Lipid Annotation by Combination of UHPLC-HRMS (MS), Molecular Networking, and Retention Time Prediction: Application to a Lipidomic Study of In Vitro Models of Dry Eye Disease
Abstract
1. Introduction
2. Results and Discussion
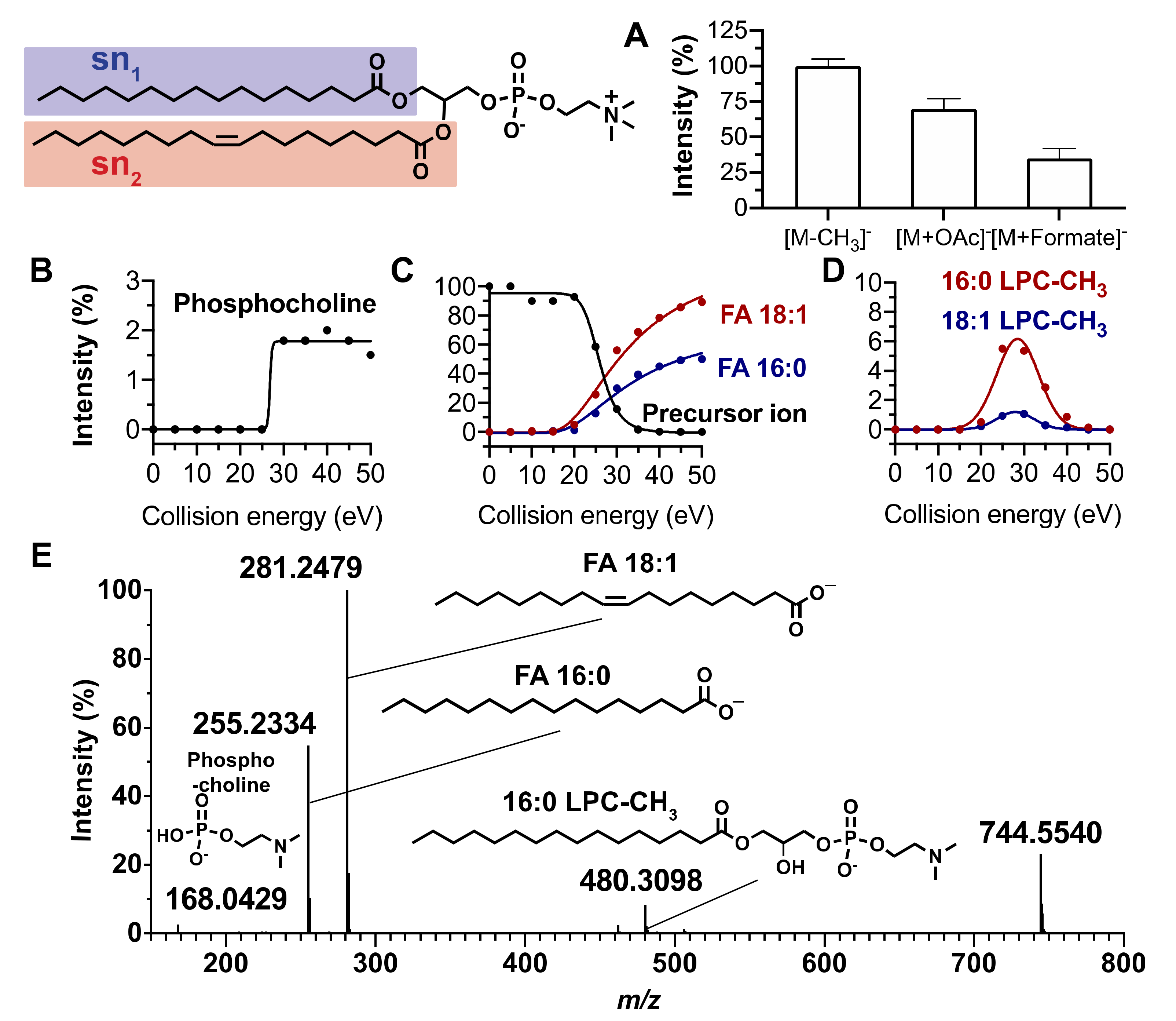
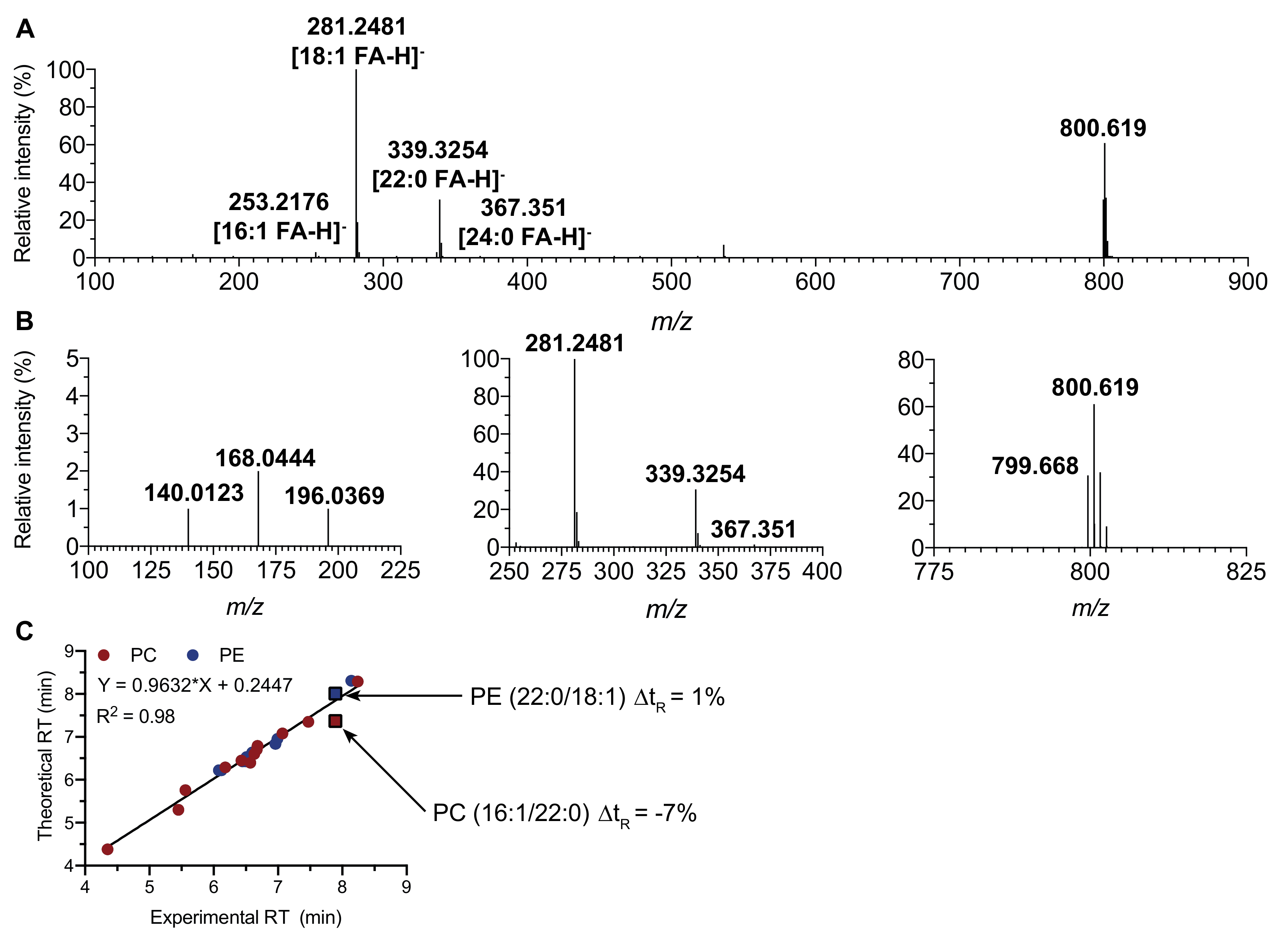
2.1. Fragmentation Patterns of Phospholipids
