Systems Biology ARDS Research with a Focus on Metabolomics
Abstract
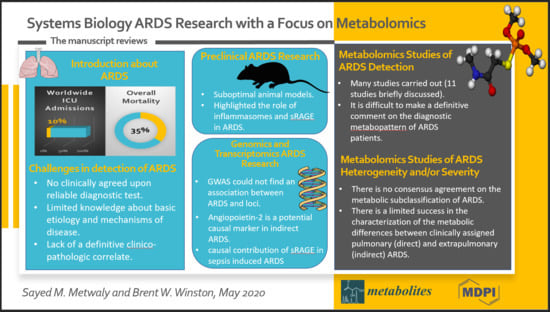
:1. Introduction
2. Challenges in the Detection of ARDS and the Characterization of Its Susceptibility
3. Pre-clinical ARDS Research
4. ARDS Genomics and Transcriptomics Research
5. Metabolomics Studies of ARDS Detection
6. Metabolomics Studies of ARDS Heterogeneity and/or Severity
7. Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Blank, R.; Napolitano, L.M. Epidemiology of ARDS and ALI. Crit. Care Clin. 2011, 27, 439–458. [Google Scholar] [CrossRef] [PubMed]
- Rubenfeld, G.D.; Caldwell, E.; Peabody, E.; Weaver, J.; Martin, D.P.; Neff, M.; Stern, E.J.; Hudson, L.D. Incidence and Outcomes of Acute Lung Injury. New Engl. J. Med. 2005, 353, 1685–1693. [Google Scholar] [CrossRef] [Green Version]
- Ranieri, V.M.; Rubenfeld, G.D.; Thompson, B.T.; Ferguson, N.D.; Caldwell, E.; Fan, E.; Camporota, L.; Slutsky, A.S. Acute Respiratory Distress Syndrome. JAMA 2012, 307, 2526–2533. [Google Scholar] [CrossRef] [PubMed]
- Parhar, K.K.S.; Zjadewicz, K.; Soo, A.; Sutton, A.; Zjadewicz, M.; Doig, L.; Lam, C.; Ferland, A.; Niven, D.J.; Fiest, K.M.; et al. Epidemiology, Mechanical Power, and 3-Year Outcomes in Acute Respiratory Distress Syndrome Patients Using Standardized Screening. An Observational Cohort Study. Ann. Am. Thorac. Soc. 2019, 16, 1263–1272. [Google Scholar] [CrossRef] [PubMed]
- Kollef, M.H.; Schuster, D.P. The Acute Respiratory Distress Syndrome. New Engl. J. Med. 1995, 332, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Thille, A.W.; Esteban, A.; Fernandez-Segoviano, P.; Rodriguez, J.-M.; Aramburu, J.-A.; Penuelas, O.; Cortés-Puch, I.; Cardinal, P.; Lorente, J.A.; Vivar, F.F. Comparison of the Berlin Definition for Acute Respiratory Distress Syndrome with Autopsy. Am. J. Respir. Crit. Care Med. 2013, 187, 761–767. [Google Scholar] [CrossRef] [Green Version]
- Levitt, J.E.; Vinayak, A.G.; Gehlbach, B.K.; Pohlman, A.; Van Cleve, W.; Hall, J.B.; Kress, J.P. Diagnostic utility of B-type natriuretic peptide in critically ill patients with pulmonary edema: A prospective cohort study. Crit. Care 2008, 12, R3. [Google Scholar] [CrossRef] [Green Version]
- Rana, R.; Vlahakis, N.E.; Daniels, C.E.; Jaffe, A.S.; Klee, G.G.; Hubmayr, R.D.; Gajic, O. B-type natriuretic peptide in the assessment of acute lung injury and cardiogenic pulmonary edema*. Crit. Care Med. 2006, 34, 1941–1946. [Google Scholar] [CrossRef]
- Luyt, C.-E.; Combes, A.; Reynaud, C.; Hekimian, G.; Nieszkowska, A.; Tonnellier, M.; Aubry, A.; Trouillet, J.-L.; Bernard, M.; Chastre, J. Usefulness of procalcitonin for the diagnosis of ventilator-associated pneumonia. Intensiv. Care Med. 2008, 34, 1434–1440. [Google Scholar] [CrossRef]
- A Matthay, M.; Zemans, R.L.; Zimmerman, G.A.; Arabi, Y.M.; Beitler, J.R.; Mercat, A.; Herridge, M.; Randolph, A.G.; Calfee, C.S. Acute respiratory distress syndrome. Nat. Rev. Dis. Prim. 2019, 5, 18. [Google Scholar] [CrossRef]
- Rubenfeld, G.D.; Caldwell, E.; Granton, J.; Hudson, L.D.; Matthay, M.A. Interobserver variability in applying a radiographic definition for ARDS. Chest 1999, 116, 1347–1353. [Google Scholar] [CrossRef] [Green Version]
