Metabolite and Phytohormone Profiling Illustrates Metabolic Reprogramming as an Escape Strategy of Deepwater Rice during Partially Submerged Stress
Abstract
:1. Introduction
2. Results
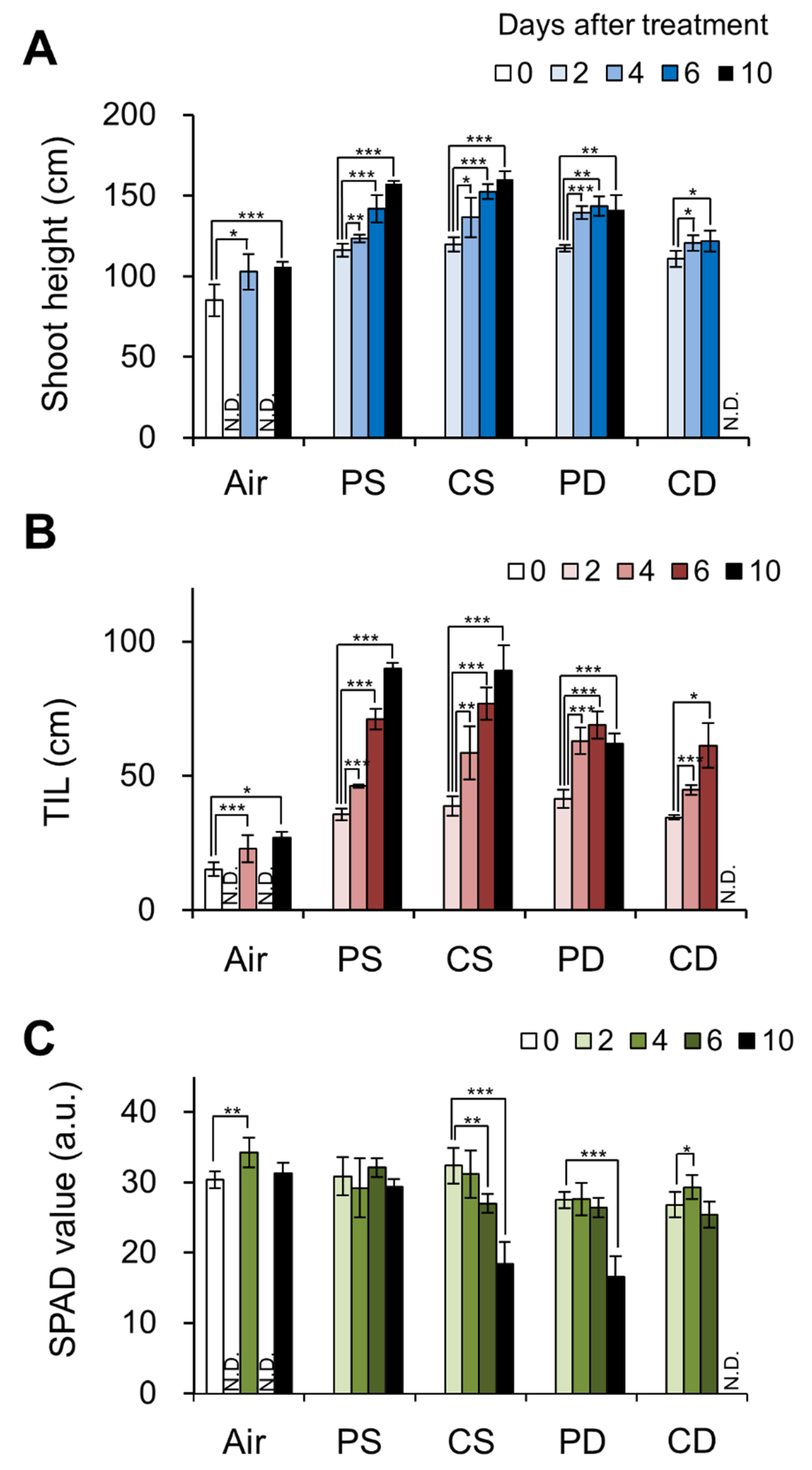
2.1. Effects of Submergence in the Presence or Absence of Light in Deepwater Cultivar C9285
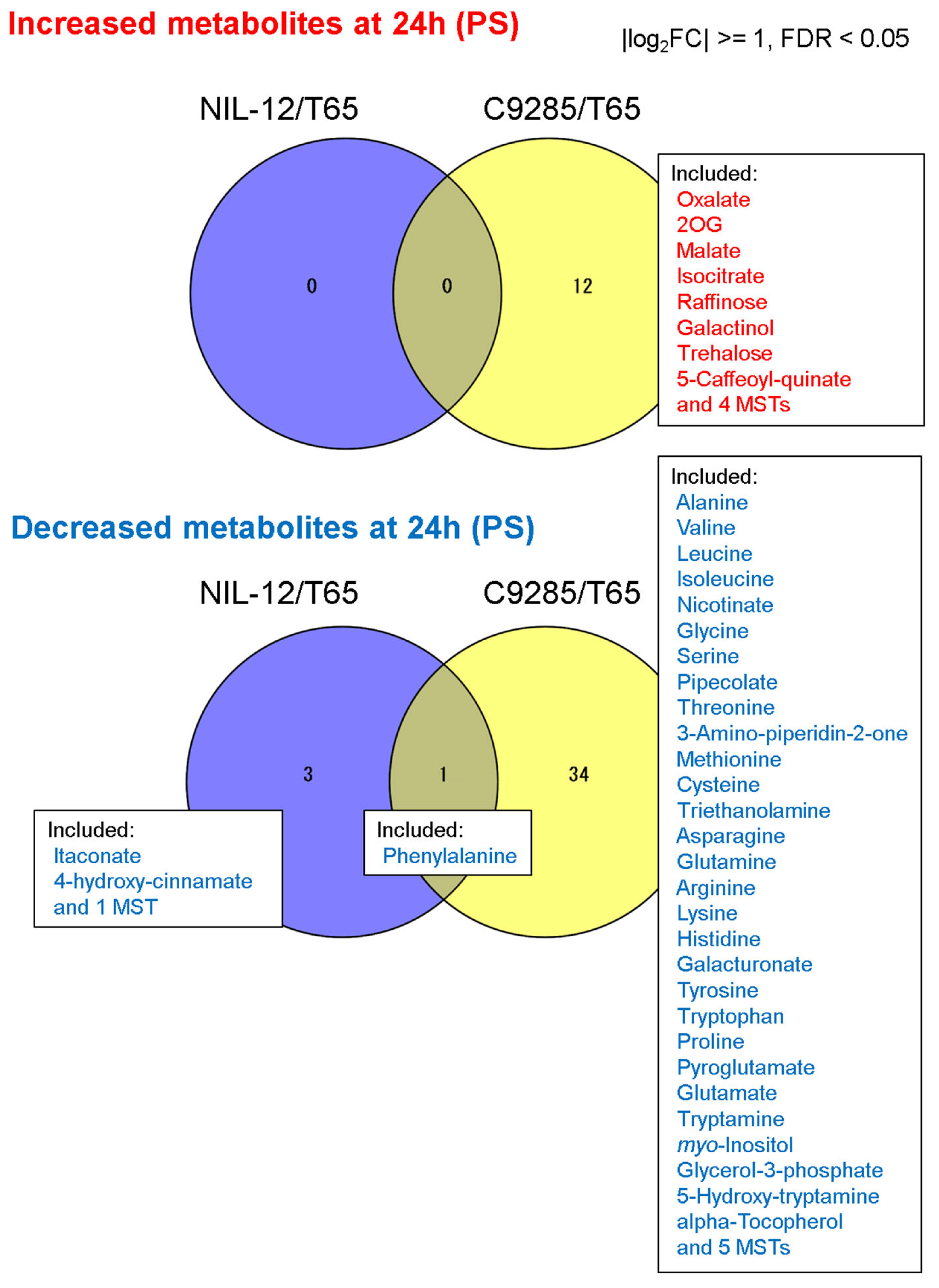
2.2. Short-Term Responses of Metabolite Levels in Deepwater Rice under the PS Condition
2.3. Metabolite and Phytohormone Profiling of NIL-12 in the Long-Term PS Treatment
3. Discussion
4. Materials and Methods
4.1. Plant Material and Cultivation
4.2. Submergence Treatments
4.3. Semi-Quantitative RT-PCR Analysis
4.4. Starch Measurement
4.5. GC-MS Analysis
4.6. CE-MS Analysis
4.7. Measurement of Hormone Levels
4.8. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bailey-Serres, J.; Voesenek, L.A. Flooding stress: Acclimations and genetic diversity. Annu. Rev. Plant Biol. 2008, 59, 313–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hattori, Y.; Nagai, K.; Ashikari, M. Rice growth adapting to deepwater. Curr. Opin. Plant Biol. 2011, 14, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Akman, M.; Bhikharie, A.V.; McLean, E.H.; Boonman, A.; Visser, E.J.; Schranz, M.E.; van Tienderen, P.H. Wait or escape? Contrasting submergence tolerance strategies of Rorippa amphibia, Rorippa sylvestris and their hybrid. Ann. Bot. 2012, 109, 1263–1276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voesenek, L.A.; Bailey-Serres, J. Flood adaptive traits and processes: An overview. New Phytol. 2015, 206, 57–73. [Google Scholar] [CrossRef]
- Loreti, E.; van Veen, H.; Perata, P. Plant responses to flooding stress. Curr. Opin. Plant Biol. 2016, 33, 64–71. [Google Scholar] [CrossRef] [Green Version]
- Nagai, K.; Hattori, Y.; Ashikari, M. Stunt or elongate? Two opposite strategies by which rice adapts to floods. J. Plant Res. 2010, 123, 303–309. [Google Scholar] [CrossRef]
- Vriezen, W.H.; Zhou, Z.; Van Der Straeten, D. Regulation of submergence-induced enhanced shoot elongation in Oryza sativa L. Ann. Bot. 2003, 91, 263–270. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Septiningsih, E.M.; Balyan, H.S.; Singh, N.K.; Rai, V. Genetics, Physiological Mechanisms and Breeding of Flood-Tolerant Rice (Oryza sativa L.). Plant Cell Physiol. 2017, 58, 185–197. [Google Scholar] [CrossRef] [Green Version]
- Vreeburg, R.A.; Benschop, J.J.; Peeters, A.J.; Colmer, T.D.; Ammerlaan, A.H.; Staal, M.; Elzenga, T.M.; Staals, R.H.; Darley, C.P.; McQueen-Mason, S.J.; et al. Ethylene regulates fast apoplastic acidification and expansin A transcription during submergence-induced petiole elongation in Rumex palustris. Plant J. 2005, 43, 597–610. [Google Scholar] [CrossRef]
