Biostimulants for Plant Growth and Mitigation of Abiotic Stresses: A Metabolomics Perspective
Abstract
1. Introduction
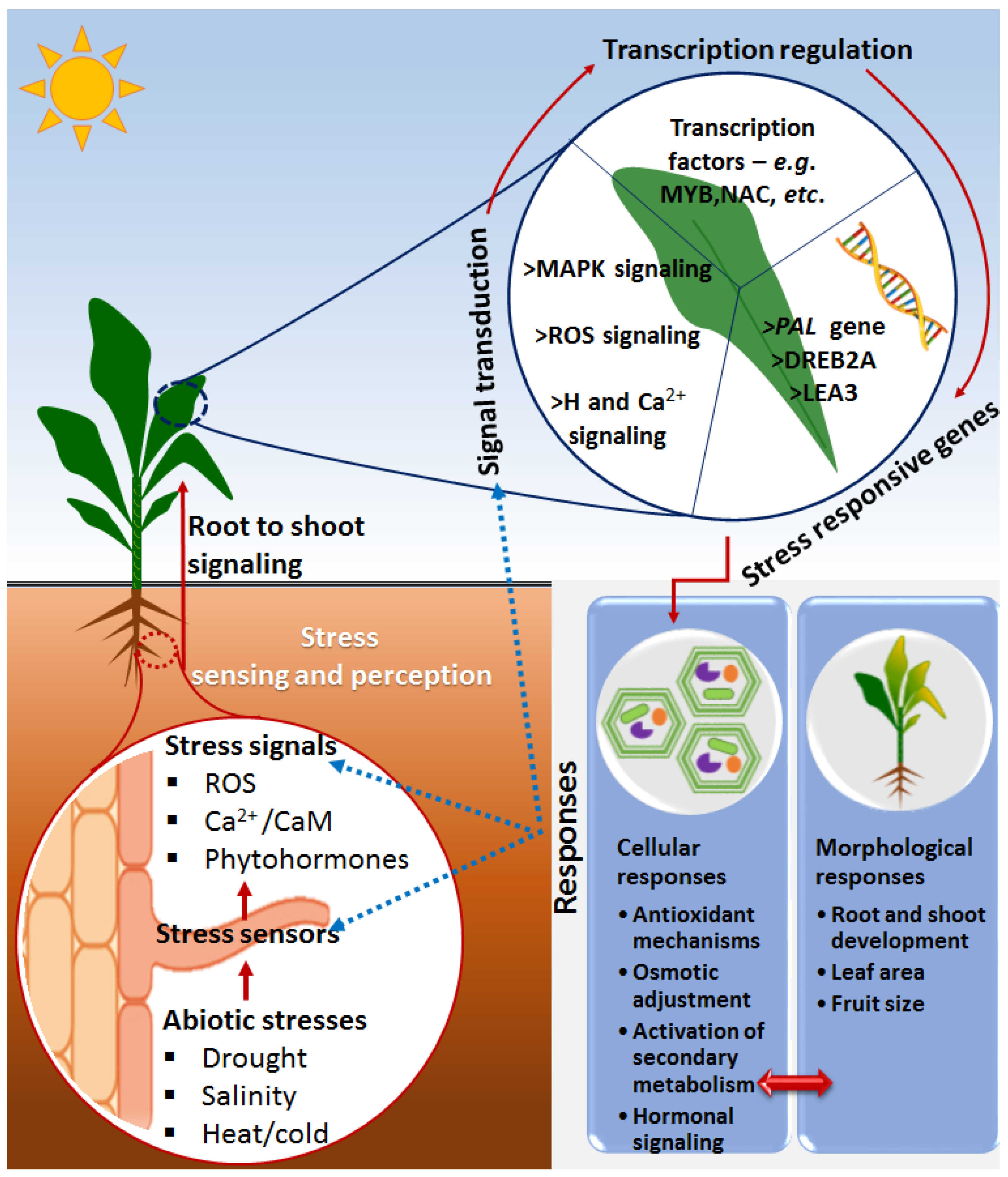
2. Basic and Overlapping Framework of Plant Immunity and Defense against Biotic and Abiotic Stresses
Priming against Abiotic Stresses
3. Biostimulants as Agronomic Tools to Promote Plant Growth and Counteract Abiotic Stresses
3.1. The (Lack of) Science of Biostimulants
3.2. The (Bio)Chemical Mechanisms Mediated by Different Classes of Biostimulants
4. Omics Sciences to Study Plant Biology in Abiotic Stress Conditions
4.1. Metabolomics in Plant Biostimulant Studies
4.1.1. An Overview of the Metabolomics Workflow
4.1.2. Application of Metabolomics in the Plant-Biostimulant-Abiotic Stress Interaction Studies
5. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Nomura, H.; Komori, T.; Uemura, S.; Kanda, Y.; Shimotani, K.; Nakai, K.; Furuichi, T.; Takebayashi, K.; Sugimoto, T.; Sano, S.; et al. Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat. Commun. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Stael, S.; Rocha, A.G.; Robinson, A.J.; Kmiecik, P.; Vothknecht, U.C.; Teige, M. Arabidopsis calcium-binding mitochondrial carrier proteins as potential facilitators of mitochondrial ATP-import and plastid SAM-import. FEBS Lett. 2011, 585, 3935–3940. [Google Scholar] [CrossRef] [PubMed]
- Balmer, A.; Pastor, V.; Glauser, G.; Mauch-Mani, B. Tricarboxylates induce defense priming against bacteria in Arab. Thaliana. Front. Plant Sci. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Balmer, A.; Pastor, V.; Gamir, J.; Flors, V.; Mauch-Mani, B. The “prime-ome”: Towards a holistic approach to priming. Trends Plant Sci. 2015, 20, 443–452. [Google Scholar] [CrossRef]
- Borges, A.A.; Jiménez-Arias, D.; Expósito-Rodríguez, M.; Sandalio, L.M.; Pérez, J.A. Priming crops against biotic and abiotic stresses: MSB as a tool for studying mechanisms. Front. Plant Sci. 2014, 5, 1–4. [Google Scholar] [CrossRef]
- Westman, S.M.; Kloth, K.J.; Hanson, J.; Ohlsson, A.B.; Albrectsen, B.R. Defence priming in Arabidopsis—A Meta-Analysis. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Martinez-medina, A.; Flors, V.; Heil, M.; Mauch-mani, B.; Pieterse, C.M.J. Recognizing plant defense priming. Trends Microbiol. 2016, 21, 818–822. [Google Scholar] [CrossRef]
- Tugizimana, F.; Djami-tchatchou, A.T.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Metabolomic analysis of defense-related reprogramming in Sorghum bicolor in response to Colletotrichum sublineolum infection reveals a functional metabolic web of phenylpropanoid and flavonoid pathways. Front. Plant Sci. 2019, 9, 1–20. [Google Scholar] [CrossRef]
- Fleming, T.R.; Fleming, C.C.; Levy, C.C.B.; Repiso, C.; Hennequart, F.; Nolasco, J.B.; Liu, F. Biostimulants enhance growth and drought tolerance in Arabidopsis thaliana and exhibit chemical priming action. Ann. Appl. Biol. 2019, 174, 153–165. [Google Scholar] [CrossRef]
- Yakhin, O.I.; Lubyanov, A.A.; Yakhin, I.A.; Brown, P.H. Biostimulants in plant science: A global perspective. Front. Plant Sci. 2017, 7, 1–32. [Google Scholar] [CrossRef]
- Paul, K.; Sorrentino, M.; Lucini, L.; Rouphael, Y.; Cardarelli, M.; Bonini, P.; Miras Moreno, M.B.; Reynaud, H.; Canaguier, R.; Trtílek, M.; et al. A combined phenotypic and metabolomic approach for elucidating the biostimulant action of a plant-derived protein hydrolysate on tomato grown under limited water availability. Front. Plant Sci. 2019, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Gao, X.; Xia, F.; Joshi, J.; Borza, T.; Wang-Pruski, G. Biostimulant and fungicidal effects of phosphite assessed by GC-TOF-MS analysis of potato leaf metabolome. Physiol. Mol. Plant Pathol. 2019, 106, 49–56. [Google Scholar] [CrossRef]
- Tugizimana, F.; Piater, L.; Dubery, I. Plant metabolomics: A new frontier in phytochemical analysis. S. Afr. J. Sci. 2013, 109, 1–11. [Google Scholar] [CrossRef]
- Rosato, A.; Tenori, L.; Cascante, M.; De Atauri Carulla, P.R.; Martins dos Santos, V.A.P.; Saccenti, E. From correlation to causation: Analysis of metabolomics data using systems biology approaches. Metabolomics 2018, 14, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Labuschagne, N.; Dubery, I.A. Unravelling the metabolic reconfiguration of the post-challenge primed state in Sorghum bicolor responding to Colletotrichum sublineolum infection. Metabolites 2019, 9, 194. [Google Scholar] [CrossRef]
- Beisken, S.; Eiden, M.; Salek, R.M. Getting the right answers: Understanding metabolomics challenges. Expert Rev. Mol. Diagn. 2015, 15, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Szecowka, M.; Heise, R.; Tohge, T.; Nunes-Nesi, A.; Vosloh, D.; Huege, J.; Feil, R.; Lunn, J.; Nikoloski, Z.; Stitt, M.; et al. Metabolic fluxes in an illuminated Arabidopsis rosette. Plant Cell 2013, 25, 694–714. [Google Scholar] [CrossRef]
