Untargeted Metabolomics and Polyamine Profiling in Serum before and after Surgery in Colorectal Cancer Patients
Abstract
:1. Introduction
2. Results
2.1. Metabolic Patterns Detected in Serum Samples of CRC Patients Using Untargeted Metabolomics
2.2. Metabolic Pathway Analysis
2.3. Quantification of Polyamines in Serum Samples of Patients with Colorectal Cancer
3. Discussion
4. Materials and Methods
4.1. Chemicals and Materials
4.2. Serum Sample Collection
4.3. Sample Preparation for Untargeted Metabolic Profiling
4.4. UPLC-MS
4.5. Sample Preparation for Targeted Profiling
4.6. LC-MS Profiling of Polyamines
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Jung, K.W.; Won, Y.J.; Kong, H.J.; Lee, E.S. Prediction of Cancer Incidence and Mortality in Korea, 2019. Cancer Res. Treat. 2019, 51, 431–437. [Google Scholar] [CrossRef]
- Milsom, J.W.; Bohm, B.; Hammerhofer, K.A.; Fazio, V.; Steiger, E.; Elson, P. A prospective, randomized trial comparing laparoscopic versus conventional techniques in colorectal cancer surgery: A preliminary report. J. Am. Coll. Surg. 1998, 187, 46–54. [Google Scholar] [CrossRef]
- Huang, Y.-M.; Huang, Y.J.; Wei, P.-L. Colorectal cancer surgery using the Da Vinci Xi and si systems: Comparison of perioperative outcomes. Surg. Innov. 2019, 26, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Liesenfeld, D.B.; Habermann, N.; Owen, R.W.; Scalbert, A.; Ulrich, C.M. Review of mass spectrometry-based metabolomics in cancer research. Cancer Epidemiol. Biomark. Prev. 2013, 22, 2182–2201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.L.; Qin, H.L.; Liu, W.J.; Peng, J.Y.; Huang, L.; Zhao, X.P.; Cheng, Y.Y. Ultra-high performance liquid chromatography-mass spectrometry for the metabolomic analysis of urine in colorectal cancer. Dig. Dis. Sci. 2009, 54, 2655–2662. [Google Scholar] [CrossRef] [PubMed]