- Pierik, R.; van Aken, J.M.; Voesenek, L.A. Is elongation-induced leaf emergence beneficial for submerged Rumex species? Ann. Bot. 2009, 103, 353–357. [Google Scholar] [CrossRef] [Green Version]
- Van Veen, H.; Mustroph, A.; Barding, G.A.; Vergeer-van Eijk, M.; Welschen-Evertman, R.A.; Pedersen, O.; Visser, E.J.; Larive, C.K.; Pierik, R.; Bailey-Serres, J.; et al. Two Rumex species from contrasting hydrological niches regulate flooding tolerance through distinct mechanisms. Plant Cell 2013, 25, 4691–4707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herzog, M.; Pedersen, O. Partial versus complete submergence: snorkelling aids root aeration in Rumex palustris but not in R. acetosa. Plant Cell Environ. 2014, 37, 2381–2390. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, R.; Mustroph, A.; Boonman, A.; Akman, M.; Ammerlaan, A.M.; Breit, T.; Schranz, M.E.; Voesenek, L.A.; van Tienderen, P.H. Root transcript profiling of two Rorippa species reveals gene clusters associated with extreme submergence tolerance. Plant Physiol. 2013, 163, 1277–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akman, M.; Bhikharie, A.; Mustroph, A.; Sasidharan, R. Extreme flooding tolerance in Rorippa. Plant Signal. Behav. 2014, 9, e27847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estioko, L.P.; Miro, B.; Baltazar, A.M.; Merca, F.E.; Ismail, A.M.; Johnson, D.E. Differences in responses to flooding by germinating seeds of two contrasting rice cultivars and two species of economically important grass weeds. AoB Plants 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Nah, G.; Im, J.H.; Kim, J.W.; Park, H.R.; Yook, M.J.; Yang, T.J.; Fischer, A.J.; Kim, D.S. Uncovering the differential molecular basis of adaptive diversity in three Echinochloa leaf transcriptomes. PLoS ONE 2015, 10, e0134419. [Google Scholar] [CrossRef] [Green Version]
- Nandi, S.; Subudhi, P.K.; Senadhira, D.; Manigbas, N.L.; Sen-Mandi, S.; Huang, N. Mapping QTLs for submergence tolerance in rice by AFLP analysis and selective genotyping. Mol. Gen. Genet. 1997, 255, 1–8. [Google Scholar] [CrossRef]
- Siangliw, M.; Toojinda, T.; Tragoonrung, S.; Vanavichit, A. Thai jasmine rice carrying QTLch9 (SubQTL) is submergence tolerant. Ann. Bot. 2003, 91, 255–261. [Google Scholar] [CrossRef] [Green Version]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 2006, 442, 705–708. [Google Scholar] [CrossRef]