- Rodríguez-Calzada, T.; Qian, M.; Strid, Å.; Neugart, S.; Schreiner, M.; Torres-Pacheco, I.; Guevara-González, R.G. Effect of UV-B radiation on morphology, phenolic compound production, gene expression, and subsequent drought stress responses in chili pepper (Capsicum annuum L.). Plant Physiol. Biochem. 2019, 134, 94–102. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Nobori, T.; Tsuda, K. The plant immune system in heterogeneous environments. Curr. Opin. Plant Biol. 2019, 50, 58–66. [Google Scholar] [CrossRef]
- Ramirez-Prado, J.S.; Abulfaraj, A.A.; Rayapuram, N.; Benhamed, M.; Hirt, H. Plant immunity: From signaling to epigenetic control of defense. Trends Plant. Sci. 2018, 23, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Gust, A.A.; Pruitt, R.; Nürnberger, T. Sensing danger: Key to activating plant immunity. Trends Plant. Sci. 2017, 22, 779–791. [Google Scholar] [CrossRef] [PubMed]
- Sanabria, N.M.; Huang, J.C.; Dubery, I.A. Self/nonself perception in plants in innate immunity and defense. Self/Nonself—Immune Recognit. Signal. 2010, 1, 40–54. [Google Scholar] [CrossRef]
- Kant, M.R.; Jonckheere, W.; Knegt, B.; Lemos, F.; Liu, J.; Schimmel, B.C.J.; Villarroel, C.A.; Ataide, L.M.S.; Dermauw, W.; Glas, J.J.; et al. Mechanisms and ecological consequences of plant defence induction and suppression in herbivore communities. Ann. Bot. 2015, 115, 1015–1051. [Google Scholar] [CrossRef] [PubMed]
- Ádám, A.L.; Nagy, Z.; Kátay, G.; Mergenthaler, E.; Viczián, O. Signals of systemic immunity in plants: Progress and open questions. Int. J. Mol. Sci. 2018, 19, 1146. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Feng, B.; He, P.; Shan, L. From chaos to harmony: Responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol. 2017, 55, 109–137. [Google Scholar] [CrossRef]
- Conrath, U. Systemic acquired resistance. Plant. Signal. Behav. 2006, 1, 179–184. [Google Scholar] [CrossRef]
- Luna, E.; Bruce, T.J.A.; Roberts, M.R.; Flors, V.; Ton, J. Next-generation systemic acquired resistance. Plant. Physiol. 2012, 158, 844–853. [Google Scholar] [CrossRef]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant. Biol. 2011, 11, 1–14. [Google Scholar] [CrossRef]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop production under drought and heat stress: Plant responses and management options. Front. Plant. Sci. 2017, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Chaves, M.M.; Maroco, J.P.; Pereira, J.S. Understanding plant responses to drought—From genes to the whole plant. Funct. Plant. Biol. 2003, 30, 239–264. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, R.A.; Gratão, P.L.; Monteiro, C.C.; Carvalho, R.F. What is new in the research on cadmium-induced stress in plants? Food Energy Secur. 2012, 1, 133–140. [Google Scholar] [CrossRef]
- Redondo-Gómez, S. Abiotic and biotic stress tolerance in plants. In Molecular Stress Physiology of Plants; Shanker, A., Venkateswarlu, B., Eds.; IntechOpen: London, UK, 2013; pp. 1–20. ISBN 9788132208075. [Google Scholar]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant. Sci. 2016, 7, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Saud, S.; Li, X.; Chen, Y.; Zhang, L.; Fahad, S.; Hussain, S.; Sadiq, A.; Chen, Y. Silicon application increases drought tolerance of Kentucky bluegrass by improving plant water relations and morphophysiological functions. Sci. World J. 2014, 2014, 368694. [Google Scholar] [CrossRef] [PubMed]
- Tátrai, Z.A.; Sanoubar, R.; Pluhár, Z.; Mancarella, S.; Orsini, F.; Gianquinto, G. Morphological and physiological plant responses to drought stress in Thymus citriodorus. Int. J. Agron. 2016, 2016, 4165750. [Google Scholar] [CrossRef]
- Hussain, H.A.; Men, S.; Hussain, S.; Chen, Y.; Ali, S.; Zhang, S.; Zhang, K.; Li, Y.; Xu, Q.; Liao, C.; et al. Interactive effects of drought and heat stresses on morpho-physiological attributes, yield, nutrient uptake and oxidative status in maize hybrids. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Nahar, K.; Hasanuzzaman, M.; Fujita, M. Roles of osmolytes in plant adaptation to drought and salinity. In Osmolytes and Plants Acclimation to Changing Environment: Emerging Omics Technologies; Iqbal, N., Nazar, R., Eds.; Springer: Chennai, India, 2015; pp. 1–170. ISBN 9788132226161. [Google Scholar]
- Hsiao, T.C.; Xu, L.K. Sensitivity of with of roots versus leaves to water stress: Biophysical analysis and relation to water. J. Exp. Bot. 2000, 51, 1595–1616. [Google Scholar] [CrossRef]
- Xiong, L.; Wang, R.G.; Mao, G.; Koczan, J.M. Identification of drought tolerance determinants by genetic analysis of root response to drought stress and abscisic acid. Plant. Physiol. 2006, 142, 1065–1074. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 1–13. [Google Scholar] [CrossRef]
