.; Lamichhane, S.; Sen, P.; Bodin, J.; Siljander, H.; Dickens, A.M.; Geng, D.; Carlsson, C.; Duberg, D.; et al. Prenatal exposure to perfluoroalkyl substances modulates neonatal serum phospholipids, increasing risk of type 1 diabetes. Environ. Int. 2020, 143, 105935. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, A.G.; Rewers, M.; Simell, O.; Simell, T.; Lempainen, J.; Steck, A.; Winkler, C.; Ilonen, J.; Veijola, R.; Knip, M.; et al. Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA 2013, 309, 2473–2479. [Google Scholar] [CrossRef] [Green Version]
- Warth, B.; Spangler, S.; Fang, M.; Johnson, C.H.; Forsberg, E.M.; Granados, A.; Martin, R.L.; Domingo-Almenara, X.; Huan, T.; Rinehart, D.; et al. Exposome-Scale Investigations Guided by Global Metabolomics, Pathway Analysis, and Cognitive Computing. Anal. Chem. 2017, 89, 11505–11513. [Google Scholar] [CrossRef]
- Escher, B.I.; Stapleton, H.M.; Schymanski, E.L. Tracking complex mixtures of chemicals in our changing environment. Science 2020, 367, 388–392. [Google Scholar] [CrossRef]
- Chadeau-Hyam, M.; Athersuch, T.J.; Keun, H.C.; De Iorio, M.; Ebbels, T.M.; Jenab, M.; Sacerdote, C.; Bruce, S.J.; Holmes, E.; Vineis, P. Meeting-in-the-middle using metabolic profiling—A strategy for the identification of intermediate biomarkers in cohort studies. Biomarkers 2011, 16, 83–88. [Google Scholar] [CrossRef]
- Hartonen, M.; Mattila, I.; Ruskeepaa, A.L.; Oresic, M.; Hyotylainen, T. Characterization of cerebrospinal fluid by comprehensive two-dimensional gas chromatography coupled to time-of-flight mass spectrometry. J. Chromatogr. A 2013, 1293, 142–149. [Google Scholar] [CrossRef]
- Hyotylainen, T.; Oresic, M. Analytical Lipidomics in Metabolic and Clinical Research. Trends Endocrinol. Metab. 2015, 26, 671–673. [Google Scholar] [CrossRef]
- Heli Nygren, T.S.-L.; Sandra, C.; Tuulia, H.; Matej, O. LC/MS-based lipidomics for studies of body fluids and tissues. Methods Mol. Biol. 2011, 708, 11. [Google Scholar]
- Hyotylainen, T.; Oresic, M. Optimizing the lipidomics workflow for clinical studies—Practical considerations. Anal. Bioanal. Chem. 2015, 407, 4973–4993. [Google Scholar] [CrossRef] [PubMed]
- Klavus, A.; Kokla, M.; Noerman, S.; Koistinen, V.M.; Tuomainen, M.; Zarei, I.; Meuronen, T.; Hakkinen, M.R.; Rummukainen, S.; Farizah Babu, A.; et al. “Notame”: Workflow for Non-Targeted LC-MS Metabolic Profiling. Metabolites 2020, 10, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Yu, N.; Yang, J.; Jin, L.; Guo, H.; Shi, W.; Zhang, X.; Yang, L.; Yu, H.; Wei, S. Suspect and non-target screening of pesticides and pharmaceuticals transformation products in wastewater using QTOF-MS. Environ. Int. 2020, 137, 105599. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yu, N.; Du, L.; Shi, W.; Yu, H.; Song, M.; Wei, S. Transplacental Transfer of Per- and Polyfluoroalkyl Substances Identified in Paired Maternal and Cord Sera Using Suspect and Nontarget Screening. Environ. Sci. Technol. 2020, 54, 3407–3416. [Google Scholar] [CrossRef]
- Wang, A.; Gerona, R.R.; Schwartz, J.M.; Lin, T.; Sirota, M.; Morello-Frosch, R.; Woodruff, T.J. A Suspect Screening Method for Characterizing Multiple Chemical Exposures among a Demographically Diverse Population of Pregnant Women in San Francisco. Environ. Health Perspect. 2018, 126, 077009. [Google Scholar] [CrossRef]
- Hollender, J.; Schymanski, E.L.; Singer, H.P.; Ferguson, P.L. Nontarget Screening with High Resolution Mass Spectrometry in the Environment: Ready to Go? Environ. Sci. Technol. 2017, 51, 11505–11512. [Google Scholar] [CrossRef] [Green Version]
- Gerona, R.R.; Schwartz, J.M.; Pan, J.; Friesen, M.M.; Lin, T.; Woodruff, T.J. Suspect screening of maternal serum to identify new environmental chemical biomonitoring targets using liquid chromatography-quadrupole time-of-flight mass spectrometry. J. Expo. Sci. Environ. Epidemiol. 2018, 28, 101–108. [Google Scholar] [CrossRef]
- Ma, Y.; Kind, T.; Yang, D.; Leon, C.; Fiehn, O. MS2Analyzer: A software for small molecule substructure annotations from accurate tandem mass spectra. Anal. Chem. 2014, 86, 10724–10731. [Google Scholar] [CrossRef] [Green Version]
- Aguilar-Mogas, A.; Sales-Pardo, M.; Navarro, M.; Guimera, R.; Yanes, O. iMet: A Network-Based Computational Tool to Assist in the Annotation of Metabolites from Tandem Mass Spectra. Anal. Chem. 2017, 89, 3474–3482. [Google Scholar] [CrossRef] [Green Version]
- Treutler, H.; Tsugawa, H.; Porzel, A.; Gorzolka, K.; Tissier, A.; Neumann, S.; Balcke, G.U. Discovering Regulated Metabolite Families in Untargeted Metabolomics Studies. Anal. Chem. 2016, 88, 8082–8090. [Google Scholar] [CrossRef]
- Tsugawa, H.; Kind, T.; Nakabayashi, R.; Yukihira, D.; Tanaka, W.; Cajka, T.; Saito, K.; Fiehn, O.; Arita, M. Hydrogen Rearrangement Rules: Computational MS/MS Fragmentation and Structure Elucidation Using MS-FINDER Software. Anal. Chem. 2016, 88, 7946–7958. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.F.; Zhang, Y.H.; Ye, J.; Jin, H.Z.; Zhang, W.D. Investigation of the chemical compounds in Pheretima aspergillum (E. Perrier) using a combination of mass spectral molecular networking and unsupervised substructure annotation topic modeling together with in silico fragmentation prediction. J. Pharm. Biomed. Anal. 2020, 184, 113197. [Google Scholar] [CrossRef] [PubMed]
- Rogers, S.; Ong, C.W.; Wandy, J.; Ernst, M.; Ridder, L.; van der Hooft, J.J.J. Deciphering complex metabolite mixtures by unsupervised and supervised substructure discovery and semi-automated annotation from MS/MS spectra. Faraday Discuss. 2019, 218, 284–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, F.; Lei, Z.; Sumner, L.W. MetExpert: An expert system to enhance gas chromatographymass spectrometry-based metabolite identifications. Anal. Chim. Acta 2018, 1037, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Ljoncheva, M.; Stepišnik, T.; Džeroski, S.; Kosjek, T. Cheminformatics in MS-based environmental exposomics: Current achievements and future directions. Trends Environ. Anal. Chem. 2020, 28, e00099. [Google Scholar] [CrossRef]
- McEachran, A.D.; Mansouri, K.; Grulke, C.; Schymanski, E.L.; Ruttkies, C.; Williams, A.J. “MS-Ready” structures for non-targeted high-resolution mass spectrometry screening studies. J. Cheminform. 2018, 10, 45. [Google Scholar] [CrossRef]
- Koelmel, J.P.; Kroeger, N.M.; Ulmer, C.Z.; Bowden, J.A.; Patterson, R.E.; Cochran, J.A.; Beecher, C.W.W.; Garrett, T.J.; Yost, R.A. LipidMatch: An automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data. BMC Bioinform. 2017, 18, 331. [Google Scholar] [CrossRef]
- Schnelle-Kreis, J.; Welthagen, W.; Sklorz, M.; Zimmermann, R. Application of direct thermal desorption gas chromatography and comprehensive two-dimensional gas chromatography coupled to time of flight mass spectrometry for analysis of organic compounds in ambient aerosol particles. J. Sep. Sci. 2005, 28, 1648–1657. [Google Scholar] [CrossRef]
- Hummel, J.; Strehmel, N.; Selbig, J.; Walther, D.; Kopka, J.J.M. Decision tree supported substructure prediction of metabolites from GC-MS profiles. Metabolomics 2010, 6, 322–333. [Google Scholar] [CrossRef] [Green Version]