- Fukao, T.; Yeung, E.; Bailey-Serres, J. The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 2011, 23, 412–427. [Google Scholar] [CrossRef] [Green Version]
- Tang, D.Q.; Kasai, Y.; Miyamoto, N.; Ukai, Y.; Nemoto, K. Comparison of QTLs for Early Elongation Ability between Two Floating Rice Cultivars with a Different Phylogenetic Origin. Breed. Sci. 2005, 55, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Hattori, Y.; Miura, K.; Asano, K.; Yamamoto, E.; Mori, H.; Kitano, H.; Matsuoka, M.; Ashikari, M. A Major QTL Confers Rapid Internode Elongation in Response to Water Rise in Deepwater Rice. Breed. Sci. 2007, 57, 305–314. [Google Scholar] [CrossRef] [Green Version]
- Hattori, Y.; Nagai, K.; Mori, H.; Kitano, H.; Matsuoka, M.; Ashikari, M. Mapping of three QTLs that regulate internode elongation in deepwater rice. Breed. Sci. 2008, 58, 39–46. [Google Scholar] [CrossRef] [Green Version]
- Kawano, R.; Doi, K.; Yasui, H.; Mochizuki, T.; Yoshimura, A. Mapping of QTLs for floating ability in rice. Breed. Sci. 2008, 58, 47–53. [Google Scholar] [CrossRef] [Green Version]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef]
- Nagai, K.; Kuroha, T.; Ayano, M.; Kurokawa, Y.; Angeles-Shim, R.B.; Shim, J.H.; Yasui, H.; Yoshimura, A.; Ashikari, M. Two novel QTLs regulate internode elongation in deepwater rice during the early vegetative stage. Breed. Sci. 2012, 62, 178–185. [Google Scholar] [CrossRef] [Green Version]
- Nagai, K.; Kondo, Y.; Kitaoka, T.; Noda, T.; Kuroha, T.; Angeles-Shim, R.B.; Yasui, H.; Yoshimura, A.; Ashikari, M. QTL analysis of internode elongation in response to gibberellin in deepwater rice. AoB Plants 2014, 6. [Google Scholar] [CrossRef] [Green Version]
- Ayano, M.; Kani, T.; Kojima, M.; Sakakibara, H.; Kitaoka, T.; Kuroha, T.; Angeles-Shim, R.B.; Kitano, H.; Nagai, K.; Ashikari, M. Gibberellin biosynthesis and signal transduction is essential for internode elongation in deepwater rice. Plant Cell Environ. 2014, 37, 2313–2324. [Google Scholar] [CrossRef]
- Fukushima, A.; Kusano, M.; Redestig, H.; Arita, M.; Saito, K. Integrated omics approaches in plant systems biology. Curr. Opin. Chem. Biol. 2009, 13, 532–538. [Google Scholar] [CrossRef]
- Rai, A.; Saito, K.; Yamazaki, M. Integrated omics analysis of specialized metabolism in medicinal plants. Plant J. 2017, 90, 764–787. [Google Scholar] [CrossRef]
