An Integrative Approach to Assessing Diet–Cancer Relationships
Abstract
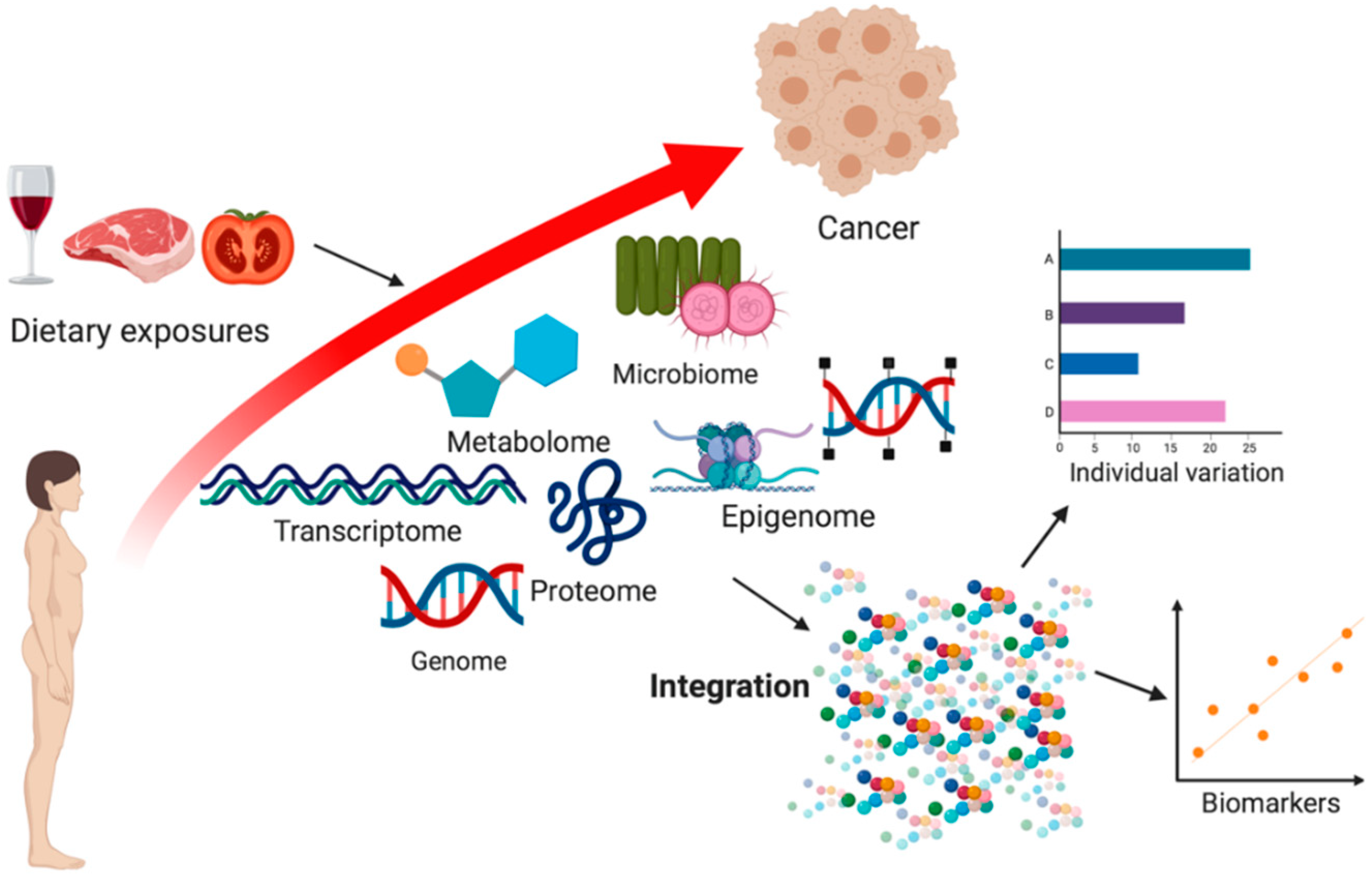
1. Introduction
Diet and Cancer
2. Omics Technologies
2.1. Genome, Epigenome and Transcriptome
2.2. Metabolome
2.3. Proteome
2.4. Microbiome
2.5. Integrative-Omics
3. Biomarkers, Diet and Cancer
4. Response to Diet
5. Challenges and Future Opportunities
6. Conclusions
Funding
Conflicts of Interest
References
- Doll, R.; Peto, R. The causes of cancer: Quantitative estimates of avoidable risks of cancer in the United States today. J. Natl. Cancer Inst. 1981, 66, 1191–1308. [Google Scholar] [CrossRef]
- Zhang, F.F.; Cudhea, F.; Shan, Z.; Michaud, D.S.; Imamura, F.; Eom, H.; Ruan, M.; Rehm, C.D.; Liu, J.; Du, M.; et al. Preventable Cancer Burden Associated with Poor Diet in the United States. JNCI Cancer Spectr. 2019, 3, pkz034. [Google Scholar] [CrossRef] [PubMed]
- Glade, M.J. Food, nutrition, and the prevention of cancer: A global perspective. American Institute for Cancer Research/World Cancer Research Fund, American Institute for Cancer Research, 1997. Nutrition 1999, 15, 523–526. [Google Scholar] [PubMed]
- Wiseman, M. The second World Cancer Research Fund/American Institute for Cancer Research expert report. Food, nutrition, physical activity, and the prevention of cancer: A global perspective. Proc. Nutr. Soc. 2008, 67, 253–256. [Google Scholar] [CrossRef]
- World Cancer Research Fund International; American Institute for Cancer Research (Eds.) Diet, Nutrition, Physical Activity and Cancer: A Global Perspective: A Summary of the Third Expert Report; World Cancer Research Fund International: London, UK, 2018; 112p. [Google Scholar]
- Vinceti, M.; Rothman, K.J. More results but no clear conclusion on selenium and cancer. Am. J. Clin. Nutr. 2016, 104, 245–246. [Google Scholar] [CrossRef] [PubMed]
- Ordovas, J.M.; Corella, D. Nutritional genomics. Annu. Rev. Genom. Hum. Genet. 2004, 5, 71–118. [Google Scholar] [CrossRef] [PubMed]
- Mayne, S.T.; Playdon, M.C.; Rock, C.L. Diet, nutrition, and cancer: Past, present and future. Nat. Rev. Clin. Oncol. 2016, 13, 504–515. [Google Scholar] [CrossRef]
- Di Sebastiano, K.M.; Murthy, G.; Campbell, K.L.; Desroches, S.; Murphy, R.A. Nutrition and Cancer Prevention: Why is the Evidence Lost in Translation? Adv. Nutr. 2019, 10, 410–418. [Google Scholar] [CrossRef]
- Guasch-Ferré, M.; Bhupathiraju, S.N.; Hu, F.B. Use of Metabolomics in Improving Assessment of Dietary Intake. Clin. Chem. 2018, 64, 82–98. [Google Scholar] [CrossRef]