- Agier, L.; Portengen, L.; Chadeau-Hyam, M.; Basagana, X.; Giorgis-Allemand, L.; Siroux, V.; Robinson, O.; Vlaanderen, J.; Gonzalez, J.R.; Nieuwenhuijsen, M.J.; et al. A Systematic Comparison of Linear Regression-Based Statistical Methods to Assess Exposome-Health Associations. Environ. Health Perspect. 2016, 124, 1848–1856. [Google Scholar] [CrossRef]
- Bobb, J.F.; Claus Henn, B.; Valeri, L.; Coull, B.A. Statistical software for analyzing the health effects of multiple concurrent exposures via Bayesian kernel machine regression. Environ. Health 2018, 17, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scrucca, L.; Fop, M.; Murphy, T.B.; Raftery, A.E. mclust 5: Clustering, Classification and Density Estimation Using Gaussian Finite Mixture Models. R J. 2016, 8, 289–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castelo, R.; Roverato, A. Reverse engineering molecular regulatory networks from microarray data with qp-graphs. J. Comp. Biol. 2009, 16, 213–227. [Google Scholar] [CrossRef] [Green Version]
- Sen, P.; Lamichhane, S.; Mathema, V.B.; McGlinchey, A.; Dickens, A.M.; Khoomrung, S.; Oresic, M. Deep learning meets metabolomics: A methodological perspective. Brief. Bioinform. 2020. [Google Scholar] [CrossRef] [PubMed]
- Mendez, K.M.; Reinke, S.N.; Broadhurst, D.I. A comparative evaluation of the generalised predictive ability of eight machine learning algorithms across ten clinical metabolomics data sets for binary classification. Metabolomics 2019, 15, 150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, D.I.; Valvi, D.; Rothman, N.; Lan, Q.; Miller, G.W.; Jones, D.P. The metabolome: A key measure for exposome research in epidemiology. Curr. Epidemiol. Rep. 2019, 6, 93–103. [Google Scholar] [CrossRef]
- Coelho, M.; Oliveira, T.; Fernandes, R. Biochemistry of adipose tissue: An endocrine organ. Arch. Med. Sci. 2013, 9, 191–200. [Google Scholar] [CrossRef] [Green Version]
- La Merrill, M.; Emond, C.; Kim, M.J.; Antignac, J.P.; Le Bizec, B.; Clement, K.; Birnbaum, L.S.; Barouki, R. Toxicological function of adipose tissue: Focus on persistent organic pollutants. Environ. Health Perspect. 2013, 121, 162–169. [Google Scholar] [CrossRef] [Green Version]
- Jackson, E.; Shoemaker, R.; Larian, N.; Cassis, L. Adipose Tissue as a Site of Toxin Accumulation. Compr. Physiol. 2017, 7, 1085–1135. [Google Scholar] [CrossRef] [Green Version]
- Heindel, J.J.; Blumberg, B.; Cave, M.; Machtinger, R.; Mantovani, A.; Mendez, M.A.; Nadal, A.; Palanza, P.; Panzica, G.; Sargis, R.; et al. Metabolism disrupting chemicals and metabolic disorders. Reprod. Toxicol. 2017, 68, 3–33. [Google Scholar] [CrossRef] [Green Version]
- Angle, B.M.; Do, R.P.; Ponzi, D.; Stahlhut, R.W.; Drury, B.E.; Nagel, S.C.; Welshons, W.V.; Besch-Williford, C.L.; Palanza, P.; Parmigiani, S.; et al. Metabolic disruption in male mice due to fetal exposure to low but not high doses of bisphenol A (BPA): Evidence for effects on body weight, food intake, adipocytes, leptin, adiponectin, insulin and glucose regulation. Reprod. Toxicol. 2013, 42, 256–268. [Google Scholar] [CrossRef] [Green Version]
- Hao, C.; Cheng, X.; Xia, H.; Ma, X. The endocrine disruptor mono-(2-ethylhexyl) phthalate promotes adipocyte differentiation and induces obesity in mice. Biosci. Rep. 2012, 32, 619–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Merrill, M.; Karey, E.; Moshier, E.; Lindtner, C.; La Frano, M.R.; Newman, J.W.; Buettner, C. Perinatal exposure of mice to the pesticide DDT impairs energy expenditure and metabolism in adult female offspring. PLoS ONE 2014, 9, e103337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mentor, A.; Brunstrom, B.; Mattsson, A.; Jonsson, M. Developmental exposure to a human relevant mixture of endocrine disruptors alters metabolism and adipogenesis in zebrafish (Danio rerio). Chemosphere 2020, 238, 124584. [Google Scholar] [CrossRef]
- Valvi, D.; Casas, M.; Mendez, M.A.; Ballesteros, G.A.; Luque, N.; Rubio, S.; Sunyer, J.; Vrijheid, M. Prenatal Bisphenol A Urine Concentrations and Early Rapid Growth and Overweight Risk in the Offspring. Epidemiology 2013, 24, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Braun, J.M.; Lanphear, B.P.; Calafat, A.M.; Deria, S.; Khoury, J.; Howe, C.J.; Venners, S.A. Early-Life Bisphenol A Exposure and Child Body Mass Index: A Prospective Cohort Study. Environ. Health Perspect. 2014, 122, 1239–1245. [Google Scholar] [CrossRef] [Green Version]
- Hoepner, L.A.; Whyatt, R.M.; Widen, E.M.; Hassoun, A.; Oberfield, S.E.; Mueller, N.T.; Diaz, D.; Calafat, A.M.; Perera, F.P.; Rundle, A.G. Bisphenol A and Adiposity in an Inner-City Birth Cohort. Environ. Health Perspect. 2016, 124, 1644–1650. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.M.; Hong, Y.-C.; Ha, M.; Kim, Y.; Park, H.; Kim, H.S.; Ha, E.-H. Prenatal Bisphenol-A exposure affects fetal length growth by maternal glutathione transferase polymorphisms, and neonatal exposure affects child volume growth by sex: From multiregional prospective birth cohort MOCEH study. Sci. Total Environ. 2018, 612, 1433–1441. [Google Scholar] [CrossRef]
- Valvi, D.; Mendez, M.A.; Garcia-Esteban, R.; Ballester, F.; Ibarluzea, J.; Goni, F.; Grimalt, J.O.; Llop, S.; Marina, L.S.; Vizcaino, E.; et al. Prenatal exposure to persistent organic pollutants and rapid weight gain and overweight in infancy. Obesity (Silver Spring) 2014, 22, 488–496. [Google Scholar] [CrossRef]
- Machala, M.; Slavik, J.; Pencikova, K.; Neca, J.; Simeckova, P.; Kulich, P.; Vondracek, J. Polychlorinated biphenyls as modulators of sphingolipid and prostaglandin metabolism, intercellular communication and cell adhesion. Toxicol. Lett. 2017, 280, S27. [Google Scholar] [CrossRef]
- Wang, X.; Xu, Y.; Song, X.; Jia, Q.; Zhang, X.; Qian, Y.; Qiu, J. Analysis of glycerophospholipid metabolism after exposure to PCB153 in PC12 cells through targeted lipidomics by UHPLC-MS/MS. Ecotoxicol. Environ. Saf. 2019, 169, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Carrizo, D.; Chevallier, O.P.; Woodside, J.V.; Brennan, S.F.; Cantwell, M.M.; Cuskelly, G.; Elliott, C.T. Untargeted metabolomic analysis of human serum samples associated with exposure levels of Persistent organic pollutants indicate important perturbations in Sphingolipids and Glycerophospholipids levels. Chemosphere 2017, 168, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Geng, N.; Zhang, H.; Zhang, B.; Wu, P.; Wang, F.; Yu, Z.; Chen, J. Effects of Short-Chain Chlorinated Paraffins Exposure on the Viability and Metabolism of Human Hepatoma HepG2 Cells. Environ. Sci. Technol. 2015, 49, 3076–3083. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Lu, Y.; Huang, S.; Gao, L.; Liang, X.; Wu, Y.; Wang, J.; Huang, Q.; Tang, L.; Wang, G.; et al. Identifying Early Urinary Metabolic Changes with Long-Term Environmental Exposure to Cadmium by Mass-Spectrometry-Based Metabolomics. Environ. Sci. Technol. 2014, 48, 6409–6418. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shen, H.; Xu, W.; Xia, Y.; Barr, D.B.; Mu, X.; Wang, X.; Liu, L.; Huang, Q.; Tian, M. Urinary Metabolomics Revealed Arsenic Internal Dose-Related Metabolic Alterations: A Proof-of-Concept Study in a Chinese Male Cohort. Environ. Sci. Technol. 2014, 48, 12265–12274. [Google Scholar] [CrossRef]