- Jung, K.H.; Seo, Y.S.; Walia, H.; Cao, P.; Fukao, T.; Canlas, P.E.; Amonpant, F.; Bailey-Serres, J.; Ronald, P.C. The submergence tolerance regulator Sub1A mediates stress-responsive expression of AP2/ERF transcription factors. Plant Physiol. 2010, 152, 1674–1692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minami, A.; Yano, K.; Gamuyao, R.; Nagai, K.; Kuroha, T.; Ayano, M.; Nakamori, M.; Koike, M.; Kondo, Y.; Niimi, Y.; et al. Time-Course Transcriptomics Analysis Reveals Key Responses of Submerged Deepwater Rice to Flooding. Plant Physiol. 2018, 176, 3081–3102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuroha, T.; Nagai, K.; Gamuyao, R.; Wang, D.R.; Furuta, T.; Nakamori, M.; Kitaoka, T.; Adachi, K.; Minami, A.; Mori, Y.; et al. Ethylene-gibberellin signaling underlies adaptation of rice to periodic flooding. Science 2018, 361, 181–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, K.; Matsuda, F. Metabolomics for functional genomics, systems biology, and biotechnology. Annu. Rev. Plant Biol. 2010, 61, 463–489. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Lei, Z.; Nikolau, B.J.; Saito, K. Modern plant metabolomics: Advanced natural product gene discoveries, improved technologies, and future prospects. Nat. Prod. Rep. 2015, 32, 212–229. [Google Scholar] [CrossRef] [PubMed]
- Alseekh, S.; Fernie, A.R. Metabolomics 20 years on: What have we learned and what hurdles remain? Plant J. 2018, 94, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Barding, G.A., Jr.; Beni, S.; Fukao, T.; Bailey-Serres, J.; Larive, C.K. Comparison of GC-MS and NMR for metabolite profiling of rice subjected to submergence stress. J. Proteome Res. 2013, 12, 898–909. [Google Scholar] [CrossRef]
- Barding, G.A., Jr.; Fukao, T.; Beni, S.; Bailey-Serres, J.; Larive, C.K. Differential metabolic regulation governed by the rice SUB1A gene during submergence stress and identification of alanylglycine by 1H NMR spectroscopy. J. Proteome Res. 2012, 11, 320–330. [Google Scholar] [CrossRef]
- Locke, A.M.; Barding, G.A., Jr.; Sathnur, S.; Larive, C.K.; Bailey-Serres, J. Rice SUB1A constrains remodelling of the transcriptome and metabolome during submergence to facilitate post-submergence recovery. Plant Cell Environ. 2018, 41, 721–736. [Google Scholar] [CrossRef]
- Herzog, M.; Fukao, T.; Winkel, A.; Konnerup, D.; Lamichhane, S.; Alpuerto, J.B.; Hasler-Sheetal, H.; Pedersen, O. Physiology, gene expression, and metabolome of two wheat cultivars with contrasting submergence tolerance. Plant Cell Environ. 2018, 41, 1632–1644. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhu, W.; Hashiguchi, A.; Nishimura, M.; Tian, J.; Komatsu, S. Metabolic profiles of flooding-tolerant mechanism in early-stage soybean responding to initial stress. Plant Mol. Biol. 2017, 94, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Kusano, M.; Baxter, I.; Fukushima, A.; Oikawa, A.; Okazaki, Y.; Nakabayashi, R.; Bouvrette, D.J.; Achard, F.; Jakubowski, A.R.; Ballam, J.M.; et al. Assessing metabolomic and chemical diversity of a soybean lineage representing 35 years of breeding. Metabolomics 2015, 11, 261–270. [Google Scholar] [CrossRef]