- Jablonska, E.; Vinceti, M. Selenium and Human Health: Witnessing a Copernican Revolution? J. Environ. Sci. Health Part C 2015, 33, 328–368. [Google Scholar] [CrossRef]
- Krumsiek, J.; Bartel, J.; Theis, F.J. Computational approaches for systems metabolomics. Curr. Opin. Biotechnol. 2016, 39, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Rochfort, S. Metabolomics Reviewed: A New “Omics” Platform Technology for Systems Biology and Implications for Natural Products Research. J. Nat. Prod. 2005, 68, 1813–1820. [Google Scholar] [CrossRef] [PubMed]
- Riekeberg, E.; Powers, R. New frontiers in metabolomics: From measurement to insight. F1000Research 2017, 6, 1148. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Giovannucci, E.; Kelsey, K.; Rimm, E.B.; Stampfer, M.J.; Colditz, G.A.; Spiegelman, D.; Willett, W.C.; Hunter, D.J. A methylenetetrahydrofolate reductase polymorphism and the risk of colorectal cancer. Cancer Res. 1996, 56, 4862–4864. [Google Scholar] [PubMed]
- Ma, J.; Stampfer, M.J.; Giovannucci, E.; Artigas, C.; Hunter, D.J.; Fuchs, C.; Willett, W.C.; Selhub, J.; Hennekens, C.H.; Rozen, R. Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Res. 1997, 57, 1098–1102. [Google Scholar] [PubMed]
- Slattery, M.L.; Potter, J.D.; Samowitz, W.; Schaffer, D.; Leppert, M. Methylenetetrahydrofolate reductase, diet, and risk of colon cancer. Cancer Epidemiol. Prev. Biomark. 1999, 8, 513–518. [Google Scholar]
- Ulrich, C.M.; Kampman, E.; Bigler, J.; Schwartz, S.M.; Chen, C.; Bostick, R.; Fosdick, L.; Beresford, S.A.; Yasui, Y.; Potter, J.D. Colorectal adenomas and the C677T MTHFR polymorphism: Evidence for gene-environment interaction? Cancer Epidemiol. Prev. Biomark. 1999, 8, 659–668. [Google Scholar]
- He, J.; Liao, X.-Y.; Zhu, J.-H.; Xue, W.-Q.; Shen, G.-P.; Huang, S.-Y.; Chen, W.; Jia, W. Association of MTHFR C677T and A1298C polymorphisms with non-Hodgkin lymphoma susceptibility: Evidence from a meta-analysis. Sci. Rep. 2014, 4, 6159. [Google Scholar] [CrossRef]
- Counts, J.L.; Goodman, J.I. Hypomethylation of DNA: A possible epigenetic mechanism involved in tumor promotion. Prog. Clin. Biol. Res. 1995, 391, 81–101. [Google Scholar] [CrossRef]
- Baylin, S.B.; Herman, J.G.; Graff, J.R.; Vertino, P.M.; Issa, J.P. Alterations in DNA methylation: A fundamental aspect of neoplasia. Adv. Cancer Res. 1998, 72, 141–196. [Google Scholar]
- Mahmoud, A.; Ali, M. Methyl Donor Micronutrients that Modify DNA Methylation and Cancer Outcome. Nutrients 2019, 11, 608. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.D.; Uthus, E.O. DNA methylation, cancer susceptibility, and nutrient interactions. Exp. Biol. Med. 2004, 229, 988–995. [Google Scholar] [CrossRef]