- Hoegl, S.; Burns, N.; Angulo, M.; Francis, D.; Osborne, C.M.; Mills, T.W.; Blackburn, M.R.; Eltzschig, H.K.; Vohwinkel, C.U. Capturing the multifactorial nature of ARDS - “Two-hit” approach to model murine acute lung injury. Physiol. Rep. 2018, 6, e13648. [Google Scholar] [CrossRef]
- Yehya, N. Lessons learned in acute respiratory distress syndrome from the animal laboratory. Ann. Transl. Med. 2019, 7, 503. [Google Scholar] [CrossRef]
- Schneemann, M.; Schoedon, G. Species differences in macrophage NO production are important. Nat. Immunol. 2002, 3, 102. [Google Scholar] [CrossRef] [PubMed]
- Rehli, M. Of mice and men: Species variations of Toll-like receptor expression. Trends Immunol. 2002, 23, 375–378. [Google Scholar] [CrossRef]
- Sone, Y.; Serikov, V.B.; Staub, N.C. Intravascular macrophage depletion attenuates endotoxin lung injury in anesthetized sheep. J. Appl. Physiol. 1999, 87, 1354–1359. [Google Scholar] [CrossRef] [PubMed]
- Schroder, K.; Tschopp, J. The inflammasomes. Cell 2010, 140, 821–832. [Google Scholar] [CrossRef] [Green Version]
- Dolinay, T.; Kim, Y.S.; Howrylak, J.; Hunninghake, G.M.; An, C.H.; Fredenburgh, L.; Massaro, A.F.; Rogers, A.; Gazourian, L.; Nakahira, K.; et al. Inflammasome-regulated Cytokines Are Critical Mediators of Acute Lung Injury. Am. J. Respir. Crit. Care Med. 2012, 185, 1225–1234. [Google Scholar] [CrossRef]
- Jones, H.D.; Crother, T.R.; Gonzalez-Villalobos, R.A.; Jupelli, M.; Chen, S.; Dagvadorj, J.; Arditi, M.; Shimada, K. The NLRP3 Inflammasome Is Required for the Development of Hypoxemia in LPS/Mechanical Ventilation Acute Lung Injury. Am. J. Respir. Cell Mol. Boil. 2013, 50, 270–280. [Google Scholar] [CrossRef]
- Grailer, J.J.; Canning, B.A.; Kalbitz, M.; Haggadone, M.D.; Dhond, R.M.; Andjelkovic, A.V.; Zetoune, F.S.; Ward, P.A. Critical role for the NLRP3 inflammasome during acute lung injury. J. Immunol. 2014, 192, 5974–5983. [Google Scholar] [CrossRef]
- Cohen, T.S.; Prince, A.S. Activation of inflammasome signaling mediates pathology of acute P. aeruginosa pneumonia. J. Clin. Investig. 2013, 123, 1630–1637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jabaudon, M.; Blondonnet, R.; Roszyk, L.; Bouvier, D.; Audard, J.; Clairefond, G.; Fournier, M.; Marceau, G.; Dechelotte, P.; Pereira, B.; et al. Soluble RAGE Predicts Impaired Alveolar Fluid Clearance in Acute Respiratory Distress Syndrome. Am. J. Respir. Crit. Care Med. 2015, 192, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Blondonnet, R.; Audard, J.; Belville, C.; Clairefond, G.; Lutz, J.; Bouvier, D.; Roszyk, L.; Gross, C.; Lavergne, M.; Fournet, M.; et al. RAGE inhibition reduces acute lung injury in mice. Sci. Rep. 2017, 7, 7208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogers, A.J. Genome-Wide Association Study in Acute Respiratory Distress Syndrome. Finding the Needle in the Haystack to Advance Our Understanding of Acute Respiratory Distress Syndrome. Am. J. Respir. Crit. Care Med. 2018, 197, 1373–1374. [Google Scholar] [CrossRef] [PubMed]