- Nishiumi, S.; Kobayashi, T.; Ikeda, A.; Yoshie, T.; Kibi, M.; Izumi, Y.; Okuno, T.; Hayashi, N.; Kawano, S.; Takenawa, T.; et al. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS ONE 2012, 7, e40459. [Google Scholar] [CrossRef] [PubMed]
- Heby, O. Role of polyamines in the control of cell proliferation and differentiation. Differentiation 1981, 19, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Manni, A.; Grove, R.; Kunselman, S.; Aldaz, C.M. Involvement of the polyamine pathway in breast cancer progression. Cancer Lett. 1995, 92, 49–57. [Google Scholar] [CrossRef]
- Suh, J.W.; Lee, S.H.; Chung, B.C.; Park, J. Urinary polyamine evaluation for effective diagnosis of various cancers. J. Chromatogr. B Biomed. Sci. Appl. 1997, 688, 179–186. [Google Scholar] [CrossRef]
- Linsalata, M.; Caruso, M.G.; Leo, S.; Guerra, V.; D’Attoma, B.; Di Leo, A. Prognostic value of tissue polyamine levels in human colorectal carcinoma. Anticancer Res. 2002, 22, 2465–2469. [Google Scholar]
- Loser, C.; Folsch, U.R.; Paprotny, C.; Creutzfeldt, W. Polyamines in colorectal cancer. Evaluation of polyamine concentrations in the colon tissue, serum, and urine of 50 patients with colorectal cancer. Cancer 1990, 65, 958–966. [Google Scholar] [CrossRef]
- Byun, J.A.; Choi, M.H.; Moon, M.H.; Kong, G.; Chul Chung, B. Serum polyamines in pre- and post-operative patients with breast cancer corrected by menopausal status. Cancer Lett. 2009, 273, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Merrill, A.H., Jr.; Schmelz, E.M.; Wang, E.; Schroeder, J.J.; Dillehay, D.L.; Riley, R.T. Role of dietary sphingolipids and inhibitors of sphingolipid metabolism in cancer and other diseases. J. Nutr. 1995, 125, S1677–S1682. [Google Scholar] [CrossRef]
- Spiegel, S.; Merrill, A.H., Jr. Sphingolipid metabolism and cell growth regulation. FASEB J. 1996, 10, 1388–1397. [Google Scholar] [CrossRef]
- Dany, M.; Ogretmen, B. Ceramide induced mitophagy and tumor suppression. Biochim. Biophys. Acta 2015, 1853, 2834–2845. [Google Scholar] [CrossRef] [Green Version]