- García-Martí, M.; Piñero, M.C.; García-Sanchez, F.; Mestre, T.C.; López-Delacalle, M.; Martínez, V.; Rivero, R.M. Amelioration of the oxidative stress generated by simple or combined abiotic stress through the K+ and Ca2+ supplementation in tomato plants. Antioxidants 2019, 8, 81. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant. J. 2017, 90, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N.; Gill, S.S.; Tuteja, R. Plant responses to abiotic stresses: Shedding light on salt, drought, cold and heavy metal stress. In Omics and plant abiotic stress tolerance; Tuteja, N., Gill, S.S., Tuteja, R., Eds.; Bentham Science Publishers: Sharjah, UAE, 2011; pp. 39–64. ISBN 9781608053841. [Google Scholar]
- He, M.; He, C.Q.; Ding, N.Z. Abiotic stresses: General defenses of land plants and chances for engineering multistress tolerance. Front. Plant. Sci. 2018, 871, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Szymańska, R.; Ślesak, I.; Orzechowska, A.; Kruk, J. Physiological and biochemical responses to high light and temperature stress in plants. Environ. Exp. Bot. 2017, 139, 165–177. [Google Scholar] [CrossRef]
- Arbona, V.; Flors, V.; Jacas, J.; García-Agustín, P.; Gómez-Cadenas, A. Enzymatic and non-enzymatic antioxidant responses of Carrizo citrange, a salt-sensitive citrus rootstock, to different levels of salinity. Plant. Cell Physiol. 2003, 44, 388–394. [Google Scholar] [CrossRef]
- De Pinto, M.C.; De Gara, L. Changes in the ascorbate metabolism of apoplastic and symplastic spaces are associated with cell differentiation. J. Exp. Bot. 2004, 55, 2559–2569. [Google Scholar] [CrossRef]
- Al Kharusi, L.; Al Yahyai, R.; Yaish, M.W. Antioxidant response to salinity in salt-tolerant and salt-susceptible cultivars of date palm. Agriculture 2019, 9, 8. [Google Scholar] [CrossRef]
- Yancey, P.H. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 2005, 208, 2819–2830. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Ascorbate and glutathione: The heart of the redox hub. Plant. Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef]
- da Silva, E.C.; de Albuquerque, M.B.; de Azevedo Neto, A.D.; da Silva Junior, C.D. Drought and its consequences to plants—From individual to ecosystem. In Responses of Organisms to Water Stress; Akıncı, S., Ed.; IntechOpen: London, UK, 2013; pp. 18–47. [Google Scholar] [CrossRef]
- Saijo, Y.; Loo, E.P. Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 2019, 225, 87–104. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and biotic stress combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Kuzyakov, Y.; Razavi, B.S. Rhizosphere size and shape: Temporal dynamics and spatial stationarity. Soil Biol. Biochem. 2019, 135, 343–360. [Google Scholar] [CrossRef]
- Sasse, J.; Martinoia, E.; Northen, T. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant. Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef]
- Conrath, U. Molecular aspects of defence priming. Trends Plant. Sci. 2011, 16, 524–531. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The chemistry of plant–microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant. Sci. 2018, 9, 1–17. [Google Scholar] [CrossRef]
- Kanjariya, K.G.; Parihar, A. Prime-ome: A molecular approach towards defence mechanisms. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 3606–3610. [Google Scholar] [CrossRef]
- Mauch-Mani, B.; Baccelli, I.; Luna, E.; Flors, V. Defense priming: An adaptive part of induced resistance. Annu. Rev. Plant. Biol. 2017, 68, 485–512. [Google Scholar] [CrossRef]
- Conrath, U.; Beckers, G.J.M.; Langenbach, C.J.G.; Jaskiewicz, M.R. Priming for enhanced defense. Annu. Rev. Phytopathol. 2015. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Chen, J.; Wang, X.; Cai, J.; Zhou, Q.; Dai, T. Parental drought-priming enhances tolerance to post-anthesis drought in offspring of wheat. Front. Plant Sci. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Azeem, M.; Iqbal, N.; Kausar, S.; Javed, M.T.; Akram, M.S.; Sajid, M.A. Efficacy of silicon priming and fertigation to modulate seedling’s vigor and ion homeostasis of wheat (Triticum aestivum L.) under saline environment. Environ. Sci. Pollut. Res. 2015, 22, 14367–14371. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X. Lipidomic reprogramming associated with drought stress priming—Enhanced heat tolerance in tall fescue (Festuca arundinacea). Plant. Cell Environ. 2019, 42, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Shehu, A.A.; Alsamadany, H.; Alzahrani, Y. β -Aminobutyric acid ( BABA ) priming and abiotic stresses: A review. Int. J. Biosci. 2019, 14, 450–459. [Google Scholar]