- Piyathilake, C.J.; Johanning, G.L.; Macaluso, M.; Whiteside, M.; Oelschlager, D.K.; Heimburger, D.C.; Grizzle, W.E. Localized folate and vitamin B-12 deficiency in squamous cell lung cancer is associated with global DNA hypomethylation. Nutr. Cancer 2000, 37, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Johanning, G.L.; Heimburger, D.C.; Piyathilake, C.J. DNA methylation and diet in cancer. J. Nutr. 2002, 132, 3814S–3818S. [Google Scholar] [CrossRef] [PubMed]
- Zeisel, S. Choline, Other Methyl-Donors and Epigenetics. Nutrients 2017, 9, 445. [Google Scholar] [CrossRef]
- Armenise, C.; Lefebvre, G.; Carayol, J.; Bonnel, S.; Bolton, J.; Di Cara, A.; Gheldof, N.; Descombes, P.; Langin, D.; Hm Saris, W.; et al. Transcriptome profiling from adipose tissue during a low-calorie diet reveals predictors of weight and glycemic outcomes in obese, nondiabetic subjects. Am. J. Clin. Nutr. 2017, 106, 736–746. [Google Scholar] [CrossRef]
- Peto, R.; Doll, R.; Buckley, J.D.; Sporn, M.B. Can dietary beta-carotene materially reduce human cancer rates? Nature 1981, 290, 201–208. [Google Scholar] [CrossRef]
- Alpha-Tocopherol, Beta Carotene Cancer Prevention Study Group. The effect of vitamin E and beta carotene on the incidence of lung cancer and other cancers in male smokers. N. Engl. J. Med. 1994, 330, 1029–1035. [Google Scholar] [CrossRef]
- Omenn, G.S.; Goodman, G.E.; Thornquist, M.D.; Balmes, J.; Cullen, M.R.; Glass, A.; Keogh, J.P.; Meyskens, F.L.; Valanis, B.; Williams, J.H.; et al. Effects of a combination of beta carotene and vitamin A on lung cancer and cardiovascular disease. N. Engl. J. Med. 1996, 334, 1150–1155. [Google Scholar] [CrossRef]
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.J.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of mammals and their gut microbes. Science 2008, 320, 1647–1651. [Google Scholar] [CrossRef]
- Maynard, C.L.; Elson, C.O.; Hatton, R.D.; Weaver, C.T. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012, 489, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Shapira, I.; Sultan, K.; Lee, A.; Taioli, E. Evolving concepts: How diet and the intestinal microbiome act as modulators of breast malignancy. ISRN Oncol. 2013, 2013, 693920. [Google Scholar] [CrossRef]
- The Intestinal Microbiome and Estrogen Receptor–Positive Female Breast Cancer. JNCI J Natl Cancer Inst. Available online: https://academic.oup.com/jnci/article/doi/10.1093/jnci/djw029/2457487/The-Intestinal-Microbiome-and-Estrogen (accessed on 4 January 2020).