- Dowdy, T.; Zhang, L.; Celiku, O.; Movva, S.; Lita, A.; Ruiz-Rodado, V.; Gilbert, M.R.; Larion, M. Sphingolipid Pathway as a Source of Vulnerability in IDH1mut Glioma. Cancers 2020, 12, 2910. [Google Scholar] [CrossRef]
- Kang, H.M.; Son, H.S.; Cui, Y.H.; Youn, B.; Son, B.; Kaushik, N.K.; Uddin, N.; Lee, J.S.; Song, J.Y.; Kaushik, N.; et al. Phytosphingosine exhibits an anti-epithelial-mesenchymal transition function by the inhibition of EGFR signaling in human breast cancer cells. Oncotarget 2017, 8, 77794–77808. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Chen, H.; Li, Y.; Li, L.; Qiu, Y.; Ren, J. Endocannabinoid and ceramide levels are altered in patients with colorectal cancer. Oncol. Rep. 2015, 34, 447–454. [Google Scholar] [CrossRef] [Green Version]
- Owen, R.W.; Dodo, M.; Thompson, M.H.; Hill, M.J. Fecal steroids and colorectal cancer. Nutr. Cancer 1987, 9, 73–80. [Google Scholar] [CrossRef]
- Di Leo, A.; Linsalata, M.; Cavallini, A.; Messa, C.; Russo, F. Sex steroid hormone receptors, epidermal growth factor receptor, and polyamines in human colorectal cancer. Dis. Colon Rectum 1992, 35, 305–309. [Google Scholar] [CrossRef]
- Amos-Landgraf, J.M.; Heijmans, J.; Wielenga, M.C.; Dunkin, E.; Krentz, K.J.; Clipson, L.; Ederveen, A.G.; Groothuis, P.G.; Mosselman, S.; Muncan, V.; et al. Sex disparity in colonic adenomagenesis involves promotion by male hormones, not protection by female hormones. Proc. Natl. Acad. Sci. USA 2014, 111, 16514–16519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bin Sayeed, M.S.; Karim, S.M.R.; Sharmin, T.; Morshed, M.M. Critical Analysis on Characterization, Systemic Effect, and Therapeutic Potential of Beta-Sitosterol: A Plant-Derived Orphan Phytosterol. Medicines 2016, 3, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Borchert, G.L.; Donald, S.P.; Surazynski, A.; Hu, C.A.; Weydert, C.J.; Oberley, L.W.; Phang, J.M. MnSOD inhibits proline oxidase-induced apoptosis in colorectal cancer cells. Carcinogenesis 2005, 26, 1335–1342. [Google Scholar] [CrossRef]