- Tugizimana, F.; Mhlongo, M.I.; Piater, L.A.; Dubery, I.A. Metabolomics in plant priming research: The way forward? Int. J. Mol. Sci. 2018, 19, 1759. [Google Scholar] [CrossRef] [PubMed]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant growth-promoting rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant. Sci. 2018, 9, 1–17. [Google Scholar] [CrossRef]
- du Jardin, P. Plant biostimulants: Definition, concept, main categories and regulation. Sci. Hortic. 2015, 196, 3–14. [Google Scholar] [CrossRef]
- Ricci, M.; Tilbury, L.; Daridon, B.; Sukalac, K. General principles to justify plant biostimulant claims. Front. Plant. Sci. 2019, 10, 1–8. [Google Scholar] [CrossRef]
- De Pascale, S.; Rouphael, Y.; Colla, G. Plant biostimulants: Innovative tool for enhancing plant nutrition in organic farming. Eur. J. Hortic. Sci. 2017, 82, 277–285. [Google Scholar] [CrossRef]
- Bulgari, R.; Cocetta, G.; Trivellini, A.; Vernieri, P.; Ferrante, A. Biostimulants and crop responses: A review. Biol. Agric. Hortic. 2015, 31, 1–17. [Google Scholar] [CrossRef]
- Le Mire, G.; Nguyen, M.L.; Fassotte, B.; Jijakli, M.H.; Du Jardin, P.; Delaplace, P.; Verheggen, F. Implementing plant biostimulants and biocontrol strategies in the agroecological management of cultivated ecosystems. A review. Biotechnol. Agron. Soc. Environ. 2016, 20, 299–313. [Google Scholar]
- Van Oosten, M.J.; Pepe, O.; De Pascale, S.; Silletti, S.; Maggio, A. The role of biostimulants and bioeffectors as alleviators of abiotic stress in crop plants. Chem. Biol. Technol. Agric. 2017, 4, 1–12. [Google Scholar] [CrossRef]
- Niu, X.; Song, L.; Xiao, Y.; Ge, W. Drought-tolerant plant growth-promoting rhizobacteria associated with foxtail millet in a semi-arid and their potential in alleviating drought stress. Front. Microbiol. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.G.; Mun, B.G.; Kang, S.M.; Hussain, A.; Shahzad, R.; Seo, C.W.; Kim, A.Y.; Lee, S.U.; Oh, K.Y.; Lee, D.Y.; et al. Bacillus aryabhattai SRB02 tolerates oxidative and nitrosative stress and promotes the growth of soybean by modulating the production of phytohormones. PLoS ONE 2017, 12, e0173203. [Google Scholar] [CrossRef] [PubMed]
- Bashan, Y.; de-Bashan, L.E.; Prabhu, S.R.; Hernandez, J.P. Advances in plant growth-promoting bacterial inoculant technology: Formulations and practical perspectives (1998–2013). Plant. Soil 2014, 378, 1–33. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Wirth, S.J.; Alqarawi, A.A.; Abd-Allah, E.F.; Hashem, A. Phytohormones and beneficial microbes: Essential components for plants to balance stress and fitness. Front. Microbiol. 2017, 8, 1–14. [Google Scholar] [CrossRef]
- Sharma, H.S.S.; Fleming, C.; Selby, C.; Rao, J.R.; Martin, T. Plant biostimulants: A review on the processing of macroalgae and use of extracts for crop management to reduce abiotic and biotic stresses. J. Appl. Phycol. 2014, 26, 465–490. [Google Scholar] [CrossRef]
- Tarakhovskaya, E.R.; Maslov, Y.I.; Shishova, M.F. Phytohormones in algae. Russ. J. Plant. Physiol. 2007, 54, 163–170. [Google Scholar] [CrossRef]
- Bradáčová, K.; Weber, N.F.; Morad-Talab, N.; Asim, M.; Imran, M.; Weinmann, M.; Neumann, G. Micronutrients (Zn/Mn), seaweed extracts, and plant growth-promoting bacteria as cold-stress protectants in maize. Chem. Biol. Technol. Agric. 2016, 3, 1–10. [Google Scholar] [CrossRef]
- Desoky, E.-S.M.; Merwad, A.-R.M.; Elrys, A.S. Response of pea plants to natural bio-stimulants under soil salinity stress. Am. J. Plant. Physiol. 2016, 12, 28–37. [Google Scholar] [CrossRef][Green Version]
- Simpson, A.J.; Kingery, W.L.; Hayes, M.H.; Spraul, M.; Humpfer, E.; Dvortsak, P.; Kerssebaum, R.; Godejohann, M.; Hofmann, M. Molecular structures and associations of humic substances in the terrestrial environment. Naturwissenschaften 2002, 89, 84–88. [Google Scholar] [CrossRef]
- Chen, Y.; Magen, H.; Clapp, C.E. Mechanisms of plant growth stimulation by humic substances: The role of organo-iron complexes. Soil Sci. Plant. Nutr. 2004, 50, 1089–1095. [Google Scholar] [CrossRef]
- Nardi, S.; Pizzeghello, D.; Muscolo, A.; Vianello, A. Physiological effects of humic substances on higher plants. Soil Biol. Biochem. 2002, 34, 1527–1536. [Google Scholar] [CrossRef]
- Çimrin, K.M.; Türkmen, Ö.; Turan, M.; Tuncer, B. Phosphorus and humic acid application alleviate salinity stress of pepper seedling. Afr. J. Biotechnol. 2010, 9, 5845–5851. [Google Scholar] [CrossRef]
- García, A.C.; Santos, L.A.; Izquierdo, F.G.; Sperandio, M.V.L.; Castro, R.N.; Berbara, R.L.L. Vermicompost humic acids as an ecological pathway to protect rice plant against oxidative stress. Ecol. Eng. 2012, 47, 203–208. [Google Scholar] [CrossRef]
- Asli, S.; Neumann, P.M. Rhizosphere humic acid interacts with root cell walls to reduce hydraulic conductivity and plant development. Plant. Soil 2010, 336, 313–322. [Google Scholar] [CrossRef]
- Colla, G.; Rouphael, Y. Biostimulants in horticulture. Sci. Hortic. 2015, 196, 1–2. [Google Scholar] [CrossRef]