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.-Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef]
- Lahti, L.; Salojärvi, J.; Salonen, A.; Scheffer, M.; de Vos, W.M. Tipping elements in the human intestinal ecosystem. Nat. Commun. 2014, 5, 4344. [Google Scholar] [CrossRef]
- Faith, J.J.; Guruge, J.L.; Charbonneau, M.; Subramanian, S.; Seedorf, H.; Goodman, A.L.; Clemente, J.C.; Knight, R.; Heath, A.C.; Leibel, R.L.; et al. The long-term stability of the human gut microbiota. Science 2013, 341, 1237439. [Google Scholar] [CrossRef]
- Zeevi, D.; Korem, T.; Zmora, N.; Israeli, D.; Rothschild, D.; Weinberger, A.; Ben-Yacov, O.; Lador, D.; Avnit-Sagi, T.; Lotan-Pompan, M.; et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell 2015, 163, 1079–1094. [Google Scholar] [CrossRef]
- Gieger, C.; Geistlinger, L.; Altmaier, E.; Hrabé de Angelis, M.; Kronenberg, F.; Meitinger, T.; Mewes, H.; Wichmann, H.; Weinberger, K.M.; Adamski, J.; et al. Genetics Meets Metabolomics: A Genome-Wide Association Study of Metabolite Profiles in Human Serum. Gibson G., editor. PLoS Genet. 2008, 4, e1000282. [Google Scholar] [CrossRef]
- Ruggles, K.V.; Wang, J.; Volkova, A.; Contreras, M.; Noya-Alarcon, O.; Lander, O.; Caballero, H.; Dominguez-Bello, M. Changes in the Gut Microbiota of Urban Subjects during an Immersion in the Traditional Diet and Lifestyle of a Rainforest Village. mSphere 2018, 3, e00193-18. [Google Scholar] [CrossRef]
- Frias-Lopez, J.; Shi, Y.; Tyson, G.W.; Coleman, M.L.; Schuster, S.C.; Chisholm, S.W.; Delong, E.F. Microbial community gene expression in ocean surface waters. Proc. Natl. Acad. Sci. USA 2008, 105, 3805–3810. [Google Scholar] [CrossRef] [PubMed]
- Learn More about Biomarkers. Available online: https://dietassessmentprimer.cancer.gov/learn/biomarkers.html (accessed on 21 December 2019).