- Kusano, M.; Redestig, H.; Hirai, T.; Oikawa, A.; Matsuda, F.; Fukushima, A.; Arita, M.; Watanabe, S.; Yano, M.; Hiwasa-Tanase, K.; et al. Covering chemical diversity of genetically-modified tomatoes using metabolomics for objective substantial equivalence assessment. PLoS ONE 2011, 6, e16989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopka, J.; Schauer, N.; Krueger, S.; Birkemeyer, C.; Usadel, B.; Bergmuller, E.; Dormann, P.; Weckwerth, W.; Gibon, Y.; Stitt, M.; et al. GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics 2005, 21, 1635–1638. [Google Scholar] [CrossRef] [Green Version]
- Schauer, N.; Steinhauser, D.; Strelkov, S.; Schomburg, D.; Allison, G.; Moritz, T.; Lundgren, K.; Roessner-Tunali, U.; Forbes, M.G.; Willmitzer, L.; et al. GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 2005, 579, 1332–1337. [Google Scholar] [CrossRef]
- Bergmeyer, H.U.; Bernt, E.; Schmidt, F.; Stork, H. D-Glucose: Determination with Hexokinase and Glucose-6-Phosphate Dehydrogenase; Academic Press: New York, NY, USA, 1974; Volume 3, pp. 1196–1201. [Google Scholar]
- Redestig, H.; Kusano, M.; Fukushima, A.; Matsuda, F.; Saito, K.; Arita, M. Consolidating metabolite identifiers to enable contextual and multi-platform metabolomics data analysis. BMC Bioinform. 2010, 11, 214. [Google Scholar] [CrossRef] [Green Version]
- Ruperti, B.; Botton, A.; Populin, F.; Eccher, G.; Brilli, M.; Quaggiotti, S.; Trevisan, S.; Cainelli, N.; Guarracino, P.; Schievano, E.; et al. Flooding Responses on Grapevine: A Physiological, Transcriptional, and Metabolic Perspective. Front. Plant Sci. 2019, 10, 339. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, O.; Rich, S.M.; Colmer, T.D. Surviving floods: Leaf gas films improve O(2) and CO(2) exchange, root aeration, and growth of completely submerged rice. Plant J. 2009, 58, 147–156. [Google Scholar] [CrossRef]
- Winkel, A.; Colmer, T.D.; Ismail, A.M.; Pedersen, O. Internal aeration of paddy field rice (Oryza sativa) during complete submergence—Importance of light and floodwater O2. New Phytol. 2013, 197, 1193–1203. [Google Scholar] [CrossRef]