- Zhu, K.; Zhou, H.; Qian, H. Antioxidant and free radical-scavenging activities of wheat germ protein hydrolysates (WGPH) prepared with alcalase. Process. Biochem. 2006, 41, 1296–1302. [Google Scholar] [CrossRef]
- Colla, G.; Nardi, S.; Cardarelli, M.; Ertani, A.; Lucini, L.; Canaguier, R.; Rouphael, Y. Protein hydrolysates as biostimulants in horticulture. Sci. Hortic. 2015, 196, 28–38. [Google Scholar] [CrossRef]
- Ertani, A.; Schiavon, M.; Muscolo, A.; Nardi, S. Alfalfa plant-derived biostimulant stimulate short-term growth of salt stressed Zea mays L. plants. Plant. Soil 2013, 364, 145–158. [Google Scholar] [CrossRef]
- Lucini, L.; Rouphael, Y.; Cardarelli, M.; Canaguier, R.; Kumar, P.; Colla, G. The effect of a plant-derived biostimulant on metabolic profiling and crop performance of lettuce grown under saline conditions. Sci. Hortic. 2015, 182, 124–133. [Google Scholar] [CrossRef]
- Ashraf, M.; Foolad, M.R. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Savvas, D.; Ntatsi, G. Biostimulant activity of silicon in horticulture. Sci. Hortic. 2015, 196, 66–81. [Google Scholar] [CrossRef]
- Petrozza, A.; Santaniello, A.; Summerer, S.; Di Tommaso, G.; Di Tommaso, D.; Paparelli, E.; Piaggesi, A.; Perata, P.; Cellini, F. Physiological responses to Megafol® treatments in tomato plants under drought stress: A phenomic and molecular approach. Sci. Hortic. 2014, 174, 185–192. [Google Scholar] [CrossRef]
- Xie, W.; Xu, P.; Liu, Q. Antioxidant activity of water-soluble chitosan derivatives. Bioorg. Med. Chem. Lett. 2001, 11, 1699–1701. [Google Scholar] [CrossRef]
- Boonlertnirun, S.; Sarobol, E.; Meechoui, S.; Sooksathan, I. Drought recovery and grain yield potential of rice after chitosan application. Kasetsart J. Nat. Sci. 2007, 41, 1–6. [Google Scholar]
- Mahdavi, B.; Sanavy, S.A.M.M.; Aghaalikhani, M.; Sharifi, M.; Dolatabadian, A. Chitosan improves osmotic potential tolerance in safflower (Carthamus tinctorius L.) seedlings. J. Crop. Improv. 2011, 25, 728–741. [Google Scholar] [CrossRef]
- Herrera-estrella, A.; Chet, I. Chitinases in biological control. In Chitin and Chitinases; Lolles, P., Muuarelli, R.A.A., Eds.; Birkhauser Verlag: Basel, Switzerland, 1999; pp. 172–184. ISBN 9783034887571. [Google Scholar]
- Jindo, K.; Martim, S.A.; Navarro, E.C.; Pérez-Alfocea, F.; Hernandez, T.; Garcia, C.; Aguiar, N.O.; Canellas, L.P. Root growth promotion by humic acids from composted and non-composted urban organic wastes. Plant. Soil 2012, 353, 209–220. [Google Scholar] [CrossRef]
- Jindo, K.; Canellas, L.P.; Albacete, A.; Dos Santos, L.F.; Frinhani Rocha, R.L.; Baia, D.C.; Aguiar Canellas, N.O.; Goron, T.L.; Olivares, F.L. Interaction between humic substances and plant hormones for phosphorous acquisition. Agronomy 2020, 10, 640. [Google Scholar] [CrossRef]
- Aydin, A.; Turan, M. Humic acid application alleviate salinity stress of bean (Phaseolus vulgaris L.) plants decreasing membrane leakage. Afr. J. Agric. Res. 2012, 7, 1073–1086. [Google Scholar] [CrossRef]
- Billard, V.; Ourry, A.; Maillard, A.; Garnica, M.; Coquet, L.; Jouenne, T.; Cruz, F.; Garcia-Mina, J.M.; Yvin, J.C.; Etienne, P. Copper-deficiency in Brassica napus induces copper remobilization, molybdenum accumulation and modification of the expression of chloroplastic proteins. PLoS ONE 2014, 9, e0109889. [Google Scholar] [CrossRef]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant. Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [PubMed]
- Pichyangkura, R.; Chadchawan, S. Biostimulant activity of chitosan in horticulture. Sci. Hortic. 2015, 196, 49–65. [Google Scholar] [CrossRef]
- El-Tanahy, A.M.M.; Mahmoud, A.R.; Abde-Mouty, M.M.; Ali, A.H. Effect of Chitosan doses and nitrogen sources on the growth, yield and seed quality of cowpea. Aust. J. Basic Appl. Sci. 2012, 6, 115–121. [Google Scholar]
- Ma, Z.; Yang, L.; Yan, H.; Kennedy, J.F.; Meng, X. Chitosan and oligochitosan enhance the resistance of peach fruit to brown rot. Carbohydr. Polym. 2013, 94, 272–277. [Google Scholar] [CrossRef]
- Dobbelaere, S.; Croonenborghs, A.; Thys, A.; Vande Broek, A.; Vanderleyden, J. Phytostimulatory effect of Azospirillum brasilense wild type and mutant strains altered in IAA production on wheat. Plant. Soil 1999, 212, 155–164. [Google Scholar] [CrossRef]
- Briglia, N.; Petrozza, A.; Hoeberichts, F.A.; Verhoef, N.; Povero, G. Investigating the impact of biostimulants on the row crops corn and soybean using high-efficiency phenotyping and next generation sequencing. Agronomy 2019, 9, 761. [Google Scholar] [CrossRef]
- Ghatak, A.; Chaturvedi, P.; Weckwerth, W. Metabolomics in plant stress physiology. In Advances in Biochemical Engineering/Biotechnology; Scheper, T., Ed.; Springer International Publishing: Cham, Switzerland, 2018; Volume 164, pp. 187–236. [Google Scholar] [CrossRef]