- Isaksson, B. Urinary nitrogen output as a validity test in dietary surveys. Am. J. Clin. Nutr. 1980, 33, 4–5. [Google Scholar] [CrossRef] [PubMed]
- Prentice, A.M.; Davies, H.L.; Black, A.E.; Ashford, J.; Coward, W.A.; Murgatroyd, P.R.; Black, A.E.; Goldberg, G.R.; Ashford, J.; Sawyer, M.; et al. Unexpectedly Low Levels of Energy Expenditure in Healthy Women. Lancet 1985, 325, 1419–1422. [Google Scholar] [CrossRef]
- Prentice, R.L.; Pettinger, M.; Tinker, L.F.; Huang, Y.; Thomson, C.A.; Johnson, K.C.; Beasley, J.; Anderson, G.; Shikany, J.M.; Chlebowski, R.T.; et al. Regression calibration in nutritional epidemiology: Example of fat density and total energy in relationship to postmenopausal breast cancer. Am. J. Epidemiol. 2013, 178, 1663–1672. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Van Horn, L.; Tinker, L.F.; Neuhouser, M.L.; Carbone, L.; Mossavar-Rahmani, Y.; Thomas, D.; Prentice, R.L. Measurement error corrected sodium and potassium intake estimation using 24-hour urinary excretion. Hypertension 2014, 63, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Bjerve, K.S.; Brubakk, A.M.; Fougner, K.J.; Johnsen, H.; Midthjell, K.; Vik, T. Omega-3 fatty acids: Essential fatty acids with important biological effects, and serum phospholipid fatty acids as markers of dietary ω3-fatty acid intake. Am. J. Clin. Nutr. 1993, 57, 801S–806S. [Google Scholar] [CrossRef] [PubMed]
- Couillard, C.; Lemieux, S.; Vohl, M.-C.; Couture, P.; Lamarche, B. Carotenoids as biomarkers of fruit and vegetable intake in men and women. Br. J. Nutr. 2016, 116, 1206–1215. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischback, M.S.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Tang, Z.-Z.; Chen, G.; Hong, Q.; Huang, S.; Smith, H.M.; Shah, R.D.; Scholz, M.; Ferguson, J.F. Multi-omic analysis of the microbiome and metabolome in healthy subjects reveals microbiome-dependent relationships between diet and metabolites. Front. Genet. 2019, 10, 454. [Google Scholar] [CrossRef]
- Bernstein, H.; Bernstein, C.; Payne, C.M.; Dvorakova, K.; Garewal, H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat. Res. 2005, 589, 47–65. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Ung, T.T.; Kim, N.H.; Jung, Y.D. Role of bile acids in colon carcinogenesis. World J. Clin. Cases 2018, 6, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Maruvada, P.; Lampe, J.W.; Wishart, D.S.; Barupal, D.; Chester, D.N.; Dodd, D.; Djoumbou-Feunang, Y.; Dorrestein, P.C.; Dragsted, L.O.; Draper, J.; et al. Perspective: Dietary Biomarkers of Intake and Exposure—Exploration with Omics Approaches. Adv. Nutr. 2020, 11, 200–215. [Google Scholar] [CrossRef] [PubMed]
- Subar, A.F.; Thompson, F.E.; Kipnis, V.; Midthune, D.; Hurwitz, P.; McNutt, S.; McIntosh, A.; Rosenfeld, S. Comparative Validation of the Block, Willett, and National Cancer Institute Food Frequency Questionnaires. Am. J. Epidemiol. 2001, 154, 1089–1099. [Google Scholar] [CrossRef] [PubMed]
- Kazemian, E.; Akbari, M.E.; Moradi, N.; Gharibzadeh, S.; Mondul, A.M.; Jamshidi-Naeini, Y.; Khademolmele, M.; Zarins, K.R.; Ghodoosi, N.; Amouzegar, A.; et al. Vitamin D Receptor Genetic Variation and Cancer Biomarkers among Breast Cancer Patients Supplemented with Vitamin D3: A Single-Arm Non-Randomized Before and After Trial. Nutrients 2019, 11, 1264. [Google Scholar] [CrossRef]
- Lowe, L.C.; Guy, M.; Mansi, J.L.; Peckitt, C.; Bliss, J.; Wilson, R.G.; Colston, K.W. Plasma 25-hydroxy vitamin D concentrations, vitamin D receptor genotype and breast cancer risk in a UK Caucasian population. Eur. J. Cancer 2005, 41, 1164–1169. [Google Scholar] [CrossRef]
- Citronberg, J.S.; Curtis, K.R.; White, E.; Newcomb, P.A.; Newton, K.; Atkinson, C.; Song, X.; Lampe, J.W.; Aj Hullar, M. Association of gut microbial communities with plasma lipopolysaccharide-binding protein (LBP) in premenopausal women. ISME J. 2018, 12, 1631–1641. [Google Scholar] [CrossRef]
- World Cancer Research Fund/American Institute for Cancer Research. Continuous Update Project Expert Report 2018. Diet, Nutrition, Physical Activity and Breast Cancer. Available online: dietandcancerreport.org (accessed on 15 January 2020).