- Dudka, I.; Kossowska, B.; Senhadri, H.; Latajka, R.; Hajek, J.; Andrzejak, R.; Antonowicz-Juchniewicz, J.; Gancarz, R. Metabonomic analysis of serum of workers occupationally exposed to arsenic, cadmium and lead for biomarker research: A preliminary study. Environ. Int. 2014, 68, 71–81. [Google Scholar] [CrossRef]
- Wang, Z.; Zheng, Y.; Zhao, B.; Zhang, Y.; Liu, Z.; Xu, J.; Chen, Y.; Yang, Z.; Wang, F.; Wang, H.; et al. Human Metabolic Responses to Chronic Environmental Polycyclic Aromatic Hydrocarbon Exposure by a Metabolomic Approach. J. Proteome Res. 2015, 14, 2583–2593. [Google Scholar] [CrossRef]
- Fader, K.A.; Nault, R.; Ammendolia, D.A.; Harkema, J.R.; Williams, K.J.; Crawford, R.B.; Kaminski, N.E.; Potter, D.; Sharratt, B.; Zacharewski, T.R. 2,3,7,8-Tetrachlorodibenzo-p-Dioxin Alters Lipid Metabolism and Depletes Immune Cell Populations in the Jejunum of C57BL/6 Mice. Toxicol. Sci. 2015, 148, 567–580. [Google Scholar] [CrossRef] [Green Version]
- Hennig, B.; Reiterer, G.; Toborek, M.; Matveev, S.V.; Daugherty, A.; Smart, E.; Robertson, L.W. Dietary fat interacts with PCBs to induce changes in lipid metabolism in mice deficient in low-density lipoprotein receptor. Environ. Health Perspect. 2005, 113, 83–87. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.S.; Furuya, S.; Soldatov, V.Y.; Kosyk, O.; Yoo, H.S.; Fukushima, H.; Lewis, L.; Iwata, Y.; Rusyn, I. Metabolism and Toxicity of Trichloroethylene and Tetrachloroethylene in Cytochrome P450 2E1 Knockout and Humanized Transgenic Mice. Toxicol. Sci. 2018, 164, 489–500. [Google Scholar] [CrossRef]
- Karami-Mohajeri, S.; Abdollahi, M. Toxic influence of organophosphate, carbamate, and organochlorine pesticides on cellular metabolism of lipids, proteins, and carbohydrates: A systematic review. Hum. Exp. Toxicol. 2011, 30, 1119–1140. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Liu, D.; Zhan, J.; Luo, M.; Han, J.; Wang, P.; Zhou, Z. New insight into the mechanism of POP-induced obesity: Evidence from DDE-altered microbiota. Chemosphere 2020, 244, 125123. [Google Scholar] [CrossRef] [PubMed]
- Bijland, S.; Rensen, P.C.; Pieterman, E.J.; Maas, A.C.; van der Hoorn, J.W.; van Erk, M.J.; Havekes, L.M.; van Dijk, K.W.; Chang, S.C.; Ehresman, D.J.; et al. Perfluoroalkyl sulfonates cause alkyl chain length-dependent hepatic steatosis and hypolipidemia mainly by impairing lipoprotein production in APOE*3-Leiden CETP mice. Toxicol. Sci. 2011, 123, 290–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.; Zitzow, J.D.; Ehresman, D.J.; Chang, S.-C.; Butenhoff, J.L.; Forster, J.; Hagenbuch, B. Na+/Taurocholate Cotransporting Polypeptide and Apical Sodium-Dependent Bile Acid Transporter are Involved in the Disposition of Perfluoroalkyl Sulfonates in Humans and Rats. Toxicol. Sci. 2015, 146, 363–373. [Google Scholar] [CrossRef] [Green Version]
- Steenland, K.; Tinker, S.; Shankar, A.; Ducatman, A. Association of perfluorooctanoic acid (PFOA) and perfluorooctane sulfonate (PFOS) with uric acid among adults with elevated community exposure to PFOA. Environ. Health Perspect. 2010, 118, 229–233. [Google Scholar] [CrossRef] [Green Version]
- Hayden, M.R.; Tyagi, S.C. Uric acid: A new look at an old risk marker for cardiovascular disease, metabolic syndrome, and type 2 diabetes mellitus: The urate redox shuttle. Nutr. Metab. 2004, 1, 10. [Google Scholar] [CrossRef] [Green Version]
- Dimitroula, H.V.; Hatzitolios, A.I.; Karvounis, H.I. The Role of Uric Acid in Stroke: The Issue Remains Unresolved. Neurologist. 2008, 14, 238–242. [Google Scholar] [CrossRef]
- Haeusler, R.A.; Astiarraga, B.; Camastra, S.; Accili, D.; Ferrannini, E. Human insulin resistance is associated with increased plasma levels of 12a-hydroxylated bile acids. Diabetes 2013, 62, 4184–4191. [Google Scholar] [CrossRef] [Green Version]
- Prawitt, J.; Caron, S.; Staels, B. Bile Acid Metabolism and the Pathogenesis of Type 2 Diabetes. Curr. Diabetes Rep. 2011, 11, 160. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Rimal, B.; Nichols, R.G.; Tian, Y.; Smith, P.B.; Hatzakis, E.; Chang, S.C.; Butenhoff, J.L.; Peters, J.M.; Patterson, A.D. Perfluorooctane sulfonate alters gut microbiota-host metabolic homeostasis in mice. Toxicology 2020, 431, 152365. [Google Scholar] [CrossRef]
- Petriello, M.C.; Hoffman, J.B.; Vsevolozhskaya, O.; Morris, A.J.; Hennig, B. Dioxin-like PCB 126 increases intestinal inflammation and disrupts gut microbiota and metabolic homeostasis. Environ. Pollut. 2018, 242, 1022–1032. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.L.; Li, X.; Lehmler, H.-J.; Phillips, B.; Shen, D.; Cui, J.Y. Gut Microbiota Modulates Interactions Between Polychlorinated Biphenyls and Bile Acid Homeostasis. Toxicol. Sci. 2018, 166, 269–287. [Google Scholar] [CrossRef] [PubMed]
- Salihović, S.; Dickens, A.M.; Schoultz, I.; Fart, F.; Sinisalu, L.; Lindeman, T.; Halfvarson, J.; Orešič, M.; Hyötyläinen, T. Simultaneous determination of perfluoroalkyl substances and bile acids in human serum using ultra-high-performance liquid chromatography–tandem mass spectrometry. Anal. Bioanal. Chem. 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeanneret, F.; Tonoli, D.; Hochstrasser, D.; Saurat, J.-H.; Sorg, O.; Boccard, J.; Rudaz, S. Evaluation and identification of dioxin exposure biomarkers in human urine by high-resolution metabolomics, multivariate analysis and in vitro synthesis. Toxicol. Lett. 2016, 240, 22–31. [Google Scholar] [CrossRef]
- Chiang, J.Y. Recent advances in understanding bile acid homeostasis. F1000Research 2017, 6, 2029. [Google Scholar] [CrossRef]
- Buhrke, T.; Kruger, E.; Pevny, S.; Rossler, M.; Bitter, K.; Lampen, A. Perfluorooctanoic acid (PFOA) affects distinct molecular signalling pathways in human primary hepatocytes. Toxicology 2015, 333, 53–62. [Google Scholar] [CrossRef]
- Iszatt, N.; Janssen, S.; Lenters, V.; Dahl, C.; Stigum, H.; Knight, R.; Mandal, S.; Peddada, S.; González, A.; Midtvedt, T.; et al. Environmental toxicants in breast milk of Norwegian mothers and gut bacteria composition and metabolites in their infants at 1 month. Microbiome 2019, 7, 34. [Google Scholar] [CrossRef] [Green Version]
- Pan, Z.; Yuan, X.; Tu, W.; Fu, Z.; Jin, Y. Subchronic exposure of environmentally relevant concentrations of F-53B in mice resulted in gut barrier dysfunction and colonic inflammation in a sex-independent manner. Environ. Pollut. 2019, 253, 268–277. [Google Scholar] [CrossRef]
- Rude, K.M.; Keogh, C.E.; Gareau, M.G. The role of the gut microbiome in mediating neurotoxic outcomes to PCB exposure. Neurotoxicology 2019, 75, 30–40. [Google Scholar] [CrossRef]