- Trivedi, D.K.; Hollywood, K.A.; Goodacre, R. Metabolomics for the masses: The future of metabolomics in a personalized world. New Horiz. Transl. Med. 2017, 3, 294–305. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.M.; Ribeiro-Barros, A.I.; António, C. Experimental design and sample preparation in forest tree metabolomics. Metabolites 2019, 9, 285. [Google Scholar] [CrossRef]
- Ivanisevic, J.; Want, E.J. From samples to insights into metabolism: Uncovering biologically relevant information in LC-HRMS metabolomics data. Metabolites 2019, 9, 308. [Google Scholar] [CrossRef] [PubMed]
- Bingol, K. Recent advances in targeted and untargeted metabolomics by NMR and MS/NMR methods. High. Throughput 2018, 7, 9. [Google Scholar] [CrossRef]
- Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Multi-platform metabolomic analyses of ergosterol-induced dynamic changes in Nicotiana tabacum cells. PLoS ONE 2014, 9, e087846. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Marsal, S.; Julià, A. Analytical methods in untargeted metabolomics: State of the art in 2015. Front. Bioeng. Biotechnol. 2015, 3, 1–20. [Google Scholar] [CrossRef]
- Beale, D.J.; Pinu, F.R.; Kouremenos, K.A.; Poojary, M.M.; Narayana, V.K.; Boughton, B.A.; Kanojia, K.; Dayalan, S.; Jones, O.A.H.; Dias, D.A. Review of recent developments in GC–MS approaches to metabolomics-based research. Metabolomics 2018, 14, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Hamany Djande, C.Y.; Pretorius, C.; Tugizimana, F.; Piater, L.A.; Dubery, I.A. Metabolomics: A tool for cultivar phenotyping and investigation of grain crops. Agronomy 2020, 10, 831. [Google Scholar] [CrossRef]
- Tugizimana, F.; Engel, J.; Salek, R.; Dubery, I.; Piater, L.; Burgess, K. The Disruptive 4IR in the Life Sciences: Metabolomics. In The Disruptive Fourth Industrial Revolution; Doorsamy, W., Sena Paul, B., Marwala, T., Eds.; 2020; Volume 674, pp. 227–255. ISBN 978-3-030-48229-9. [Google Scholar]
- Broadhurst, D.; Goodacre, R.; Reinke, S.N.; Kuligowski, J.; Wilson, I.D.; Lewis, M.R.; Dunn, W.B. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics 2018, 14, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D.; Dunn, W.B.; Bandukwala, A.; Bethan, B.; Broadhurst, D.; Clish, C.B.; Dasari, S.; Derr, L.; Evans, A.; Fischer, S.; et al. Towards quality assurance and quality control in untargeted metabolomics studies. Metabolomics 2019, 15, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O.; Robertson, D.; Griffin, J.; vab der Werf, M.; Nikolau, B.; Morrison, N.; Sumner, L.W.; Goodacre, R.; Hardy, N.W.; Taylor, C.; et al. The metabolomics standards initiative (MSI). Metabolomics 2007, 3, 175–178. [Google Scholar] [CrossRef]
- Gomez-Casati, D.F.; Zanor, M.I.; Busi, M.V. Metabolomics in plants and humans: Applications in the prevention and diagnosis of diseases. Biomed. Res. Int. 2013, 2013, 1–11. [Google Scholar] [CrossRef]
- Miggiels, P.; Wouters, B.; van Westen, G.J.P.; Dubbelman, A.C.; Hankemeier, T. Novel technologies for metabolomics: More for less. TrAC Trends Anal. Chem. 2019, 120, 1–9. [Google Scholar] [CrossRef]
- Pezzatti, J.; Bergé, M.; Boccard, J.; Codesido, S.; Gagnebin, Y.; Viollier, P.H.; González-Ruiz, V.; Rudaz, S. Choosing an optimal sample preparation in Caulobacter crescentus for untargeted metabolomics approaches. Metabolites 2019, 9, 193. [Google Scholar] [CrossRef]
- Razzaq, A.; Sadia, B.; Raza, A.; Khalid Hameed, M.; Saleem, F. Metabolomics: A way forward for crop improvement. Metabolites 2019, 9, 303. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Verpoorte, R. Sample preparation for plant metabolomics. Phytochem. Anal. 2010, 21, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Salem, M.A.; De Souza, L.P.; Serag, A.; Fernie, A.R.; Farag, M.A.; Ezzat, S.M.; Alseekh, S. Metabolomics in the context of plant natural products research: From sample preparation to metabolite analysis. Metabolites 2020, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Vuckovic, D. Improving metabolome coverage and data quality: Advancing metabolomics and lipidomics for biomarker discovery. Chem. Commun. 2018, 54, 6728–6749. [Google Scholar] [CrossRef] [PubMed]
- Maria John, K.M.; Harnly, J.; Luthria, D. Influence of direct and sequential extraction methodology on metabolic profiling. J. Chromatogr. B 2018, 1073, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.R.; Goldansaz, S.A.; Jaine, J. Translational metabolomics: Current challenges and future opportunities. Metabolites 2019, 9, 108. [Google Scholar] [CrossRef]
- Haggarty, J.; Burgess, K.E. Recent advances in liquid and gas chromatography methodology for extending coverage of the metabolome. Curr. Opin. Biotechnol. 2017, 43, 77–85. [Google Scholar] [CrossRef]
- Wishart, D.S. Metabolomics for investigating physiological and pathophysiological processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef]