- Erickson, S.E.; Shlipak, M.G.; Martin, G.S.; Wheeler, A.P.; Ancukiewicz, M.; Matthay, M.A.; Eisner, M.D. National Institutes of Health National Heart, Lung, and Blood Institute Acute Respiratory Distress Syndrome Network Racial and ethnic disparities in mortality from acute lung injury*. Crit. Care Med. 2009, 37, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ryb, G.E.; Cooper, C. Race/ethnicity and acute respiratory distress syndrome: A National Trauma Data Bank study. J. Natl. Med Assoc. 2010, 102, 865–869. [Google Scholar] [CrossRef]
- Jones, T.K.; Feng, R.; Kerchberger, V.E.; Reilly, J.P.; Anderson, B.J.; Shashaty, M.G.S.; Wang, F.; Dunn, T.G.; Riley, T.R.; Abbott, J.; et al. Plasma sRAGE Acts as a Genetically Regulated Causal Intermediate in Sepsis-associated Acute Respiratory Distress Syndrome. Am. J. Respir. Crit. Care Med. 2020, 201, 47–56. [Google Scholar] [CrossRef]
- Agrawal, A.; Matthay, M.A.; Kangelaris, K.N.; Stein, J.; Chu, J.C.; Imp, B.M.; Cortez, A.; Abbott, J.; Liu, K.D.; Calfee, C.S. Plasma Angiopoietin-2 Predicts the Onset of Acute Lung Injury in Critically Ill Patients. Am. J. Respir. Crit. Care Med. 2013, 187, 736–742. [Google Scholar] [CrossRef] [Green Version]
- Bos, L.D.; Schouten, L.R.; A Van Vught, L.; A Wiewel, M.; Ong, D.S.Y.; Cremer, O.; Artigas, A.; Martin-Loeches, I.; Hoogendijk, A.J.; Van Der Poll, T.; et al. Identification and validation of distinct biological phenotypes in patients with acute respiratory distress syndrome by cluster analysis. Thorax 2017, 72, 876–883. [Google Scholar] [CrossRef]
- Calfee, C.S.; Delucchi, K.; Parsons, P.E.; Thompson, B.T.; Ware, L.B.; Matthay, M.A.; Network, N.A. Subphenotypes in acute respiratory distress syndrome: Latent class analysis of data from two randomised controlled trials. Lancet Respir. Med. 2014, 2, 611–620. [Google Scholar] [CrossRef] [Green Version]
- Calfee, C.S.; Network, A.T.N.A.R.D.S.C.T.; Eisner, M.D.; Parsons, P.E.; Thompson, B.T.; Conner, E.R.; Matthay, M.A.; Ware, L.B. Soluble intercellular adhesion molecule-1 and clinical outcomes in patients with acute lung injury. Intensiv. Care Med. 2008, 35, 248–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calfee, C.S.; Gallagher, D.; Abbott, J.; Thompson, B.T.; Matthay, M.A.; Network, N.A. Plasma angiopoietin-2 in clinical acute lung injury. Crit. Care Med. 2012, 40, 1731–1737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calfee, C.S.; Ware, L.B.; Eisner, M.D.; E Parsons, P.; Thompson, B.T.; Wickersham, N.; A Matthay, M.; Network, N.A. Plasma receptor for advanced glycation end products and clinical outcomes in acute lung injury. Thorax 2008, 63, 1083–1089. [Google Scholar] [CrossRef] [Green Version]
- Famous, K.R.; Delucchi, K.; Ware, L.B.; Kangelaris, K.N.; Liu, K.D.; Thompson, B.T.; Calfee, C.S. Acute Respiratory Distress Syndrome Subphenotypes Respond Differently to Randomized Fluid Management Strategy. Am. J. Respir. Crit. Care Med. 2017, 195, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Bos, L.D.; Scicluna, B.P.; Ong, D.S.Y.; Cremer, O.; Van Der Poll, T.; Schultz, M.J. On behalf of the MARS consortium Understanding Heterogeneity in Biologic Phenotypes of Acute Respiratory Distress Syndrome by Leukocyte Expression Profiles. Am. J. Respir. Crit. Care Med. 2019, 200, 42–50. [Google Scholar] [CrossRef]