- Roomi, M.W.; Ivanov, V.; Kalinovsky, T.; Niedzwiecki, A.; Rath, M. In vivo antitumor effect of ascorbic acid, lysine, proline and green tea extract on human colon cancer cell HCT 116 xenografts in nude mice: Evaluation of tumor growth and immunohistochemistry. Oncol. Rep. 2005, 13, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Phang, J.M.; Liu, W. Proline metabolism and cancer. Front. Biosci. 2012, 17, 1835–1845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simoneau, A.R.; Gerner, E.W.; Nagle, R.; Ziogas, A.; Fujikawa-Brooks, S.; Yerushalmi, H.; Ahlering, T.E.; Lieberman, R.; McLaren, C.E.; Anton-Culver, H.; et al. The effect of difluoromethylornithine on decreasing prostate size and polyamines in men: Results of a year-long phase IIb randomized placebo-controlled chemoprevention trial. Cancer Epidemiol. Biomark. Prev. 2008, 17, 292–299. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Chen, Y.; Hao, L.; Hou, A.; Chen, X.; Li, Y.; Wang, R.; Luo, P.; Ruan, Z.; Ou, J.; et al. Macrophages induce resistance to 5-fluorouracil chemotherapy in colorectal cancer through the release of putrescine. Cancer Lett. 2016, 381, 305–313. [Google Scholar] [CrossRef]
- Harik, S.I.; Sutton, C.H. Putrescine as a biochemical marker of malignant brain tumors. Cancer Res. 1979, 39, 5010–5015. [Google Scholar]
- Brachet, P.; Quemener, V.; Havouis, R.; Tome, D.; Moulinoux, J.P. Alterations in intestinal uptake of putrescine and tissue polyamine concentrations in tumor-bearing rats. Biochim. Biophys. Acta 1994, 1227, 161–170. [Google Scholar] [CrossRef]
- Farriol, M.; Segovia-Silvestre, T.; Castellanos, J.M.; Venereo, Y.; Orta, X. Role of putrescine in cell proliferation in a colon carcinoma cell line. Nutrition 2001, 17, 934–938. [Google Scholar] [CrossRef]