- De Aguiar Vallim, T.Q.; Tarling, E.J.; Edwards, P.A. Pleiotropic Roles of Bile Acids in Metabolism. Cell Metab. 2013, 17, 657–669. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Rang, O.; Liu, F.; Xia, W.; Li, Y.; Zhang, Y.; Lu, S.; Xu, S.J.M. A systematic review of metabolomics biomarkers for Bisphenol A exposure. Metabolomics 2018, 14, 45. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Park, H.; Lee, H.A.; Park, B.; Gwak, H.S.; Lee, H.-R.; Jee, S.H.; Park, Y.H. Elevated Metabolites of Steroidogenesis and Amino Acid Metabolism in Preadolescent Female Children with High Urinary Bisphenol A Levels: A High-Resolution Metabolomics Study. Toxicol. Sci. 2017, 160, 371–385. [Google Scholar] [CrossRef] [PubMed]
- Alderete, T.L.; Jin, R.; Walker, D.I.; Valvi, D.; Chen, Z.; Jones, D.P.; Peng, C.; Gilliland, F.D.; Berhane, K.; Conti, D.V.; et al. Perfluoroalkyl substances, metabolomic profiling, and alterations in glucose homeostasis among overweight and obese Hispanic children: A proof-of-concept analysis. Environ. Int. 2019, 126, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; McConnell, R.; Catherine, C.; Xu, S.; Walker, D.I.; Stratakis, N.; Jones, D.P.; Miller, G.W.; Peng, C.; Conti, D.V.; et al. Perfluoroalkyl substances and severity of nonalcoholic fatty liver in Children: An untargeted metabolomics approach. Environ. Int. 2020, 134, 105220. [Google Scholar] [CrossRef] [PubMed]
- Arneth, B.; Arneth, R.; Shams, M. Metabolomics of Type 1 and Type 2 Diabetes. Int. J. Mol. Sci. 2019, 20, 2467. [Google Scholar] [CrossRef] [Green Version]
- Chuang, W.-H.; Arundhathi, A.; Lu, C.; Chen, C.-C.; Wu, W.-C.; Susanto, H.; Purnomo, J.D.T.; Wang, C.-H.J.M. Altered plasma acylcarnitine and amino acid profiles in type 2 diabetic kidney disease. Metabolomics 2016, 12, 108. [Google Scholar] [CrossRef]
- Floegel, A.; Stefan, N.; Yu, Z.; Mühlenbruch, K.; Drogan, D.; Joost, H.-G.; Fritsche, A.; Häring, H.-U.; Hrabě de Angelis, M.; Peters, A.; et al. Identification of Serum Metabolites Associated With Risk of Type 2 Diabetes Using a Targeted Metabolomic Approach. Diabetes 2013, 62, 639–648. [Google Scholar] [CrossRef] [Green Version]
- Gar, C.; Rottenkolber, M.; Prehn, C.; Adamski, J.; Seissler, J.; Lechner, A. Serum and plasma amino acids as markers of prediabetes, insulin resistance, and incident diabetes. Crit. Rev. Clin. Lab. Sci. 2018, 55, 21–32. [Google Scholar] [CrossRef]
- Wang, X.; Liu, L.; Zhang, W.; Zhang, J.; Du, X.; Huang, Q.; Tian, M.; Shen, H. Serum metabolome biomarkers associate low-level environmental perfluorinated compound exposure with oxidative/nitrosative stress in humans. Environ. Pollut. 2017, 229, 168–176. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, X.; He, B.; Guo, J.; Zhao, B.; Zhang, Y.; Zhou, Z.; Zhou, X.; Zhang, R.; Abliz, Z. The impact of chronic environmental metal and benzene exposure on human urinary metabolome among Chinese children and the elderly population. Ecotoxicol. Environ. Saf. 2019, 169, 232–239. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, H.; Geng, N.; Ren, X.; Zhang, B.; Gong, Y.; Chen, J. A metabolomics strategy to assess the combined toxicity of polycyclic aromatic hydrocarbons (PAHs) and short-chain chlorinated paraffins (SCCPs). Environ. Pollut. 2018, 234, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Moutinho, J.L.; Golan, R.; Yu, T.; Ladva, C.N.; Niedzwiecki, M.; Walker, D.I.; Sarnat, S.E.; Chang, H.H.; Greenwald, R.; et al. Use of high-resolution metabolomics for the identification of metabolic signals associated with traffic-related air pollution. Environ. Int. 2018, 120, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Caserta, D.; Ciardo, F.; Bordi, G.; Guerranti, C.; Fanello, E.; Perra, G.; Borghini, F.; La Rocca, C.; Tait, S.; Bergamasco, B.; et al. Correlation of endocrine disrupting chemicals serum levels and white blood cells gene expression of nuclear receptors in a population of infertile women. Int. J. Endocrinol. 2013, 2013, 510703. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Huang, Y.; Li, J.; Yang, K.; Yang, W.; Shen, G.; Li, X.; Lei, Y.; Pang, S.; Wang, C.; et al. New insights into the mechanism of phthalate-induced developmental effects. Environ. Pollut. 2018, 241, 674–683. [Google Scholar] [CrossRef]
- Xia, B.; Zhu, Q.; Zhao, Y.; Ge, W.; Zhao, Y.; Song, Q.; Zhou, Y.; Shi, H.; Zhang, Y. Phthalate exposure and childhood overweight and obesity: Urinary metabolomic evidence. Environ. Int. 2018, 121, 159–168. [Google Scholar] [CrossRef]
- Jin, Y.; Wu, S.; Zeng, Z.; Fu, Z. Effects of environmental pollutants on gut microbiota. Environ. Pollut. 2017, 222, 1–9. [Google Scholar] [CrossRef]
- Zhang, L.; Nichols, R.G.; Correll, J.; Murray, I.A.; Tanaka, N.; Smith, P.B.; Hubbard, T.D.; Sebastian, A.; Albert, I.; Hatzakis, E.; et al. Persistent Organic Pollutants Modify Gut Microbiota-Host Metabolic Homeostasis in Mice Through Aryl Hydrocarbon Receptor Activation. Environ. Health Perspect. 2015, 123, 679–688. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Keerthisinghe, T.P.; Han, Y.; Liu, M.; Wanjaya, E.R.; Fang, M. “Cocktail” of Xenobiotics at Human Relevant Levels Reshapes the Gut Bacterial Metabolome in a Species-Specific Manner. Environ. Sci. Technol. 2018, 52, 11402–11410. [Google Scholar] [CrossRef]
- Yuan, X.; Pan, Z.; Jin, C.; Ni, Y.; Fu, Z.; Jin, Y. Gut microbiota: An underestimated and unintended recipient for pesticide-induced toxicity. Chemosphere 2019, 227, 425–434. [Google Scholar] [CrossRef]
- Bi, Y.; Wang, W.; Xu, M.; Wang, T.; Lu, J.; Xu, Y.; Dai, M.; Chen, Y.; Zhang, D.; Sun, W.; et al. Diabetes Genetic Risk Score Modifies Effect of Bisphenol A Exposure on Deterioration in Glucose Metabolism. J. Clin. Endocrinol. Metab. 2016, 101, 143–150. [Google Scholar] [CrossRef]
- Bodin, J.; Groeng, E.-C.; Andreassen, M.; Dirven, H.; Nygaard, U.C. Exposure to perfluoroundecanoic acid (PFUnDA) accelerates insulitis development in a mouse model of type 1 diabetes. Toxicol. Rep. 2016, 3, 664–672. [Google Scholar] [CrossRef] [Green Version]
- Bodin, J.; Stene, L.C.; Nygaard, U.C. Can Exposure to Environmental Chemicals Increase the Risk of Diabetes Type 1 Development? Biomed Res. Int. 2015, 2015, 208947. [Google Scholar] [CrossRef]
- Cui, J.; Fu, Y.; Lu, R.; Bi, Y.; Zhang, L.; Zhang, C.; Aschner, M.; Li, X.; Chen, R. Metabolomics analysis explores the rescue to neurobehavioral disorder induced by maternal PM2.5 exposure in mice. Ecotoxicol. Environ. Saf. 2019, 169, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Domazet, S.L.; Grøntved, A.; Timmermann, A.G.; Nielsen, F.; Jensen, T.K. Longitudinal Associations of Exposure to Perfluoroalkylated Substances in Childhood and Adolescence and Indicators of Adiposity and Glucose Metabolism 6 and 12 Years Later: The European Youth Heart Study. Diabetes Care 2016, 39, 1745–1751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giulivo, M.; de Alda, M.L.; Capri, E.; Barcelo, D. Human exposure to endocrine disrupting compounds: Their role in reproductive systems, metabolic syndrome and breast cancer. A review. Environ. Res. 2016, 151, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Glynn, A.; Igra, A.M.; Sand, S.; Ilbäck, N.G.; Hellenäs, K.E.; Rosén, J.; Aspenström-Fagerlund, B. Are additive effects of dietary surfactants on intestinal tight junction integrity an overlooked human health risk?—A mixture study on Caco-2 monolayers. Food Chem. Toxicol. 2017, 106, 314–323. [Google Scholar] [CrossRef]