- Ordovas, J.M.; Ferguson, L.R.; Tai, E.S.; Mathers, J.C. Personalised nutrition and health. BMJ 2018, 13, 2173. [Google Scholar] [CrossRef]
- Giovannucci, E.; Harlan, D.M.; Archer, M.C.; Bergenstal, R.M.; Gapstur, S.M.; Habel, L.A.; Pollak, M.; Regensteiner, J.G.; Yee, D. Diabetes and Cancer: A consensus report. Diabetes Care 2010, 33, 1674–1685. [Google Scholar] [CrossRef]
- Prentice, R.L.; Tinker, L.F.; Huang, Y.; Neuhouser, M.L. Calibration of self-reported dietary measures using biomarkers: An approach to enhancing nutritional epidemiology reliability. Curr. Atheroscler. Rep. 2013, 15, 353. [Google Scholar] [CrossRef]
- Thompson, F.E.; Kirkpatrick, S.I.; Subar, A.F.; Reedy, J.; Schap, T.E.; Wilson, M.M. The National Cancer Institute’s Dietary Assessment Primer: A Resource for Diet Research. J. Acad. Nutr. Diet. 2015, 115, 1986–1995. [Google Scholar] [CrossRef]
- Lo Siou, G.; Csizmadi, I.; Boucher, B.; Akawung, A.; Whelan, H.; Sharma, M.; Al Rajabi, A.; Vena, J.E.; Kirkpatric, S.I.; Koushik, A.; et al. The Comparative Reliability and Feasibility of the Past-Year Canadian Diet History Questionnaire II: Comparison of the Paper and Web Versions. Nutrients 2017, 9, 133. [Google Scholar] [CrossRef]
- Hoffmann, I. Transcending reductionism in nutrition research. Am. J. Clin. Nutr. 2003, 78, 514S–516S. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.H.; Huang, M.; Kelly, R.S.; Benedetti, E.; Siddiqui, J.K.; Zeleznik, O.A.; Pereira, A.; Herrington, D.; Wheelock, C.E.; Krumsiek, J.; et al. Integration of Metabolomic and Other Omics Data in Population-Based Study Designs: An Epidemiological Perspective. Metabolites 2019, 9, 117. [Google Scholar] [CrossRef] [PubMed]
- Mathieson, T.; Franken, H.; Kosinski, J.; Kurzawa, N.; Zinn, N.; Sweetman, G.; Poeckel, D.; Ratnu, V.S.; Schramm, M.; Becher, I.; et al. Systematic analysis of protein turnover in primary cells. Nat. Commun. 2018, 9, 689. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Schloss, P.D. Dynamics and associations of microbial community types across the human body. Nature 2014, 509, 357–360. [Google Scholar] [CrossRef]
- Yu, B.; Zanetti, K.A.; Temprosa, M.; Albanes, D.; Appel, N.; Barrera, C.B.; Ben-Shlomo, Y.; Boerwinkle, E.; Casas, J.P.; Clish, C.; et al. The Consortium of Metabolomics Studies (COMETS): Metabolomics in 47 Prospective Cohort Studies. Am. J. Epidemiol. 2019, 188, 991–1012. [Google Scholar] [CrossRef]
- Swerdlow, A.J.; Harvey, C.E.; Milne, R.L.; Pottinger, C.A.; Vachon, C.M.; Wilkens, L.R.; Gapstur, S.M.; Johansson, M.; Weiderpass, E.; Winn, D.M. The National Cancer Institute Cohort Consortium: An International Pooling Collaboration of 58 Cohorts from 20 Countries. Cancer Epidemiol. Biomark. Prev. 2018, 27, 1307–1319. [Google Scholar] [CrossRef]
- Kelly, R.S.; Croteau-Chonka, D.C.; Dahlin, A.; Mirzakhani, H.; Wu, A.C.; Wan, E.S.; McGeachie, M.J.; Qiu, W.; Sordillo, J.E.; Al-Garawi, A.; et al. Integration of metabolomic and transcriptomic networks in pregnant women reveals biological pathways and predictive signatures associated with preeclampsia. Metabolomics 2017, 13, 7. [Google Scholar] [CrossRef]
- Liu, B.; Pop, M. MetaPath: Identifying differentially abundant metabolic pathways in metagenomic datasets. BMC Proc. 2011, 5, S9. [Google Scholar] [CrossRef]
- Li, C.-X.; Wheelock, C.E.; Sköld, C.M.; Wheelock, Å.M. Integration of multi-omics datasets enables molecular classification of COPD. Eur. Respir. J. 2018, 51, 1701930. [Google Scholar] [CrossRef]
- Kirkpatrick, S.I.; Reedy, J.; Butler, E.N.; Dodd, K.W.; Subar, A.F.; Thompson, F.E.; McKinnon, R.A. Dietary assessment in food environment research: A systematic review. Am. J. Prev. Med. 2014, 46, 94–102. [Google Scholar] [CrossRef] [PubMed]
| Author | Population | Aim | Omics or Biomarkers Assessed | Dietary Measures | Key Findings |
|---|---|---|---|---|---|