- Lee, Y.R.; Im, E.; Kim, H.; Lew, B.L.; Sim, W.Y.; Lee, J.; Oh, H.B.; Paeng, K.J.; Hong, J.; Chung, B.C. Untargeted Metabolomics and Steroid Signatures in Urine of Male Pattern Baldness Patients after Finasteride Treatment for a Year. Metabolites 2020, 10, 131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westerhuis, J.A.; Hoefsloot, H.C.J.; Smit, S.; Vis, D.J.; Smilde, A.K.; van Velzen, E.J.J.; van Duijnhoven, J.P.M.; van Dorsten, F.A. Assessment of PLSDA cross validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef] [Green Version]
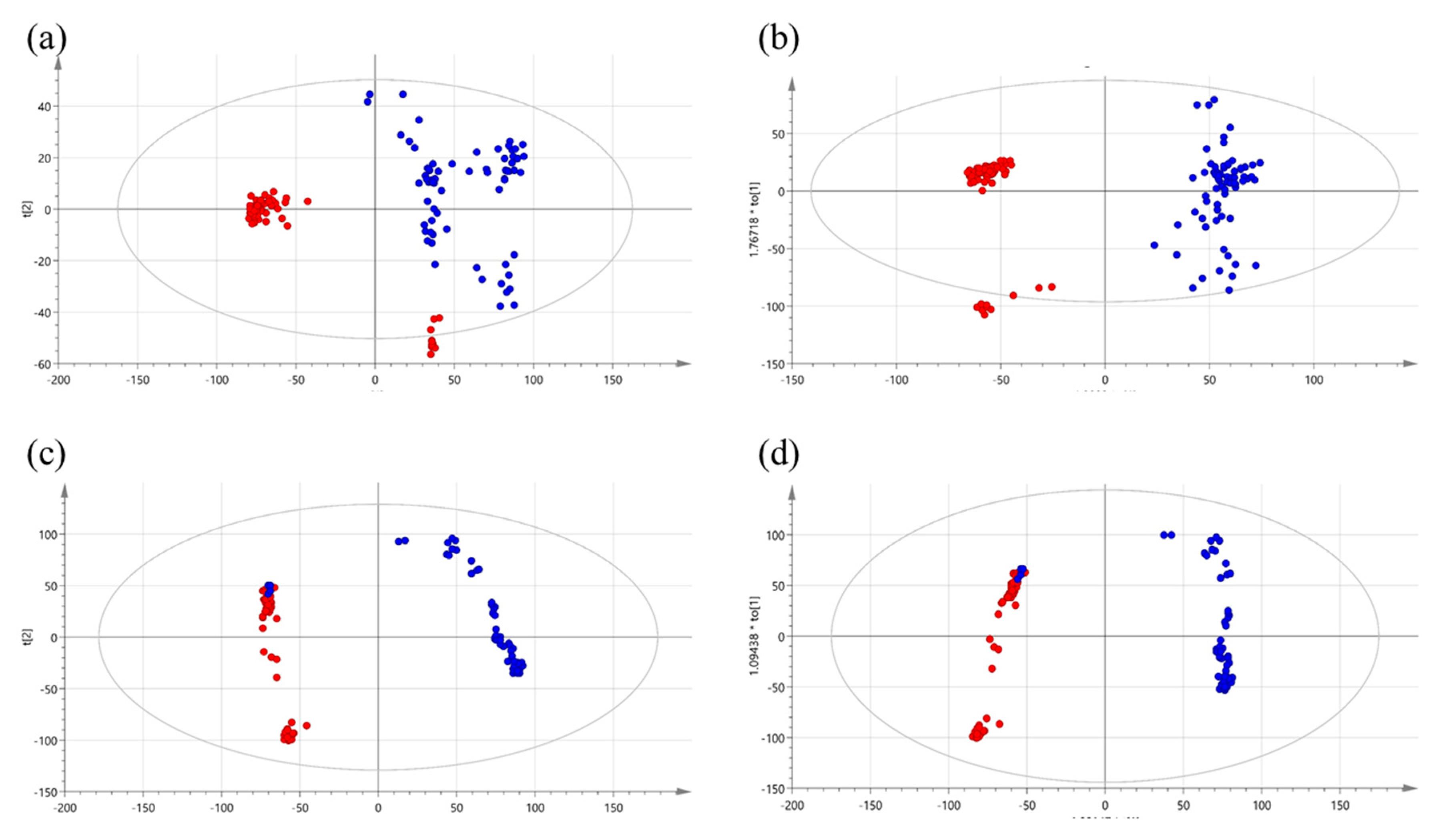
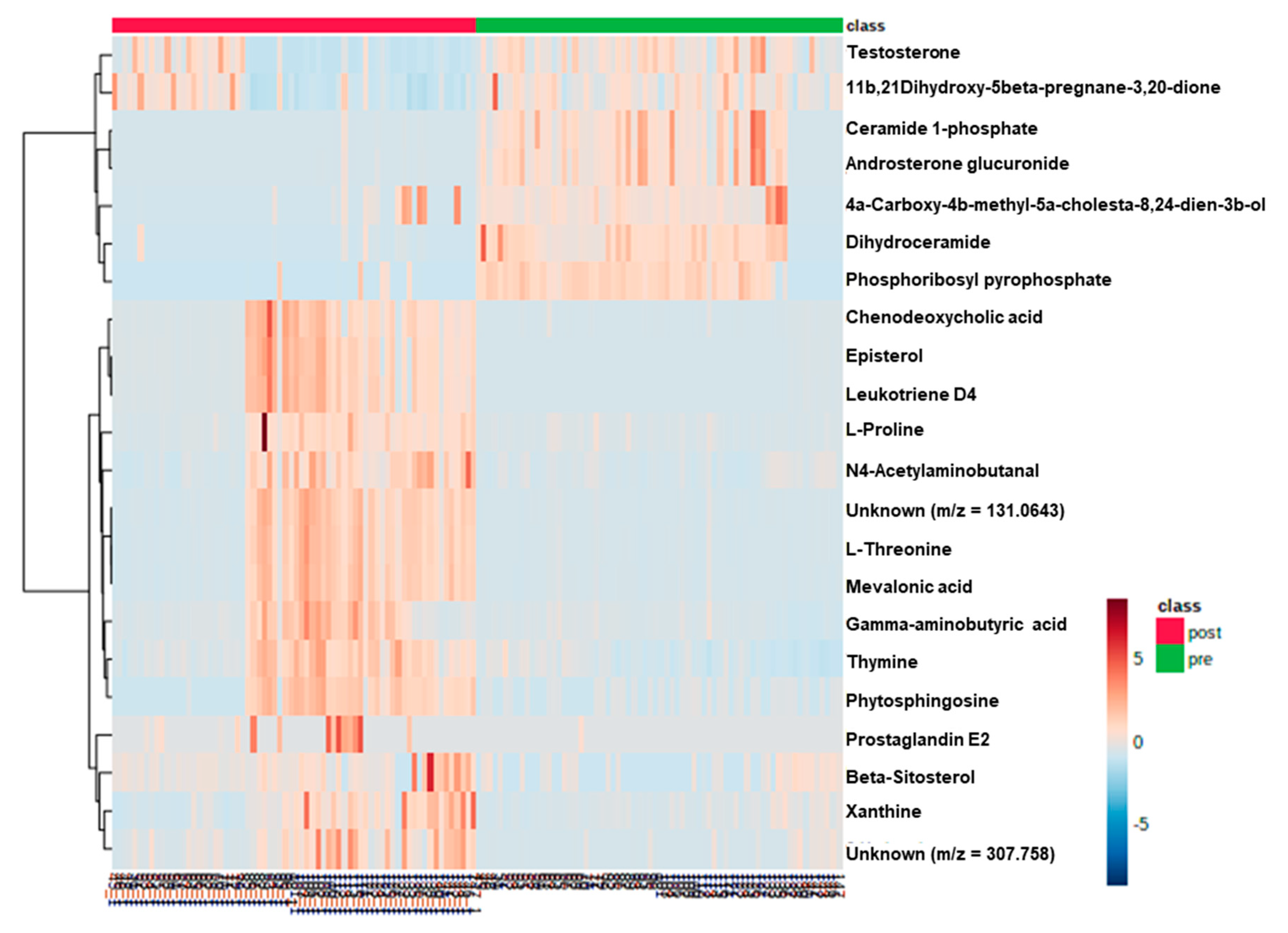
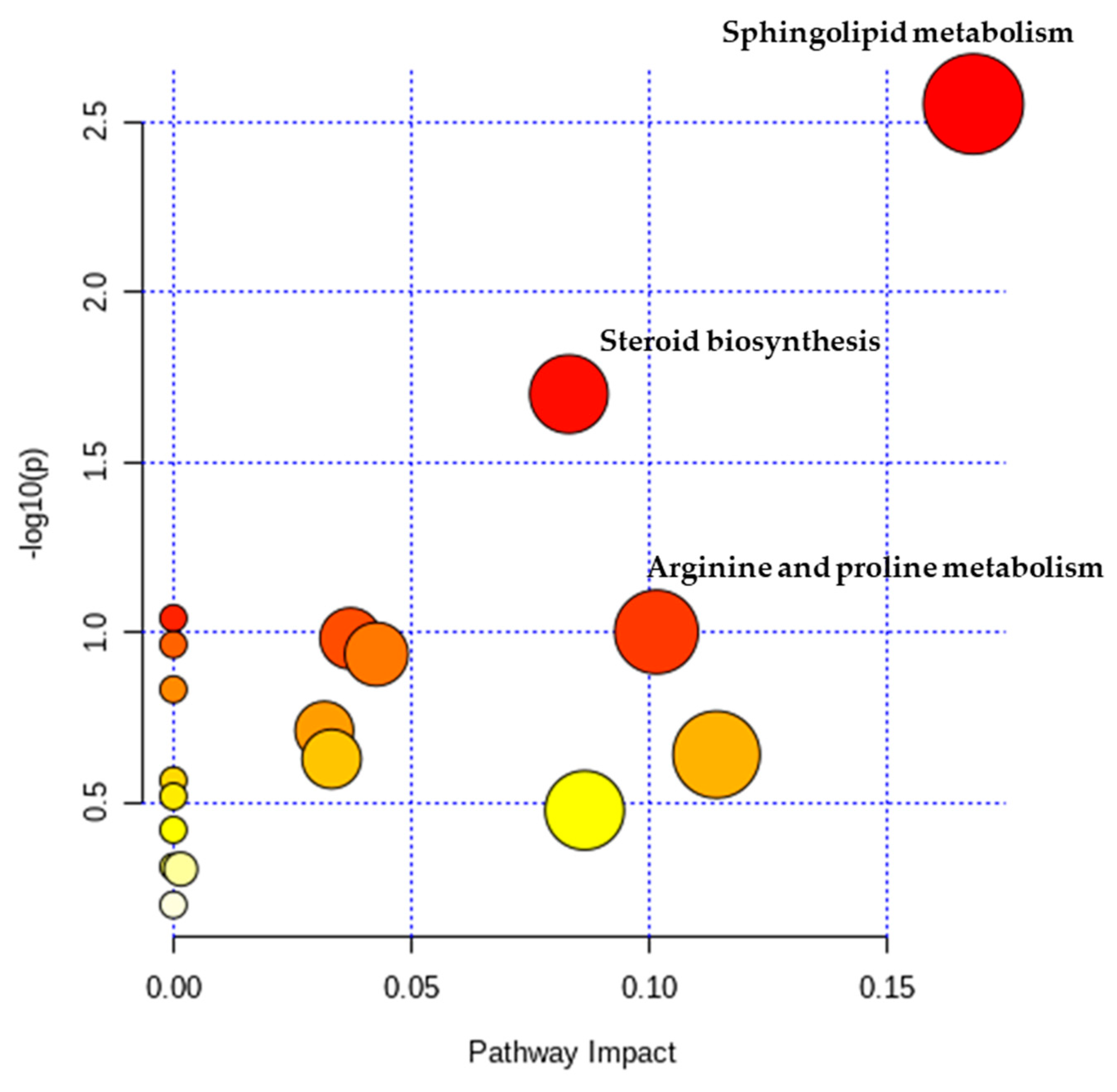

| Metabolites | Related Pathways | VIP | p-Value | Fold Change | Correlation Coefficient | Regulation |
|---|---|---|---|---|---|---|
| Gamma-aminobutyric acid | Arginine and proline metabolism | 1.0023 | <0.001 | 0.145828341 | 4.117358559 | Up |
| l-Proline | Arginine and proline metabolism | 1.71621 | <0.001 | 0.248728569 | 3.038287404 | Up |
| l-Threonine | Glycine, serine, and threonine metabolism | 4.84114 | <0.001 | −0.053349474 | 6.563445768 | Up |
| N4-Acetylaminobutanal | Lysine degradation | 1.62713 | <0.001 | 0.261500031 | 2.625861377 | Up |
| Unknown (m/z = 131.0643) | NA | 1.06877 | <0.001 | −0.080578542 | 6.860433801 | Up |
| Thymine | pyrimidine metabolism | 1.50327 | <0.001 | −0.37427256 | 1.606954027 | Up |
| Mevalonic acid | Terpenoid backbone biosynthesis | 1.01718 | <0.001 | −0.093645679 | 6.286436005 | Up |
| Xanthine | Purine metabolism | 1.00114 | <0.001 | 0.516947844 | 2.518903854 | Up |
| Unknown (m/z = 307.0758) | NA | 1.42141 | <0.001 | 0.423472971 | 7.681495389 | Up |
| Testosterone | Steroid hormone biosynthesis | 1.0227 | <0.001 | −0.181019259 | 0.467837133 | Down |
| Phytosphingosine | Sphingolipid metabolism | 1.02506 | <0.001 | 0.283161742 | 4.211946967 | Up |