- Govarts, E.; Iszatt, N.; Trnovec, T.; de Cock, M.; Eggesbø, M.; Palkovicova Murinova, L.; van de Bor, M.; Guxens, M.; Chevrier, C.; Koppen, G.; et al. Prenatal exposure to endocrine disrupting chemicals and risk of being born small for gestational age: Pooled analysis of seven European birth cohorts. Environ. Int. 2018, 115, 267–278. [Google Scholar] [CrossRef] [Green Version]
- Granum, B.; Haug, L.S.; Namork, E.; Stolevik, S.B.; Thomsen, C.; Aaberge, I.S.; van Loveren, H.; Lovik, M.; Nygaard, U.C. Pre-natal exposure to perfluoroalkyl substances may be associated with altered vaccine antibody levels and immune-related health outcomes in early childhood. J. Immunotoxicol. 2013, 10, 373–379. [Google Scholar] [CrossRef]
- Kharrazian, D. The Potential Roles of Bisphenol A (BPA) Pathogenesis in Autoimmunity. Autoimmune Dis. 2014, 2014, 743616. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Eom, S.; Kim, H.J.; Lee, J.J.; Choi, G.; Choi, S.; Kim, S.; Kim, S.Y.; Cho, G.; Kim, Y.D.; et al. Association between maternal exposure to major phthalates, heavy metals, and persistent organic pollutants, and the neurodevelopmental performances of their children at 1 to 2years of age- CHECK cohort study. Sci. Total Environ. 2018, 624, 377–384. [Google Scholar] [CrossRef]
- Manzano-Salgado, C.B.; Casas, M.; Lopez-Espinosa, M.-J.; Ballester, F.; Iñiguez, C.; Martinez, D.; Costa, O.; Santa-Marina, L.; Pereda-Pereda, E.; Schettgen, T.; et al. Prenatal exposure to perfluoroalkyl substances and birth outcomes in a Spanish birth cohort. Environ. Int. 2017, 108, 278–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, J.W.; Hatch, E.E.; Webster, T.F. Exposure to polyfluoroalkyl chemicals and cholesterol, body weight, and insulin resistance in the general U.S. population. Environ. Health Perspect. 2010, 118, 197–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nisticò, L.; Iafusco, D.; Galderisi, A.; Fagnani, C.; Cotichini, R.; Toccaceli, V.; Stazi, M.A.; the Study Group on Diabetes of the Italian Society of Pediatric Endocrinology and Diabetology. Emerging Effects of Early Environmental Factors over Genetic Background for Type 1 Diabetes Susceptibility: Evidence from a Nationwide Italian Twin Study. J. Clin. Endocrinol. Metab. 2012, 97, E1483–E1491. [Google Scholar] [CrossRef] [Green Version]
- Predieri, B.; Iughetti, L.; Guerranti, C.; Bruzzi, P.; Perra, G.; Focardi, S.E. High Levels of Perfluorooctane Sulfonate in Children at the Onset of Diabetes. Int. J. Endocrinol. 2015, 2015, 234358. [Google Scholar] [CrossRef] [PubMed]
- Sears, M.E.; Genuis, S.J. Environmental determinants of chronic disease and medical approaches: Recognition, avoidance, supportive therapy, and detoxification. J. Environ. Public Health 2012, 2012, 356798. [Google Scholar] [CrossRef] [Green Version]
- Spratlen, M.J.; Grau-Perez, M.; Umans, J.G.; Yracheta, J.; Best, L.G.; Francesconi, K.; Goessler, W.; Bottiglieri, T.; Gamble, M.V.; Cole, S.A.; et al. Targeted metabolomics to understand the association between arsenic metabolism and diabetes-related outcomes: Preliminary evidence from the Strong Heart Family Study. Environ. Res. 2019, 168, 146–157. [Google Scholar] [CrossRef]
- Timmermann, C.A.G.; Rossing, L.I.; Grøntved, A.; Ried-Larsen, M.; Dalgård, C.; Andersen, L.B.; Grandjean, P.; Nielsen, F.; Svendsen, K.D.; Scheike, T.; et al. Adiposity and Glycemic Control in Children Exposed to Perfluorinated Compounds. J. Clin. Endocrinol. Metab. 2014, 99, E608–E614. [Google Scholar] [CrossRef] [Green Version]
- Yu, N.; Wei, S.; Li, M.; Yang, J.; Li, K.; Jin, L.; Xie, Y.; Giesy, J.P.; Zhang, X.; Yu, H. Effects of Perfluorooctanoic Acid on Metabolic Profiles in Brain and Liver of Mouse Revealed by a High-throughput Targeted Metabolomics Approach. Sci. Rep. 2016, 6, 23963. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Hatzakis, E.; Nichols, R.G.; Hao, R.; Correll, J.; Smith, P.B.; Chiaro, C.R.; Perdew, G.H.; Patterson, A.D. Metabolomics Reveals that Aryl Hydrocarbon Receptor Activation by Environmental Chemicals Induces Systemic Metabolic Dysfunction in Mice. Environ. Sci. Technol. 2015, 49, 8067–8077. [Google Scholar] [CrossRef] [Green Version]
- Carlsten, C. Synergistic Environmental Exposures and the Airways Capturing Complexity in Humans: An Underappreciated World of Complex Exposures. Chest 2018, 154, 918–924. [Google Scholar] [CrossRef]
- Leite, D.F.B.; Morillon, A.-C.; Melo Júnior, E.F.; Souza, R.T.; McCarthy, F.P.; Khashan, A.; Baker, P.; Kenny, L.C.; Cecatti, J.G. Examining the predictive accuracy of metabolomics for small-for-gestational-age babies: A systematic review. BMJ Open 2019, 9, e031238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maitre, L.; Villanueva, C.M.; Lewis, M.R.; Ibarluzea, J.; Santa-Marina, L.; Vrijheid, M.; Sunyer, J.; Coen, M.; Toledano, M.B. Maternal urinary metabolic signatures of fetal growth and associated clinical and environmental factors in the INMA study. BMC Med. 2016, 14, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellmuth, C.; Uhl, O.; Standl, M.; Demmelmair, H.; Heinrich, J.; Koletzko, B.; Thiering, E. Cord Blood Metabolome is Highly Associated with Birth Weight, but Less Predictive for Later Weight Development. Obes. Facts 2017, 10, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.P.; Reichetzeder, C.; Prehn, C.; Yin, L.H.; Yun, C.; Zeng, S.; Chu, C.; Adamski, J.; Hocher, B. Cord Blood Lysophosphatidylcholine 16: 1 is Positively Associated with Birth Weight. Cell. Physiol. Biochem. 2018, 45, 614–624. [Google Scholar] [CrossRef]
- Rolle-Kampczyk, U.E.; Krumsiek, J.; Otto, W.; Röder, S.W.; Kohajda, T.; Borte, M.; Theis, F.; Lehmann, I.; von Bergen, M.J.M. Metabolomics reveals effects of maternal smoking on endogenous metabolites from lipid metabolism in cord blood of newborns. Metabolomics 2016, 12, 76. [Google Scholar] [CrossRef] [Green Version]
- Meng, Z.; Tian, S.; Yan, J.; Jia, M.; Yan, S.; Li, R.; Zhang, R.; Zhu, W.; Zhou, Z. Effects of perinatal exposure to BPA, BPF and BPAF on liver function in male mouse offspring involving in oxidative damage and metabolic disorder. Environ. Pollut. 2019, 247, 935–943. [Google Scholar] [CrossRef]
- Meng, Z.; Wang, D.; Liu, W.; Li, R.; Yan, S.; Jia, M.; Zhang, L.; Zhou, Z.; Zhu, W. Perinatal exposure to Bisphenol S (BPS) promotes obesity development by interfering with lipid and glucose metabolism in male mouse offspring. Environ. Res. 2019, 173, 189–198. [Google Scholar] [CrossRef]
- Susiarjo, M.; Xin, F.; Stefaniak, M.; Mesaros, C.; Simmons, R.A.; Bartolomei, M.S. Bile Acids and Tryptophan Metabolism Are Novel Pathways Involved in Metabolic Abnormalities in BPA-Exposed Pregnant Mice and Male Offspring. Endocrinology 2017, 158, 2533–2542. [Google Scholar] [CrossRef]