- Wilson, J.G.; Calfee, C.S. ARDS Subphenotypes: Understanding a Heterogeneous Syndrome. Crit. Care 2020, 24, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Fermier, B.; Blasco, H.; Godat, E.; Bocca, C.; Moënne-Loccoz, J.; Emond, P.; Andres, C.R.; Laffon, M.; Ferrandière, M. Specific Metabolome Profile of Exhaled Breath Condensate in Patients with Shock and Respiratory Failure: A Pilot Study. Metab. 2016, 6, 26. [Google Scholar] [CrossRef] [Green Version]
- Rogers, A.J.; Matthay, M.A. Applying metabolomics to uncover novel biology in ARDS. Am. J. Physiol. Cell. Mol. Physiol. 2014, 306, L957–L961. [Google Scholar] [CrossRef] [Green Version]
- Banoei, M.M.; Donnelly, S.J.; Mickiewicz, B.; Weljie, A.; Vogel, H.J.; Winston, B.W. Metabolomics in critical care medicine: A new approach to biomarker discovery. Clin. Investig. Med. 2014, 37, E363–E376. [Google Scholar] [CrossRef] [Green Version]
- Metwaly, S.; Côté, A.; Donnelly, S.J.; Banoei, M.M.; Mourad, A.I.; Winston, B.W. Evolution of ARDS biomarkers: Will metabolomics be the answer? Am. J. Physiol. Cell. Mol. Physiol. 2018, 315, L526–L534. [Google Scholar] [CrossRef]
- Bowler, R.P.; Wendt, C.H.; Fessler, M.B.; Foster, M.W.; Kelly, R.; Lasky-Su, J.; Rogers, A.J.; Stringer, K.A.; Winston, B.W. New Strategies and Challenges in Lung Proteomics and Metabolomics. An Official American Thoracic Society Workshop Report. Ann. Am. Thorac. Soc. 2017, 14, 1721–1743. [Google Scholar] [CrossRef] [PubMed]
- Schubert, J.; Müller, W.P.E.; Benzing, A.; Geiger, K. Application of a new method for analysis of exhaled gas in critically ill patients. Intensiv. Care Med. 1998, 24, 415–421. [Google Scholar] [CrossRef] [PubMed]
- King, J.; Koc, H.; Unterkofler, K.; Mochalski, P.; Kupferthaler, A.; Teschl, G.; Teschl, S.; Hinterhuber, H.; Amann, A. Physiological modeling of isoprene dynamics in exhaled breath. J. Theor. Boil. 2010, 267, 626–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, J.; Mochalski, P.; Unterkofler, K.; Teschl, G.; Klieber, M.; Stein, M.; Amann, A.; Baumann, M. Breath isoprene: Muscle dystrophy patients support the concept of a pool of isoprene in the periphery of the human body. Biochem. Biophys. Res. Commun. 2012, 423, 526–530. [Google Scholar] [CrossRef] [Green Version]
- Bos, L.D.; Weda, H.; Wang, Y.; Knobel, H.; Nijsen, T.M.; Vink, T.J.; Zwinderman, A.H.; Sterk, P.J.; Schultz, M.J. Exhaled breath metabolomics as a noninvasive diagnostic tool for acute respiratory distress syndrome. Eur. Respir. J. 2014, 44, 188–197. [Google Scholar] [CrossRef] [Green Version]
- Bos, L.D.; Schultz, M.J.; Sterk, P.J. Exhaled breath profiling for diagnosing acute respiratory distress syndrome. BMC Pulm. Med. 2014, 14, 72. [Google Scholar] [CrossRef] [Green Version]
- Stringer, K.A.; McKay, R.; Karnovsky, A.; Quémerais, B.; Lacy, P. Metabolomics and Its Application to Acute Lung Diseases. Front. Immunol. 2016, 7, 263. [Google Scholar] [CrossRef] [Green Version]