- Kurokawa, Y.; Nagai, K.; Huan, P.D.; Shimazaki, K.; Qu, H.; Mori, Y.; Toda, Y.; Kuroha, T.; Hayashi, N.; Aiga, S.; et al. Rice leaf hydrophobicity and gas films are conferred by a wax synthesis gene (LGF1) and contribute to flood tolerance. New Phytol. 2018, 218, 1558–1569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, Y.; Kurokawa, Y.; Koike, M.; Malik, A.I.; Colmer, T.D.; Ashikari, M.; Pedersen, O.; Nagai, K. Diel O2 Dynamics in Partially and Completely Submerged Deepwater Rice: Leaf Gas Films Enhance Internodal O2 Status, Influence Gene Expression and Accelerate Stem Elongation for ‘Snorkelling’ during Submergence. Plant Cell Physiol. 2019, 60, 973–985. [Google Scholar] [CrossRef] [PubMed]
- Obata, T.; Fernie, A.R. The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci. 2012, 69, 3225–3243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mackill, D.; Ismail, A.; Singh, U.; Labios, R.; Paris, T. Development and Rapid Adoption of Submergence-Tolerant (Sub1) Rice Varieties. Adv. Agron. 2012, 115, 299–352. [Google Scholar] [CrossRef]
- Van Dongen, J.T.; Frohlich, A.; Ramirez-Aguilar, S.J.; Schauer, N.; Fernie, A.R.; Erban, A.; Kopka, J.; Clark, J.; Langer, A.; Geigenberger, P. Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of arabidopsis plants. Ann. Bot. 2009, 103, 269–280. [Google Scholar] [CrossRef]
- Rocha, M.; Licausi, F.; Araujo, W.L.; Nunes-Nesi, A.; Sodek, L.; Fernie, A.R.; van Dongen, J.T. Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol. 2010, 152, 1501–1513. [Google Scholar] [CrossRef] [Green Version]
- Fukao, T.; Xu, K.; Ronald, P.C.; Bailey-Serres, J. A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 2006, 18, 2021–2034. [Google Scholar] [CrossRef] [Green Version]
- Alpuerto, J.B.; Hussain, R.M.F.; Fukao, T. The key regulator of submergence tolerance, SUB1A, promotes photosynthetic and metabolic recovery from submergence damage in rice leaves. Plant Cell Environ. 2016, 39, 672–684. [Google Scholar] [CrossRef] [Green Version]
- Kende, H.; van der Knaap, E.; Cho, H.T. Deepwater rice: A model plant to study stem elongation. Plant Physiol. 1998, 118, 1105–1110. [Google Scholar] [CrossRef] [Green Version]
- Bailey-Serres, J.; Voesenek, L.A. Life in the balance: A signaling network controlling survival of flooding. Curr. Opin. Plant Biol. 2010, 13, 489–494. [Google Scholar] [CrossRef]
- Woodward, A.W.; Bartel, B. Auxin: Regulation, action, and interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludwig-Muller, J. Auxin conjugates: Their role for plant development and in the evolution of land plants. J. Exp. Bot. 2011, 62, 1757–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rampey, R.A.; LeClere, S.; Kowalczyk, M.; Ljung, K.; Sandberg, G.; Bartel, B. A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 2004, 135, 978–988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staswick, P.E. The tryptophan conjugates of jasmonic and indole-3-acetic acids are endogenous auxin inhibitors. Plant Physiol. 2009, 150, 1310–1321. [Google Scholar] [CrossRef] [Green Version]