- Mamsen, L.S.; Jönsson, B.A.G.; Lindh, C.H.; Olesen, R.H.; Larsen, A.; Ernst, E.; Kelsey, T.W.; Andersen, C.Y. Concentration of perfluorinated compounds and cotinine in human foetal organs, placenta, and maternal plasma. Sci. Total Environ. 2017, 596–597, 97–105. [Google Scholar] [CrossRef] [Green Version]
- Kishi, R.; Araki, A.; Minatoya, M.; Hanaoka, T.; Miyashita, C.; Itoh, S.; Kobayashi, S.; Ait Bamai, Y.; Yamazaki, K.; Miura, R.; et al. The Hokkaido Birth Cohort Study on Environment and Children’s Health: Cohort profile-updated 2017. Environ. Health Prev. Med. 2017, 22, 46. [Google Scholar] [CrossRef]
- Achenbach, P.; Bonifacio, E.; Koczwara, K.; Ziegler, A.-G. Natural History of Type 1 Diabetes. Diabetes 2005, 54, S25–S31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Starling, A.P.; Adgate, J.L.; Hamman, R.F.; Kechris, K.; Calafat, A.M.; Ye, X.; Dabelea, D. Perfluoroalkyl Substances during Pregnancy and Offspring Weight and Adiposity at Birth: Examining Mediation by Maternal Fasting Glucose in the Healthy Start Study. Environ. Health Perspect. 2017, 125, 067016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashley-Martin, J.; Dodds, L.; Arbuckle, T.E.; Bouchard, M.F.; Fisher, M.; Morriset, A.-S.; Monnier, P.; Shapiro, G.D.; Ettinger, A.S.; Dallaire, R.; et al. Maternal Concentrations of Perfluoroalkyl Substances and Fetal Markers of Metabolic Function and Birth Weight: The Maternal-Infant Research on Environmental Chemicals (MIREC) Study. Am. J. Epidemiol. 2017, 185, 185–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marks, K.J.; Cutler, A.J.; Jeddy, Z.; Northstone, K.; Kato, K.; Hartman, T.J. Maternal serum concentrations of perfluoroalkyl substances and birth size in British boys. Int. J. Hyg. Environ. Health 2019, 222, 889–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maisonet, M.; Terrell, M.L.; McGeehin, M.A.; Christensen, K.Y.; Holmes, A.; Calafat, A.M.; Marcus, M. Maternal concentrations of polyfluoroalkyl compounds during pregnancy and fetal and postnatal growth in British girls. Environ. Health Perspect. 2012, 120, 1432–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashino, I.; Sasaki, S.; Okada, E.; Matsuura, H.; Goudarzi, H.; Miyashita, C.; Okada, E.; Ito, Y.M.; Araki, A.; Kishi, R. Prenatal exposure to 11 perfluoroalkyl substances and fetal growth: A large-scale, prospective birth cohort study. Environ. Int. 2020, 136, 105355. [Google Scholar] [CrossRef] [PubMed]
- Darrow, L.A.; Stein, C.R.; Steenland, K. Serum Perfluorooctanoic Acid and Perfluorooctane Sulfonate Concentrations in Relation to Birth Outcomes in the Mid-Ohio Valley, 2005–2010. Environ. Health Perspect. 2013, 121, 1207–1213. [Google Scholar] [CrossRef] [Green Version]
- Kishi, R.; Nakajima, T.; Goudarzi, H.; Kobayashi, S.; Sasaki, S.; Okada, E.; Miyashita, C.; Itoh, S.; Araki, A.; Ikeno, T.; et al. The Association of Prenatal Exposure to Perfluorinated Chemicals with Maternal Essential and Long-Chain Polyunsaturated Fatty Acids during Pregnancy and the Birth Weight of Their Offspring: The Hokkaido Study. Environ. Health Perspect. 2015, 123, 1038–1045. [Google Scholar] [CrossRef] [Green Version]
- Bach, C.C.; Bech, B.H.; Brix, N.; Nohr, E.A.; Bonde, J.P.E.; Henriksen, T.B. Perfluoroalkyl and polyfluoroalkyl substances and human fetal growth: A systematic review. Crit. Rev. Toxicol. 2015, 45, 53–67. [Google Scholar] [CrossRef]
- Halldorsson, T.I.; Rytter, D.; Haug, L.S.; Bech, B.H.; Danielsen, I.; Becher, G.; Henriksen, T.B.; Olsen, S.F. Prenatal exposure to perfluorooctanoate and risk of overweight at 20 years of age: A prospective cohort study. Environ. Health Perspect. 2012, 120, 668–673. [Google Scholar] [CrossRef] [Green Version]
- Fleisch, A.F.; Rifas-Shiman, S.L.; Mora, A.M.; Calafat, A.M.; Ye, X.; Luttmann-Gibson, H.; Gillman, M.W.; Oken, E.; Sagiv, S.K. Early-Life Exposure to Perfluoroalkyl Substances and Childhood Metabolic Function. Environ. Health Perspect. 2017, 125, 481–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Misra, B.B.; Misra, A. The chemical exposome of type 2 diabetes mellitus: Opportunities and challenges in the omics era. Diabetes Metab. Syndr. 2020, 14, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Rantakokko, P.; Männistö, V.; Airaksinen, R.; Koponen, J.; Viluksela, M.; Kiviranta, H.; Pihlajamäki, J. Persistent organic pollutants and non-alcoholic fatty liver disease in morbidly obese patients: A cohort study. Environ. Health 2015, 14, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunderland, E.M.; Hu, X.C.; Dassuncao, C.; Tokranov, A.K.; Wagner, C.C.; Allen, J.G. A review of the pathways of human exposure to poly- and perfluoroalkyl substances (PFASs) and present understanding of health effects. J. Expo. Sci. Environ. Epidemiol. 2019, 29, 131–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.; Liu, Y.; Xu, B.; Gu, L.; Tang, W. PFOA is associated with diabetes and metabolic alteration in US men: National Health and Nutrition Examination Survey 2003–2012. Sci. Total Environ. 2018, 625, 566–574. [Google Scholar] [CrossRef]
- Cardenas, A.; Gold, D.R.; Hauser, R.; Kleinman, K.P.; Hivert, M.-F.; Calafat, A.M.; Ye, X.; Webster, T.F.; Horton, E.S.; Oken, E. Plasma Concentrations of Per- and Polyfluoroalkyl Substances at Baseline and Associations with Glycemic Indicators and Diabetes Incidence among High-Risk Adults in the Diabetes Prevention Program Trial. Environ. Health Perspect. 2017, 125, 107001. [Google Scholar] [CrossRef] [Green Version]
- Bach, J.F. The effect of infections on susceptibility to autoimmune and allergic diseases. N. Engl. J. Med. 2002, 347, 911–920. [Google Scholar] [CrossRef]
- Patterson, C.C.; Dahlquist, G.G.; Gyürüs, E.; Green, A.; Soltész, G. Incidence trends for childhood type 1 diabetes in Europe during 1989–2003 and predicted new cases 2005–20: A multicentre prospective registration study. Lancet 2009, 373, 2027–2033. [Google Scholar] [CrossRef]
- Atkinson, M.A.; Eisenbarth, G.S.; Michels, A.W. Type 1 diabetes. Lancet 2014, 383, 69–82. [Google Scholar] [CrossRef] [Green Version]
- Knip, M.; Veijola, R.; Virtanen, S.M.; Hyöty, H.; Vaarala, O.; Åkerblom, H.K. Environmental triggers and determinants of type 1 diabetes. Diabetes 2005, 54, S125–S136. [Google Scholar] [CrossRef] [Green Version]
- Harjutsalo, V.; Sund, R.; Knip, M.; Groop, P.H. Incidence of type 1 diabetes in Finland. JAMA 2013, 310, 427–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaikkonen, R.; Mäki, P.; Hakulinen-Viitanen, T.; Markkula, J.; Wikström, K.; Ovaskainen, M.-L.; Virtanen, S.; Laatikainen, T. Health and Well-Being Inequalities Among Children and Their Families; National Institute for Health and Wellfare: Helsinki, Finland, 2012. [Google Scholar]
- Corsini, E.; Luebke, R.W.; Germolec, D.R.; DeWitt, J.C. Perfluorinated compounds: Emerging POPs with potential immunotoxicity. Toxicol. Lett. 2014, 230, 263–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- National Toxicology Program. Systematic Review of Immunotoxicity Associated with Exposure to PFOA or PFOS; National Toxicology Program: Research Triangle Park, NC, USA, 2016. Available online: https://ntp.niehs.nih.gov/ntp/ohat/pfoa_pfos/pfoa_pfosmonograph_508.pdf (accessed on 8 November 2020).