- Stringer, K.A.; Serkova, N.J.; Karnovsky, A.; Guire, K.; Paine, R.; Standiford, T.J. Metabolic consequences of sepsis-induced acute lung injury revealed by plasma 1H-nuclear magnetic resonance quantitative metabolomics and computational analysis. Am. J. Physiol. Cell. Mol. Physiol. 2011, 300, L4–L11. [Google Scholar] [CrossRef] [Green Version]
- Biswas, S.K.; Rahman, I. Environmental toxicity, redox signaling and lung inflammation: The role of glutathione. Mol. Asp. Med. 2008, 30, 60–76. [Google Scholar] [CrossRef] [Green Version]
- Brealey, D.; Brand, M.; Hargreaves, I.; Heales, S.; Land, J.; Smoleński, R.; A Davies, N.; Cooper, C.; Singer, M. Association between mitochondrial dysfunction and severity and outcome of septic shock. Lancet 2002, 360, 219–223. [Google Scholar] [CrossRef] [Green Version]
- Lucas, R.; Verin, A.D.; Black, S.M.; Catravas, J.D. Regulators of endothelial and epithelial barrier integrity and function in acute lung injury. Biochem. Pharmacol. 2009, 77, 1763–1772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stringer, K.A.; Jones, A.E.; Puskarich, M.A.; Karnovsky, A.; Serkova, N.J. 1H-Nuclear Magnetic Resonance (NMR)-Detected Lipids Associated with Apoptosis Differentiate Early Acute Respiratory Distress Syndrome (ARDS) from Sepsis. Am. J. Respir. Crit. Care Med. 2014, 189, A5000. [Google Scholar]
- Owens, R.L.; Stigler, W.S.; Hess, D.R. Do Newer Monitors of Exhaled Gases, Mechanics, and Esophageal Pressure Add Value? Clin. Chest Med. 2008, 29, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo-García, J.L.; Naz, S.; Nin, N.; Rojas, Y.; Erazo, M.; Martínez-Caro, L.; Garcia, A.; De Paula, M.; Fernandez-Segoviano, P.; Casals, C.; et al. A Metabolomic Approach to the Pathogenesis of Ventilator-induced Lung Injury. Anesthesiol. 2014, 120, 694–702. [Google Scholar] [CrossRef]
- Rai, R.; Azim, A.; Sinha, N.; Sahoo, J.N.; Singh, C.; Ahmed, A.; Saigal, S.; Baronia, A.K.; Gupta, D.; Gurjar, M.; et al. Metabolic profiling in human lung injuries by high-resolution nuclear magnetic resonance spectroscopy of bronchoalveolar lavage fluid (BALF). Metabolomics 2012, 9, 667–676. [Google Scholar] [CrossRef]
- Evans, C.; Karnovsky, A.; Kovach, M.A.; Standiford, T.J.; Burant, C.; Stringer, K.A. Untargeted LC–MS Metabolomics of Bronchoalveolar Lavage Fluid Differentiates Acute Respiratory Distress Syndrome from Health. J. Proteome Res. 2013, 13, 640–649. [Google Scholar] [CrossRef]
- Singh, C.; Rai, R.; Azim, A.; Sinha, N.; Ahmed, A.; Singh, K.; Kayastha, A.M.; Baronia, A.K.; Gurjar, M.; Poddar, B.; et al. Metabolic profiling of human lung injury by 1H high-resolution nuclear magnetic resonance spectroscopy of blood serum. Metabolomics 2014, 11, 166–174. [Google Scholar] [CrossRef]
- Rogers, A.J.; Contrepois, K.; Wu, M.; Zheng, M.; Peltz, G.; Ware, L.B.; Matthay, M.A. Profiling of ARDS pulmonary edema fluid identifies a metabolically distinct subset. Am. J. Physiol. Cell. Mol. Physiol. 2017, 312, L703–L709. [Google Scholar] [CrossRef]
- Izquierdo-García, J.L.; Nin, N.; Jimenez-Clemente, J.; Horcajada, J.P.; Arenas-Miras, M.D.M.; Gea, J.; Esteban, A.; Ruíz-Cabello, J.; Lorente, J.A. Metabolomic Profile of ARDS by Nuclear Magnetic Resonance Spectroscopy in Patients with H1N1 Influenza Virus Pneumonia. Shock. 2018, 50, 504–510. [Google Scholar] [CrossRef]