| Dihydroceramide | Sphingolipid metabolism | 2.90611 | <0.001 | 0.089880246 | 0.05540464 | Down |
| Prostaglandin E2 | Arachidonic acid metabolism | 1.44895 | 0.001 | −0.052474587 | 46.02889156 | Up |
| 11β,21-Dihydroxy-5β-pregnane-3,20-dione | Steroid hormone biosynthesis | 1.02444 | 0.002 | −0.002157481 | 0.726773548 | Down |
| Chenodeoxycholic acid | Primary bile acid biosynthesis | 1.01772 | <0.001 | −0.197870962 | 31.81241423 | Up |
| Phosphoribosyl pyrophosphate | pyrimidine metabolism | 1.05047 | <0.001 | 0.039117952 | 0.084114079 | Down |
| Episterol | Steroid biosynthesis | 1.58977 | <0.001 | −0.01518394 | 65.07462767 | Up |
| Beta-Sitosterol | Steroid biosynthesis | 1.34755 | <0.001 | 0.548996098 | 2.874787603 | Up |
| 4α-Carboxy-4β-methyl-5α-cholesta-8,24-dien-3β-ol | Steroid biosynthesis | 1.08986 | <0.001 | 0.317512451 | 0.329377471 | Down |
| Leukotriene D4 | Arachidonic acid metabolism | 2.41915 | <0.001 | −0.017846652 | 85.53840187 | Up |
| Ceramide 1-phosphate | Sphingolipid metabolism | 1.03921 | <0.001 | −0.167097911 | 0.016887882 | Down |
| Androsterone glucuronide | Steroid hormone biosynthesis | 1.023 | <0.001 | −0.164779789 | 0.022209325 | Down |
| Compound | Pre-Operative Patients (n = 68) | Post-Operative Patients (n = 68) | p-Value | ||
|---|---|---|---|---|---|
| Mean ± SD | Median, Range | Mean ± SD | Median, Range | ||
| N-PUT | 157.49 ± 82.84 | 137.38, 34.62–368.14 | 154.86 ± 76.96 | 131.01, 43.88–346.82 | 0.858 |
| N-CAD | 28.41 ± 12.97 | 26.38, 8.88–82.7 | 28.47 ± 15.27 | 23.62, 9.02–63.32 | 0.981 |
| DAP | 14.62 ± 10 | 12.24, 2.53–47.05 | 15 ± 12.16 | 11.21, 2.55–46.28 | 0.858 |
| PUT | 28.45 ± 16.4 | 30.22, 2.29–63.93 | 18.18 ± 11.67 | 15.63, 4.76–80.91 | 4.97−05 |
| CAD | 245.37 ± 304.23 | 28.21, 11.4–803.81 | 210.92 ± 193.49 | 200.65, 5.94–665.98 | 0.482 |
| N-SPD | 58.08 ± 20.65 | 53.61, 27.38–137.7 | 64.93 ± 18.44 | 67.43, 19.22–97.34 | 0.166 |
| SPD | 111.11 ± 78.11 | 98.5, 4.61–457.76 | 95.72 ± 78.62 | 64.23, 13.44–326.68 | 0.274 |