- Liu, S.; Yin, N.; Faiola, F. PFOA and PFOS Disrupt the Generation of Human Pancreatic Progenitor Cells. Environ. Sci. Technol. Lett. 2018, 5, 237–242. [Google Scholar] [CrossRef]
- Conway, B.; Innes, K.E.; Long, D. Perfluoroalkyl substances and beta cell deficient diabetes. J. Diabetes Complicat. 2016, 30, 993–998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeWitt, J.C.; Shnyra, A.; Badr, M.Z.; Loveless, S.E.; Hoban, D.; Frame, S.R.; Cunard, R.; Anderson, S.E.; Meade, B.J.; Peden-Adams, M.M.; et al. Immunotoxicity of Perfluorooctanoic Acid and Perfluorooctane Sulfonate and the Role of Peroxisome Proliferator-Activated Receptor Alpha. Crit. Rev. Toxicol. 2009, 39, 76–94. [Google Scholar] [CrossRef]
- Salo, H.M.; Koponen, J.; Kiviranta, H.; Rantakokko, P.; Honkanen, J.; Härkönen, T.; Ilonen, J.; Virtanen, S.M.; Tillmann, V.; Knip, M.; et al. No evidence of the role of early chemical exposure in the development of β-cell autoimmunity. Env. Sci. Pollu. Res. Int. 2018. [Google Scholar] [CrossRef] [Green Version]
- Fang, X.; Zhang, L.; Feng, Y.; Zhao, Y.; Dai, J. Immunotoxic Effects of Perfluorononanoic Acid on BALB/c Mice. Toxicol. Sci. 2008, 105, 312–321. [Google Scholar] [CrossRef]
- Oresic, M.; Simell, S.; Sysi-Aho, M.; Nanto-Salonen, K.; Seppanen-Laakso, T.; Parikka, V.; Katajamaa, M.; Hekkala, A.; Mattila, I.; Keskinen, P.; et al. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J. Exp. Med. 2008, 205, 2975–2984. [Google Scholar] [CrossRef] [Green Version]
- Overgaard, A.J.; Kaur, S.; Pociot, F. Metabolomic Biomarkers in the Progression to Type 1 Diabetes. Curr. Diabetes Rep. 2016, 16, 127. [Google Scholar] [CrossRef]
- Johnson, R.K.; Vanderlinden, L.; DeFelice, B.C.; Kechris, K.; Uusitalo, U.; Fiehn, O.; Sontag, M.; Crume, T.; Beyerlein, A.; Lernmark, A.; et al. Metabolite-related dietary patterns and the development of islet autoimmunity. Sci. Rep. 2019, 9, 14819. [Google Scholar] [CrossRef] [Green Version]
- Oresic, M.; Gopalacharyulu, P.; Mykkanen, J.; Lietzen, N.; Makinen, M.; Nygren, H.; Simell, S.; Simell, V.; Hyoty, H.; Veijola, R.; et al. Cord serum lipidome in prediction of islet autoimmunity and type 1 diabetes. Diabetes 2013, 62, 3268–3274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Torre, D.; Seppanen-Laakso, T.; Larsson, H.E.; Hyotylainen, T.; Ivarsson, S.A.; Lernmark, A.; Oresic, M.; DiPi, S.S.G. Decreased cord-blood phospholipids in young age-at-onset type 1 diabetes. Diabetes 2013, 62, 3951–3956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alkotob, S.S.; Cannedy, C.; Harter, K.; Movassagh, H.; Paudel, B.; Prunicki, M.; Sampath, V.; Schikowski, T.; Smith, E.; Zhao, Q.; et al. Advances and novel developments in environmental influences on the development of atopic diseases. Allergy 2020. [Google Scholar] [CrossRef]
- Eguiluz-Gracia, I.; Mathioudakis, A.G.; Bartel, S.; Vijverberg, S.J.H.; Fuertes, E.; Comberiati, P.; Cai, Y.S.; Tomazic, P.V.; Diamant, Z.; Vestbo, J.; et al. The need for clean air: The way air pollution and climate change affect allergic rhinitis and asthma. Allergy 2020, 75, 2170–2184. [Google Scholar] [CrossRef]
- Guarnieri, M.; Balmes, J.R. Outdoor air pollution and asthma. Lancet 2014, 383, 1581–1592. [Google Scholar] [CrossRef] [Green Version]
- Burney, P.; Amaral, A.F.S. Air pollution and chronic airway disease: Is the evidence always clear? Lancet 2019, 394, 2198–2200. [Google Scholar] [CrossRef]
- Miller, R.L.; Peden, D.B. Environmental effects on immune responses in patients with atopy and asthma. J. Allergy Clin. Immunol. 2014, 134, 1001–1008. [Google Scholar] [CrossRef] [Green Version]
- Cecchi, L.; D’Amato, G.; Annesi-Maesano, I. External exposome and allergic respiratory and skin diseases. J. Allergy Clin. Immunol. 2018, 141, 846–857. [Google Scholar] [CrossRef] [Green Version]
- Burbank, A.J.; Sood, A.K.; Kesic, M.J.; Peden, D.B.; Hernandez, M.L. Environmental determinants of allergy and asthma in early life. J. Allergy Clin. Immunol. 2017, 140, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Kantor, R.; Silverberg, J.I. Environmental risk factors and their role in the management of atopic dermatitis. Expert Rev. Clin. Immunol. 2017, 13, 15–26. [Google Scholar] [CrossRef]
- Kim, D.; Seo, S.; Min, S.; Simoni, Z.; Kim, S.; Kim, M. A Closer Look at the Bivariate Association between Ambient Air Pollution and Allergic Diseases: The Role of Spatial Analysis. Int. J. Environ. Res. Public Health 2018, 15, 1625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlsten, C.; Rider, C.F. Traffic-related air pollution and allergic disease: An update in the context of global urbanization. Curr. Opin. Allergy Clin. Immunol. 2017, 17, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.K.; Bing, R.; Kiang, J.; Bashir, S.; Spath, N.; Stelzle, D.; Mortimer, K.; Bularga, A.; Doudesis, D.; Joshi, S.S.; et al. Adverse health effects associated with household air pollution: A systematic review, meta-analysis, and burden estimation study. Lancet Glob. Health 2020, 8, e1427–e1434. [Google Scholar] [CrossRef]
- Bowatte, G.; Lodge, C.; Lowe, A.J.; Erbas, B.; Perret, J.; Abramson, M.J.; Matheson, M.; Dharmage, S.C. The influence of childhood traffic-related air pollution exposure on asthma, allergy and sensitization: A systematic review and a meta-analysis of birth cohort studies. Allergy 2015, 70, 245–256. [Google Scholar] [CrossRef]
- Lloyd, C.M.; Marsland, B.J. Lung Homeostasis: Influence of Age, Microbes, and the Immune System. Immunity 2017, 46, 549–561. [Google Scholar] [CrossRef] [Green Version]
- Agache, I.; Miller, R.; Gern, J.E.; Hellings, P.W.; Jutel, M.; Muraro, A.; Phipatanakul, W.; Quirce, S.; Peden, D. Emerging concepts and challenges in implementing the exposome paradigm in allergic diseases and asthma: A Practall document. Allergy 2019, 74, 449–463. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Ryu, M.H.; Rider, C.F.; Tse, W.; Clifford, R.L.; Aristizabal, M.J.; Wen, W.; Carlsten, C. Predominant DNMT and TET mediate effects of allergen on the human bronchial epithelium in a controlled air pollution exposure study. J. Allergy Clin. Immunol. 2020. [Google Scholar] [CrossRef]