- Park, J.E.; Park, J.Y.; Kim, Y.S.; Staswick, P.E.; Jeon, J.; Yun, J.; Kim, S.Y.; Kim, J.; Lee, Y.H.; Park, C.M. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 2007, 282, 10036–10046. [Google Scholar] [CrossRef] [Green Version]
- Ostrowski, M.; Ciarkowska, A.; Jakubowska, A. The auxin conjugate indole-3-acetyl-aspartate affects responses to cadmium and salt stress in Pisum sativum L. J. Plant Physiol. 2016, 191, 63–72. [Google Scholar] [CrossRef]
- Kusano, M.; Fukushima, A.; Kobayashi, M.; Hayashi, N.; Jonsson, P.; Moritz, T.; Ebana, K.; Saito, K. Application of a metabolomic method combining one-dimensional and two-dimensional gas chromatography-time-of-flight/mass spectrometry to metabolic phenotyping of natural variants in rice. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007, 855, 71–79. [Google Scholar] [CrossRef]
- Kusano, M.; Fukushima, A.; Arita, M.; Jonsson, P.; Moritz, T.; Kobayashi, M.; Hayashi, N.; Tohge, T.; Saito, K. Unbiased characterization of genotype-dependent metabolic regulations by metabolomic approach in Arabidopsis thaliana. BMC Syst. Biol. 2007, 1, 53. [Google Scholar] [CrossRef] [Green Version]
- Jonsson, P.; Johansson, A.I.; Gullberg, J.; Trygg, J.; A, J.; Grung, B.; Marklund, S.; Sjostrom, M.; Antti, H.; Moritz, T. High-throughput data analysis for detecting and identifying differences between samples in GC/MS-based metabolomic analyses. Anal. Chem. 2005, 77, 5635–5642. [Google Scholar] [CrossRef]
- Jonsson, P.; Johansson, E.S.; Wuolikainen, A.; Lindberg, J.; Schuppe-Koistinen, I.; Kusano, M.; Sjostrom, M.; Trygg, J.; Moritz, T.; Antti, H. Predictive metabolite profiling applying hierarchical multivariate curve resolution to GC-MS data—A potential tool for multi-parametric diagnosis. J. Proteome Res. 2006, 5, 1407–1414. [Google Scholar] [CrossRef]
- Redestig, H.; Fukushima, A.; Stenlund, H.; Moritz, T.; Arita, M.; Saito, K.; Kusano, M. Compensation for systematic cross-contribution improves normalization of mass spectrometry based metabolomics data. Anal. Chem. 2009, 81, 7974–7980. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Kusano, M.; Oikawa, A.; Fukushima, A.; Noji, M.; Saito, K. Physiological roles of the beta-substituted alanine synthase gene family in Arabidopsis. Plant Physiol. 