- Sousa Silva, M.; Cordeiro, C.; Roessner, U.; Figueiredo, A. Editorial: Metabolomics in crop research—Current and emerging methodologies. Front. Plant. Sci. 2019, 10, 1–3. [Google Scholar] [CrossRef]
- Arroussi, H.E.L.; Benhima, R.; Elbaouchi, A.; Sijilmassi, B.; Mernissi, N.E.L.; Aafsar, A. Dunaliella salina exopolysaccharides: A promising biostimulant for salt stress tolerance in tomato (Solanum lycopersicum). J. Appl. Phycol. 2018, 30, 2929–2941. [Google Scholar] [CrossRef]
- Ertani, A.; Pizzeghello, D.; Francioso, O.; Sambo, P.; Sanchez-cortes, S.; Nardi, S. Capsicum chinensis L. growth and nutraceutical properties are enhanced by biostimulants in a long-term period: Chemical and metabolomic approaches. Front. Plant. Sci. 2014, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Conselvan, G.B.; Fuentes, D.; Merchant, A.; Peggion, C.; Francioso, O.; Carletti, P. Effects of humic substances and indole-3-acetic acid on Arabidopsis sugar and amino acid metabolic profile. Plant. Soil 2018, 426, 17–32. [Google Scholar] [CrossRef]
- Lucini, L.; Rouphael, Y.; Cardarelli, M.; Bonini, P.; Baffi, C.; Colla, G.; Lucia, B. De A vegetal biopolymer-based biostimulant promoted root growth in melon while triggering brassinosteroids and stress-related compounds. Front. Plant. Sci. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Paul, K.; Sorrentino, M.; Lucini, L.; Rouphael, Y.; Cardarelli, M.; Bonini, P.; Reynaud, H.; Canaguier, R.; Trtílek, M.; Panzarová, K.; et al. Understanding the biostimulant action of vegetal-derived protein hydrolysates by high-throughput plant phenotyping and metabolomics: A case study on tomato. Front. Plant. Sci. 2019, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Barrajón-Catalán, E.; Álvarez-Martínez, F.J.; Borrás, F.; Pérez, D.; Herrero, N.; Ruiz, J.J.; Micol, V. Metabolomic analysis of the effects of a commercial complex biostimulant on pepper crops. Food Chem. 2020, 310, 125818. [Google Scholar] [CrossRef]
- Farid, R.; Mutale-joan, C.; Redouane, B.; Mernissi Najib, E.; Abderahime, A.; Laila, S.; Arroussi Hicham, E. Effect of microalgae polysaccharides on biochemical and metabolomics pathways related to plant defense in Solanum lycopersicum. Appl. Biochem. Biotechnol. 2018, 188, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Abou Chehade, L.; Al Chami, Z.; De Pascali, S.A.; Cavoski, I.; Fanizzi, F.P. Biostimulants from food processing by-products: Agronomic, quality and metabolic impacts on organic tomato (Solanum lycopersicum L.). J. Sci. Food Agric. 2017, 98, 1426–1436. [Google Scholar] [CrossRef]
- Lucini, L.; Colla, G.; Miras Moreno, M.B.; Bernardo, L.; Cardarelli, M.; Terzi, V.; Bonini, P.; Rouphael, Y. Inoculation of Rhizoglomus irregulare or Trichoderma atroviride differentially modulates metabolite profiling of wheat root exudates. Phytochemistry 2019, 157, 158–167. [Google Scholar] [CrossRef]
- Kim, H.J.; Ku, K.M.; Choi, S.; Cardarelli, M. Vegetal-derived biostimulant enhances adventitious rooting in cuttings of basil, tomato, and chrysanthemum via brassinosteroid-mediated processes. Agronomy 2019, 9, 74. [Google Scholar] [CrossRef]
- Omidbakhshfard, M.A.; Sujeeth, N.; Gupta, S.; Omranian, N.; Guinan, K.J.; Brotman, Y.; Nikoloski, Z.; Fernie, A.R.; Mueller-Roeber, B.; Gechev, T.S. A biostimulant obtained from the seaweed Ascophyllum nodosum protects Arabidopsis thaliana from severe oxidative stress. Int. J. Mol. Sci. 2020, 21, 474. [Google Scholar] [CrossRef]



| Main Biostimulant Classes | Cellular Mechanism (i.e., Interaction with Cellular Components and Processes) | Physiological Function (i.e., Action on Whole-Plant Processes) | Agricultural/Horticultural Function (i.e., Output Traits Relevant for Crop Performance) |
|---|---|---|---|
| Humic substances | Activate plasma membrane proton-pumping ATPases, promote cell wall loosening and cell elongation in maize roots [102]. Increases antioxidation capacity under several abiotic stresses, upregulates the biosynthesis of defense-related secondary metabolites [103]. | Increased linear growth of roots, root biomass [103]. | Increased root foraging capacity, enhanced nutrient use efficiency [103,104]. |
| Seaweed extracts | Stimulates expression of genes encoding transporters of micronutrients (e.g., Cu, Fe, Zn) in Brassica napus [105]. | Increased tissue concentrations and root to shoot transport of micronutrients | Improved mineral composition of plant tissues |
| Protein hydrolysates | Stimulation of phenylalanine ammonia-lyase (PAL) enzyme and gene expression, and production of flavonoids under salt stress [93]. | Protection by flavonoids against UV and oxidative damage [106]. | Increased crop tolerance to abiotic (e.g., salt) stress |