| N-SPM | 139.7 ± 98.81 | 120.77, 13.87–426.94 | 109.82 ± 83.62 | 72.08, 18.75–353.23 | 0.107 |
| SPM | 1040.28 ± 1040.5 | 669.95, 10.89–3764.49 | 849.93 ± 843.35 | 709.89, 9.8–3683.69 | 0.262 |
| Characteristic | Value |
|---|---|
| Patients (n) | 68 |
| Sex | |
| Male | 49 |
| Female | 19 |
| Age | |
| Mean ± SD | 57.52 ± 9.71 |
| Range | 20–71 |
| Nationality, n (%) | Korean (100) |
| Body mass index | 23.2 |
| Stage | |
| 0 | 5 |
| Ι | 21 |
| II | 21 |
| III | 1 |
| IIIA | 5 |
| IIIB | 9 |
| IIIC | 2 |
| n/a | 4 |
| Method of operation | |
| Laparoscopy | 55 |
| Da Vinci | 6 |
| Low anterior | 4 |
| Left hemicolectomy | 1 |
| Right hemicolectomy | 2 |
| Other chronic diseases, n (%) | |
| None | 54 |
| Hypertension | 10 |
| Diabetes | 3 |
| Crohn’s disease | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.R.; An, K.-Y.; Jeon, J.; Kim, N.K.; Lee, J.W.; Hong, J.; Chung, B.C. Untargeted Metabolomics and Polyamine Profiling in Serum before and after Surgery in Colorectal Cancer Patients. Metabolites 2020, 10, 487. https://doi.org/10.3390/metabo10120487
Lee YR, An K-Y, Jeon J, Kim NK, Lee JW, Hong J, Chung BC. Untargeted Metabolomics and Polyamine Profiling in Serum before and after Surgery in Colorectal Cancer Patients. Metabolites. 2020; 10(12):487. https://doi.org/10.3390/metabo10120487
Chicago/Turabian StyleLee, Yu Ra, Ki-Yong An, Justin Jeon, Nam Kyu Kim, Ji Won Lee, Jongki Hong, and Bong Chul Chung. 2020. "Untargeted Metabolomics and Polyamine Profiling in Serum before and after Surgery in Colorectal Cancer Patients" Metabolites 10, no. 12: 487. https://doi.org/10.3390/metabo10120487
APA StyleLee, Y. R., An, K.-Y., Jeon, J., Kim, N. K., Lee, J. W., Hong, J., & Chung, B. C. (2020). Untargeted Metabolomics and Polyamine Profiling in Serum before and after Surgery in Colorectal Cancer Patients. Metabolites, 10(12), 487. https://doi.org/10.3390/metabo10120487