2008, 146, 310–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kojima, M.; Kamada-Nobusada, T.; Komatsu, H.; Takei, K.; Kuroha, T.; Mizutani, M.; Ashikari, M.; Ueguchi-Tanaka, M.; Matsuoka, M.; Suzuki, K.; et al. Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: An application for hormone profiling in Oryza sativa. Plant Cell Physiol. 2009, 50, 1201–1214. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, Article3. [Google Scholar] [CrossRef]
- Smyth, G.K. Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions using R and Bioconductor; Gentleman, R., Carey, V., Dudoit, S., Irizarry, R., Huber, W., Eds.; Springer: New York, NY, USA, 2005; pp. 397–420. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Galili, T.; O’Callaghan, A.; Sidi, J.; Sievert, C. heatmaply: An R package for creating interactive cluster heatmaps for online publishing. Bioinformatics 2018, 34, 1600–1602. [Google Scholar] [CrossRef]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef]







© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fukushima, A.; Kuroha, T.; Nagai, K.; Hattori, Y.; Kobayashi, M.; Nishizawa, T.; Kojima, M.; Utsumi, Y.; Oikawa, A.; Seki, M.; et al. Metabolite and Phytohormone Profiling Illustrates Metabolic Reprogramming as an Escape Strategy of Deepwater Rice during Partially Submerged Stress. Metabolites 2020, 10, 68. https://doi.org/10.3390/metabo10020068
Fukushima A, Kuroha T, Nagai K, Hattori Y, Kobayashi M, Nishizawa T, Kojima M, Utsumi Y, Oikawa A, Seki M, et al. Metabolite and Phytohormone Profiling Illustrates Metabolic Reprogramming as an Escape Strategy of Deepwater Rice during Partially Submerged Stress. Metabolites. 2020; 10(2):68. https://doi.org/10.3390/metabo10020068
Chicago/Turabian StyleFukushima, Atsushi, Takeshi Kuroha, Keisuke Nagai, Yoko Hattori, Makoto Kobayashi, Tomoko Nishizawa, Mikiko Kojima, Yoshinori Utsumi, Akira Oikawa, Motoaki Seki, and et al. 2020. "Metabolite and Phytohormone Profiling Illustrates Metabolic Reprogramming as an Escape Strategy of Deepwater Rice during Partially Submerged Stress" Metabolites 10, no. 2: 68. https://doi.org/10.3390/metabo10020068
APA StyleFukushima, A., Kuroha, T., Nagai, K., Hattori, Y., Kobayashi, M., Nishizawa, T., Kojima, M., Utsumi, Y., Oikawa, A., Seki, M., Sakakibara, H., Saito, K., Ashikari, M., & Kusano, M. (2020). Metabolite and Phytohormone Profiling Illustrates Metabolic Reprogramming as an Escape Strategy of Deepwater Rice during Partially Submerged Stress. Metabolites, 10(2), 68. https://doi.org/10.3390/metabo10020068