| Chitosan | Induce the accumulation of reactive oxygen species (ROS) and phenolic compounds. enhances the ROS scavenging system, regulates the stomatal conductance [107]. | Increased all vegetative growth and yield [108]. | Protection against brown rot, delayed fruit softening and senescence [109]. |
| Microorganisms (e.g., PGPR) | Azospirillum brasilense releases auxins and activates auxin signaling pathways involved in root morphogenesis wheat [110]. | Increased lateral root density and surface of root hairs | Increased root foraging capacity, enhanced nutrient use efficiency |
| Fungi | Enhance phosphate (P) availability under nutrient deficiency via excretion of P-solubilizing substances [72] | Increased root growth and activity | Increased nutrient availability in the soil |
| Study | Biostimulant | Effects | Plant | Mechanism of Action—Metabolic Changes | References |
|---|---|---|---|---|---|
| Capsicum chinensis L. growth and nutraceutical properties are enhanced by biostimulants in a long-term period: Chemical and metabolomic approaches | Plant-based biostimulants: one derived from AH and RG | Growth promotion | Pepper |
| [138] |
| The effect of a plant-derived biostimulant on metabolic profiling and crop performance of lettuce grown under saline conditions | Plant-derived protein hydrolysates | Growth promotion and resistance to salt stress | Lettuce |
| [94] |
| Dunaliella salina exopolysaccharides: a promising biostimulant for salt stress tolerance in tomato (Solanum lycopersicum) | Microalgal exopolysaccharides | Resistance to salt stress | Tomato |
| [137] |
| Effects of humic substances and indole-3-acetic acidon Arabidopsis sugar and amino acid metabolic profile | Humic substances | Growth promotion | Arabidopsis |
| [139] |
| A vegetal biopolymer-basedbiostimulant promoted root growth in melon while triggeringbrassinosteroids and stress-related compounds | Vegetal biopolymer-basedbiostimulant | Growth promotion | Melon |
| [140] |
| Understanding the biostimulant action of vegetal-derived protein hydrolysates by high-throughput plant phenotyping and metabolomics: A Case Study on Tomato | Vegetal-derived protein hydrolysates | Growth promotion | Tomato |
| [141] |
| A combined phenotypic and metabolomic approach for elucidating the biostimulant action of a plant-derived protein hydrolysate on tomato grown under limited water availability | Plant-derived protein hydrolysate | Resistance against drought | Tomato |
| [11] |
| Metabolomic analysis of the effects of a commercial complex biostimulant on pepper crops | Commercial biostimulant, Actium | Growth promotion | Pepper |
| [142] |
| Effect of microalgae polysaccharides on biochemical and metabolomics pathways related to plant defense in Solanum lycopersicum | Microalgae polysaccharides | Plant defense | Tomato |
| [143] |
| Biostimulants from food processing by-products: agronomic, quality and metabolic impacts on organic tomato (Solanum lycopersicum L.) | Biostimulants from food processing by-products | Growth promotion | Tomato |
| [144] |
| Inoculation of Rhizoglomus irregulare or Trichoderma atroviride differentially modulates metabolite profiling of wheat root exudates | T. atroviride and R. irregulare | Growth promotion | Wheat |
| [145] |
| Vegetal-derived biostimulant enhances adventitious rooting in cuttings of basil, tomato, and Chrysanthemum via brassinosteroid-mediated processes | Vegetal-derived biostimulant | Growth promotion | Basil, Tomato, and Chrysanthemum |
| [146] |
| A biostimulant obtained from the seaweed Ascophyllum nodosum protects Arabidopsis thaliana from severe oxidative stress | Seaweed extracts | Oxidative stress | Arabidopsis |
| [147] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nephali, L.; Piater, L.A.; Dubery, I.A.; Patterson, V.; Huyser, J.; Burgess, K.; Tugizimana, F. Biostimulants for Plant Growth and Mitigation of Abiotic Stresses: A Metabolomics Perspective. Metabolites 2020, 10, 505. https://doi.org/10.3390/metabo10120505
Nephali L, Piater LA, Dubery IA, Patterson V, Huyser J, Burgess K, Tugizimana F. Biostimulants for Plant Growth and Mitigation of Abiotic Stresses: A Metabolomics Perspective. Metabolites. 2020; 10(12):505. https://doi.org/10.3390/metabo10120505
Chicago/Turabian StyleNephali, Lerato, Lizelle A. Piater, Ian A. Dubery, Veronica Patterson, Johan Huyser, Karl Burgess, and Fidele Tugizimana. 2020. "Biostimulants for Plant Growth and Mitigation of Abiotic Stresses: A Metabolomics Perspective" Metabolites 10, no. 12: 505. https://doi.org/10.3390/metabo10120505
APA StyleNephali, L., Piater, L. A., Dubery, I. A., Patterson, V., Huyser, J., Burgess, K., & Tugizimana, F. (2020). Biostimulants for Plant Growth and Mitigation of Abiotic Stresses: A Metabolomics Perspective. Metabolites, 10(12), 505. https://doi.org/10.3390/metabo10120505