- Maestre-Batlle, D.; Huff, R.D.; Schwartz, C.; Alexis, N.E.; Tebbutt, S.J.; Turvey, S.; Bolling, A.K.; Carlsten, C. Dibutyl Phthalate Augments Allergen-induced Lung Function Decline and Alters Human Airway Immunology. A Randomized Crossover Study. Am. J. Respir Crit. Care Med. 2020, 202, 672–680. [Google Scholar] [CrossRef]
- Murrison, L.B.; Brandt, E.B.; Myers, J.B.; Hershey, G.K.K. Environmental exposures and mechanisms in allergy and asthma development. J. Clin. Investig. 2019, 129, 1504–1515. [Google Scholar] [CrossRef] [Green Version]
- Schultz, E.S.; Litonjua, A.A.; Melen, E. Effects of Long-Term Exposure to Traffic-Related Air Pollution on Lung Function in Children. Curr. Allergy Asthma Rep. 2017, 17, 41. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.; Bai, L.; Burnett, R.T.; Kwong, J.C.; Hystad, P.; van Donkelaar, A.; Lavigne, E.; Weichenthal, S.; Copes, R.; Martin, R.V.; et al. Air Pollution as a Risk Factor for Incident COPD and Asthma: 15-Year Population-Based Cohort Study. Am. J. Respir Crit. Care Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Granum, B.; Oftedal, B.; Agier, L.; Siroux, V.; Bird, P.; Casas, M.; Warembourg, C.; Wright, J.; Chatzi, L.; de Castro, M.; et al. Multiple environmental exposures in early-life and allergy-related outcomes in childhood. Environ. Int. 2020, 144, 106038. [Google Scholar] [CrossRef] [PubMed]
- Ait Bamai, Y.; Miyashita, C.; Araki, A.; Nakajima, T.; Sasaki, S.; Kishi, R. Effects of prenatal di(2-ethylhexyl) phthalate exposure on childhood allergies and infectious diseases: The Hokkaido Study on Environment and Children’s Health. Sci. Total Environ. 2018, 618, 1408–1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gascon, M.; Casas, M.; Morales, E.; Valvi, D.; Ballesteros-Gomez, A.; Luque, N.; Rubio, S.; Monfort, N.; Ventura, R.; Martinez, D.; et al. Prenatal exposure to bisphenol A and phthalates and childhood respiratory tract infections and allergy. J. Allergy Clin. Immunol. 2015, 135, 370–378. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Contaminants in the Food Chain (EFSA CONTAM, Panel); Schrenk, D.; Bignami, M.; Bodin, L.; Chipman, J.K.; Del Mazo, J.; Grasl-Kraupp, B.; Hogstrand, C.; Hoogenboom, L.R.; Leblanc, J.C.; et al. Risk to human health related to the presence of perfluoroalkyl substances in food. Efsa J. 2020, 18, e06223. [Google Scholar] [CrossRef]
- Jackson-Browne, M.S.; Eliot, M.; Patti, M.; Spanier, A.J.; Braun, J.M. PFAS (per- and polyfluoroalkyl substances) and asthma in young children: NHANES 2013–2014. Int. J. Hyg. Environ. Health 2020, 229, 113565. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Deji, Z.; Huang, Z. Exposure to perfluoroalkyl substances and allergic outcomes in children: A systematic review and meta-analysis. Environ. Res. 2020, 191, 110145. [Google Scholar] [CrossRef]
- Moran, T.P. The External Exposome and Food Allergy. Curr. Allergy Asthma Rep. 2020, 20, 37. [Google Scholar] [CrossRef]
- Savage, J.H.; Matsui, E.C.; Wood, R.A.; Keet, C.A. Urinary levels of triclosan and parabens are associated with aeroallergen and food sensitization. J. Allergy Clin. Immunol. 2012, 130, 453–460. [Google Scholar] [CrossRef]
- Bertelsen, R.J.; Longnecker, M.P.; Lovik, M.; Calafat, A.M.; Carlsen, K.H.; London, S.J.; Lodrup Carlsen, K.C. Triclosan exposure and allergic sensitization in Norwegian children. Allergy 2013, 68, 84–91. [Google Scholar] [CrossRef]
- Lee-Sarwar, K.; Hauser, R.; Calafat, A.M.; Ye, X.; O’Connor, G.T.; Sandel, M.; Bacharier, L.B.; Zeiger, R.S.; Laranjo, N.; Gold, D.R.; et al. Prenatal and early-life triclosan and paraben exposure and allergic outcomes. J. Allergy Clin. Immunol. 2018, 142, 269–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herberth, G.; Pierzchalski, A.; Feltens, R.; Bauer, M.; Roder, S.; Olek, S.; Hinz, D.; Borte, M.; von Bergen, M.; Lehmann, I.; et al. Prenatal phthalate exposure associates with low regulatory T-cell numbers and atopic dermatitis in early childhood: Results from the LINA mother-child study. J. Allergy Clin. Immunol. 2017, 139, 1376–1379. [Google Scholar] [CrossRef] [Green Version]
- Podlecka, D.; Gromadzinska, J.; Mikolajewska, K.; Fijalkowska, B.; Stelmach, I.; Jerzynska, J. Longitudinal effect of phthalates exposure on allergic diseases in children. Ann. Allergy Asthma Immunol. 2020, 125, 84–89. [Google Scholar] [CrossRef]
- Renz, H.; Skevaki, C. Early life microbial exposures and allergy risks: Opportunities for prevention. Nat. Rev. Immunol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.E.; Mallapaty, A.; Miller, R.L. It’s not just the food you eat: Environmental factors in the development of food allergies. Environ. Res. 2018, 165, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Rappaport, S.M. Genetic Factors Are Not the Major Causes of Chronic Diseases. PLoS ONE 2016, 11, e0154387. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orešič, M.; McGlinchey, A.; Wheelock, C.E.; Hyötyläinen, T. Metabolic Signatures of the Exposome—Quantifying the Impact of Exposure to Environmental Chemicals on Human Health. Metabolites 2020, 10, 454. https://doi.org/10.3390/metabo10110454
Orešič M, McGlinchey A, Wheelock CE, Hyötyläinen T. Metabolic Signatures of the Exposome—Quantifying the Impact of Exposure to Environmental Chemicals on Human Health. Metabolites. 2020; 10(11):454. https://doi.org/10.3390/metabo10110454
Chicago/Turabian StyleOrešič, Matej, Aidan McGlinchey, Craig E. Wheelock, and Tuulia Hyötyläinen. 2020. "Metabolic Signatures of the Exposome—Quantifying the Impact of Exposure to Environmental Chemicals on Human Health" Metabolites 10, no. 11: 454. https://doi.org/10.3390/metabo10110454
APA StyleOrešič, M., McGlinchey, A., Wheelock, C. E., & Hyötyläinen, T. (2020). Metabolic Signatures of the Exposome—Quantifying the Impact of Exposure to Environmental Chemicals on Human Health. Metabolites, 10(11), 454. https://doi.org/10.3390/metabo10110454