- Izquierdo-García, J.L.; Nin, N.; Cardinal-Fernandez, P.; Ruiz-Cabello, J.; Lorente, J. Ángel Metabolomic profile of acute respiratory distress syndrome of different etiologies. Intensiv. Care Med. 2019, 45, 1318–1320. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.; Yue, X.; Wu, H.; Han, T.-L.; Zhu, J.; Wang, C.; Lei, M.; Zhang, M.; Liu, Q.; Xu, F. Explore potential plasma biomarkers of acute respiratory distress syndrome (ARDS) using GC-MS metabolomics analysis. Clin. Biochem. 2019, 66, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Barnett, N.; Ware, L.B. Biomarkers in Acute Lung Injury—Marking Forward Progress. Crit. Care Clin. 2011, 27, 661–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viswan, A.; Singh, C.; Rai, R.; Azim, A.; Sinha, N.; Baronia, A.K. Metabolomics based predictive biomarker model of ARDS: A systemic measure of clinical hypoxemia. PLOS ONE 2017, 12, e0187545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viswan, A.; Ghosh, P.; Gupta, D.; Azim, A.; Sinha, N. Distinct Metabolic Endotype Mirroring Acute Respiratory Distress Syndrome (ARDS) Subphenotype and its Heterogeneous Biology. Sci. Rep. 2019, 9, 2108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thille, A.W.; Richard, J.-C.M.; Maggiore, S.M.; Ranieri, V.M.; Brochard, L. Alveolar recruitment in pulmonary and extrapulmonary acute respiratory distress syndrome: Comparison using pressure-volume curve or static compliance. Anesthesiology 2007, 106, 212–217. [Google Scholar] [CrossRef]

| Technique | NMR | GC-MS | LC-MS |
|---|---|---|---|
| Strengths |
|
|
|
| Weaknesses |
|
|
|
| Authors | Study | Cases | Controls | Sample Type | Analytical Platform | Metabolites Profiled | ARDS Associated Metabolites |
|---|---|---|---|---|---|---|---|
| Schubert et al., 1998 [42] | ARDS detection | n = 19 ARDS | n = 18 ventilated SICU | Exhaled breath | GC-MS (targeted) | 9 | Isoprene |
| Stringer et al., 2011 [48] | ARDS detection and severity | n = 13 sepsis-induced ALI | n = 6 healthy | Plasma | 1-H NMR | 40 | Total glutathione, adenosine, phosphatidylserine, sphingomyelin |
| Rai et al., 2012 [55] | ARDS detection | n = 21 ARDS | n = 9 ventilated ICU | mBALF | 1-H NMR | >100 | BCA, arginine, glycine, aspartic acid, succinate, glutamate, lactate, ethanol, acetate, proline |
| Evans et al., 2014 [56] | ARDS detection | n = 18 ARDS | n = 8 healthy | BALF | LC-MS | >500 | Guanosine, xanthine, hypoxanthine, lactate, phosphatidylcholines |
| Bos et al., 2014 [45] | ARDS detection, heterogeneity and severity | n = 42 ARDS | n = 59 ventilated ICU | Exhaled breath | GC-MS (untargeted) | >500 (untargeted for test group) 5 for training and validation groups | 3-methylheptane, octane, acetaldehyde |
| Singh et al., 2014 [57] | ARDS detection | n = 26 ARDS | n = 19 ventilated non-ARDS | Serum | 1-H NMR | >100 | N-acetylglycoproteins, acetoacetate, lactate, creatinine, histidine, formate, branched-chain amino acids |