| Zeevi et al. [39] | 800 healthy subjects aged 18–70 | To measure individualized post prandial glucose response, variability in response and factors related to variability | Microbiome in stool (16S rRNA), blood glucose | Food frequency questionnaires at baseline, food logs, and standardized meals provided to subjects | High interpersonal variability in post prandial glucose response to the same meal (self-reported and standardized). Variability was associated with microbiome taxa and phylum. Microbiome factors were used (along with other factors) to predict post prandial glucose response |
| Tang et al. [51] | 136 healthy subjects | To determine associations between microbiome composition and habitual diet | Microbiome (16S rRNA) in stool and saliva, metabolomics (plasma and stool) on a subset (N = 75) | 3-day food records and food frequency questionnaire (NCI’s DHQ I) [55] | On a global level, long-term diet was associated with the gut microbiome while short-term diet was associated with the gut and plasma metabolome. 61 dietary nutrients, predominately plant and dairy derived were associated with at least 1 bacterial genus. Metabolic flux through plant-derived nutrients and metals were susceptible to interactions between diet and microbiome composition |
| Kazemian et al. [56] | Single-arm non-randomized pre-and post-trial in 176 breast cancer survivors who received vitamin D supplementation for 12 weeks | To study if polymorphisms in vitamin D receptor (VDR) are associated with change in biomarkers known to relate to breast cancer risk and survival | VDR polymorphisms, Biomarkers: E-cadherin, MMP9, interferon β, s-ICAM-1, VCAM-1, TNFα, IL6, PAI-1, hs-CRP | 4000 IUD vitamin D3 supplement, 3 X 24-hr food records | Variation in the response to vitamin D supplementation was observed. Changes in cancer biomarkers pre and post vitamin D supplementation differed by genotype and haplotype, e.g., women with AA and GA genotypes of cdx2 had greater increase in MMP9 levels. Genotype differences were also observed for TNFα, suggesting potential relevance for cancer risk and survival. |
| Lowe et al. [57] | Breast cancer patients (n = 179) and control women (n = 179) in the United Kingdom | To determine whether low 25(OH)D levels in combination with VDR polymorphisms are associated with breast cancer risk | VDR polymorphisms, Biomarker: plasma 25(OH)D levels measured by ELISA | None in addition to plasma 25(OH)D | 25(OH)D levels were lower in breast cancer patients. Increased odds of breast cancer among people with the BsmI polymorphism in the VDR. People with both both low 25(OH)D and BsmI polymorphism had the greatest risk of breast cancer while those with either low 25(OH)D or BsmI had intermediate risk. |
| Citronberg et al. [58] | 110 premenopausal women in the United States | To dermine associations between gut microbial communities, inflammation and dietary intake | Microbiome in stool (16S rNA), Biomarkers: plasma LPS-binding protein and CRP | 3-day food records | Dietary fat intake, particularly saturated fat intake and CRP were positively associated with LBP. The abundance of actinobacteria and lipopolysaccharide synthesis differed by LPS tertile. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murphy, R. An Integrative Approach to Assessing Diet–Cancer Relationships. Metabolites 2020, 10, 123. https://doi.org/10.3390/metabo10040123
Murphy R. An Integrative Approach to Assessing Diet–Cancer Relationships. Metabolites. 2020; 10(4):123. https://doi.org/10.3390/metabo10040123
Chicago/Turabian StyleMurphy, Rachel. 2020. "An Integrative Approach to Assessing Diet–Cancer Relationships" Metabolites 10, no. 4: 123. https://doi.org/10.3390/metabo10040123
APA StyleMurphy, R. (2020). An Integrative Approach to Assessing Diet–Cancer Relationships. Metabolites, 10(4), 123. https://doi.org/10.3390/metabo10040123