| Stringer et al., 2014 [52] | ARDS detection | n = 14 ARDS | n = 33 unventilated sepsis | Serum | 1-H NMR | 51 | Phosphatidylserine, total lipids, total methylene lipids, total cholines (in ARDS compared to sepsis) |
| Rogers et al. 2017 [58] | ARDS detection and heterogeneity | n = 16 ARDS | n = 13 hydrostatic pulmonary edema | Pulmonary edema fluid | UHLC/MS/MS2 for basic species, acidic species, and lipids. | 760 | 235 were significantly higher in a subset of 6 ARDS patients (hypermetabolic) |
| Viswan et al., 2017 [63] | ARDS severity | n = 36 ARDS (23 moderate/severe ARDS and 13 mild ARDS) | None | mBALF | 1-H NMR (high resolution, 800 MHz) | 29 | A proposed biomarker composed of six metabolites was identified. Proline, lysine/arginine, taurine and threonine were correlated to moderate/severe ARDS while glutamate was found to be characteristic of mild ARDS. |
| Izquierdo-García et al., [59] | ARDS detection | -Derivation set: n = 12 * -Validation set: n = 13 * | -Derivation set: n = 18 ** -Validation set: n = 13 ** | Serum | 1-H NMR (500 MHz) | N/A (spectral binning was applied, and only the significantly different bins were profiled) | ARDS patients have low serum glucose, alanine, glutamine, methylhistidine and fatty acid concentrations, and high phenylalanine and methylguanidine. |
| Izquierdo-García et al., [60] | ARDS detection | n = 13 # | n = 17 ## | Serum | 1-H NMR (500 MHz) | N/A (spectral binning was applied, and only the significantly different bins were profiled) | ARDS patients have low serum glucose, alanine, methylhistidine, fatty acids, citrate, creatine, creatinine and valine. Acetone levels are higher. |
| Viswan et al., 2019 [64] | ARDS heterogeneity and severity | Severity: - n = 176 serum - n = 146 mBALF Heterogeneity: - n = 147 serum - n = 128 mBALF | - n = 68 serum - n = 40 mBALF | Serum and mBALF | 1-H NMR (high resolution, 800 MHz) | -54 in serum -52 in mBALF | -Serum: proline, glutamate, phenylalanine, valine -mBALF: isoleucine, leucine, valine, lysine/arginine, tyrosine, threonine |
| Lin et al., 2019 [61] | ARDS detection and heterogeneity | n = 37 ARDS | n = 28 healthy controls | Plasma | GC-MS | 222 | 128 metabolites were significantly different in ARDS patients. There is a correlation between phenylalanine, aspartic acid and carbamic acid levels and ARDS severity grades. Ornithine, caprylic acid, azetidine and iminodiacetic acid, is a potential biomarker for ARDS severity. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Metwaly, S.M.; Winston, B.W. Systems Biology ARDS Research with a Focus on Metabolomics. Metabolites 2020, 10, 207. https://doi.org/10.3390/metabo10050207
Metwaly SM, Winston BW. Systems Biology ARDS Research with a Focus on Metabolomics. Metabolites. 2020; 10(5):207. https://doi.org/10.3390/metabo10050207
Chicago/Turabian StyleMetwaly, Sayed M., and Brent W. Winston. 2020. "Systems Biology ARDS Research with a Focus on Metabolomics" Metabolites 10, no. 5: 207. https://doi.org/10.3390/metabo10050207
APA StyleMetwaly, S. M., & Winston, B. W. (2020). Systems Biology ARDS Research with a Focus on Metabolomics. Metabolites, 10(5), 207. https://doi.org/10.3390/metabo10050207