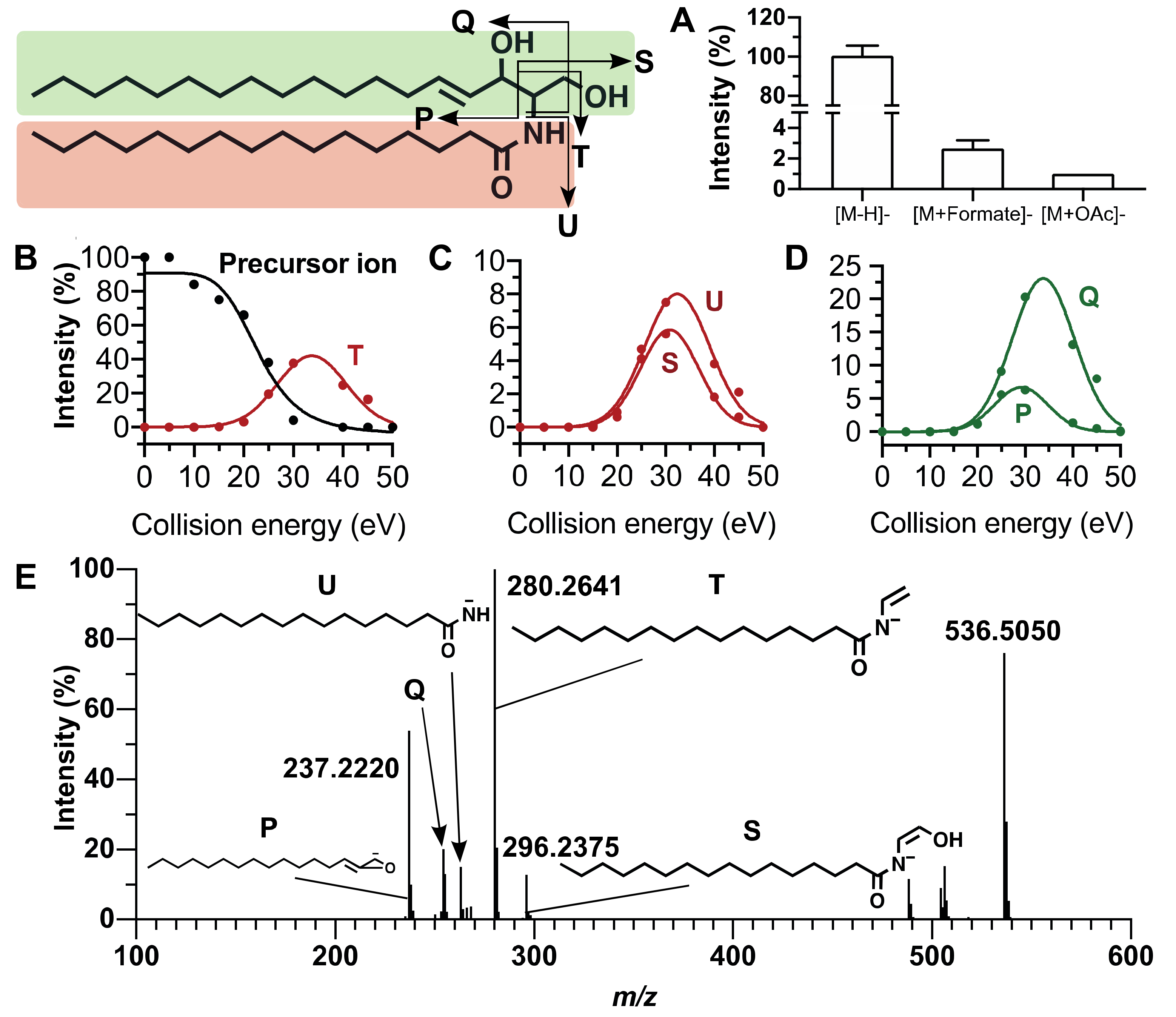
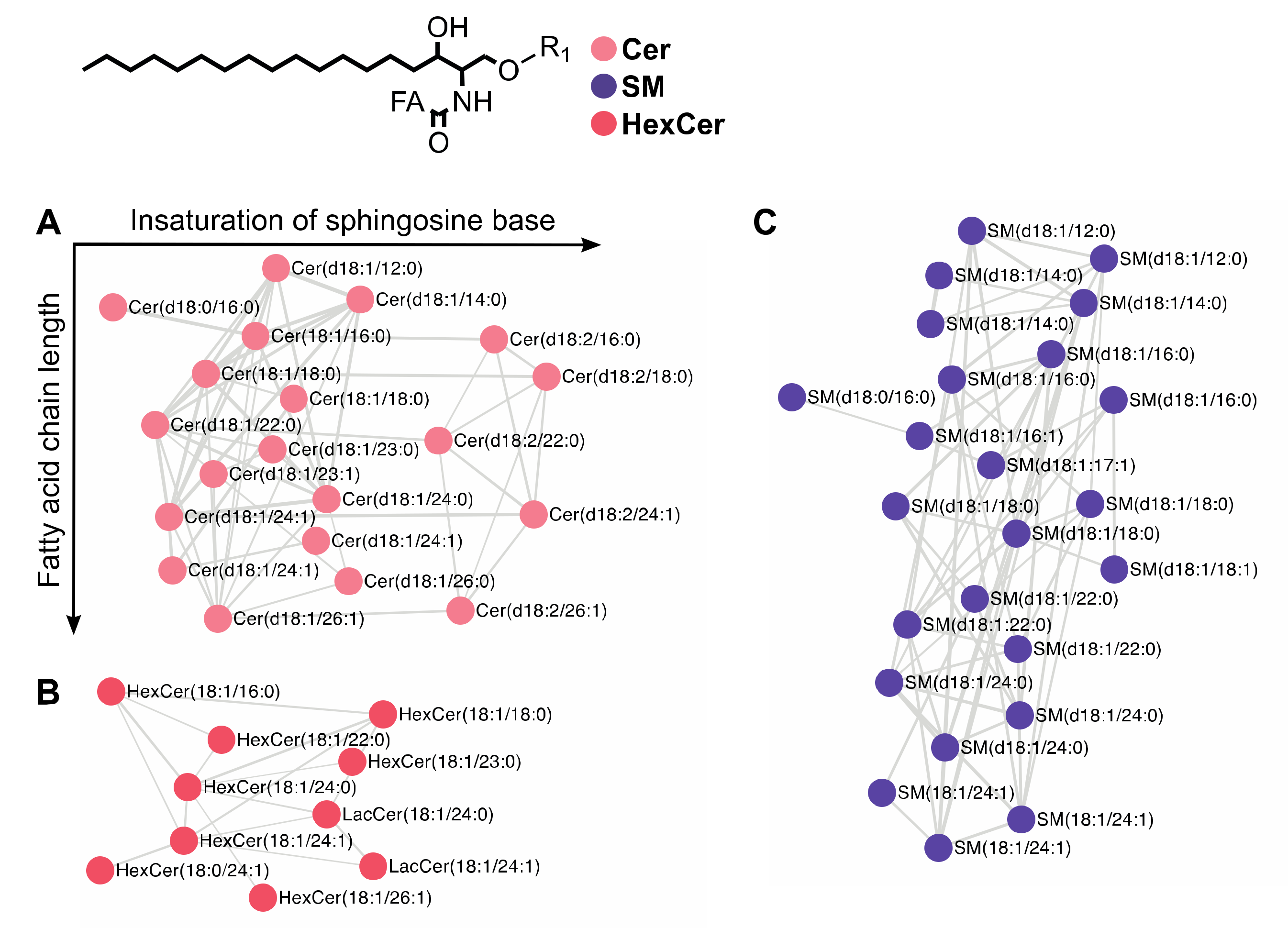
2.2. Fragmentation Patterns of Sphingolipids
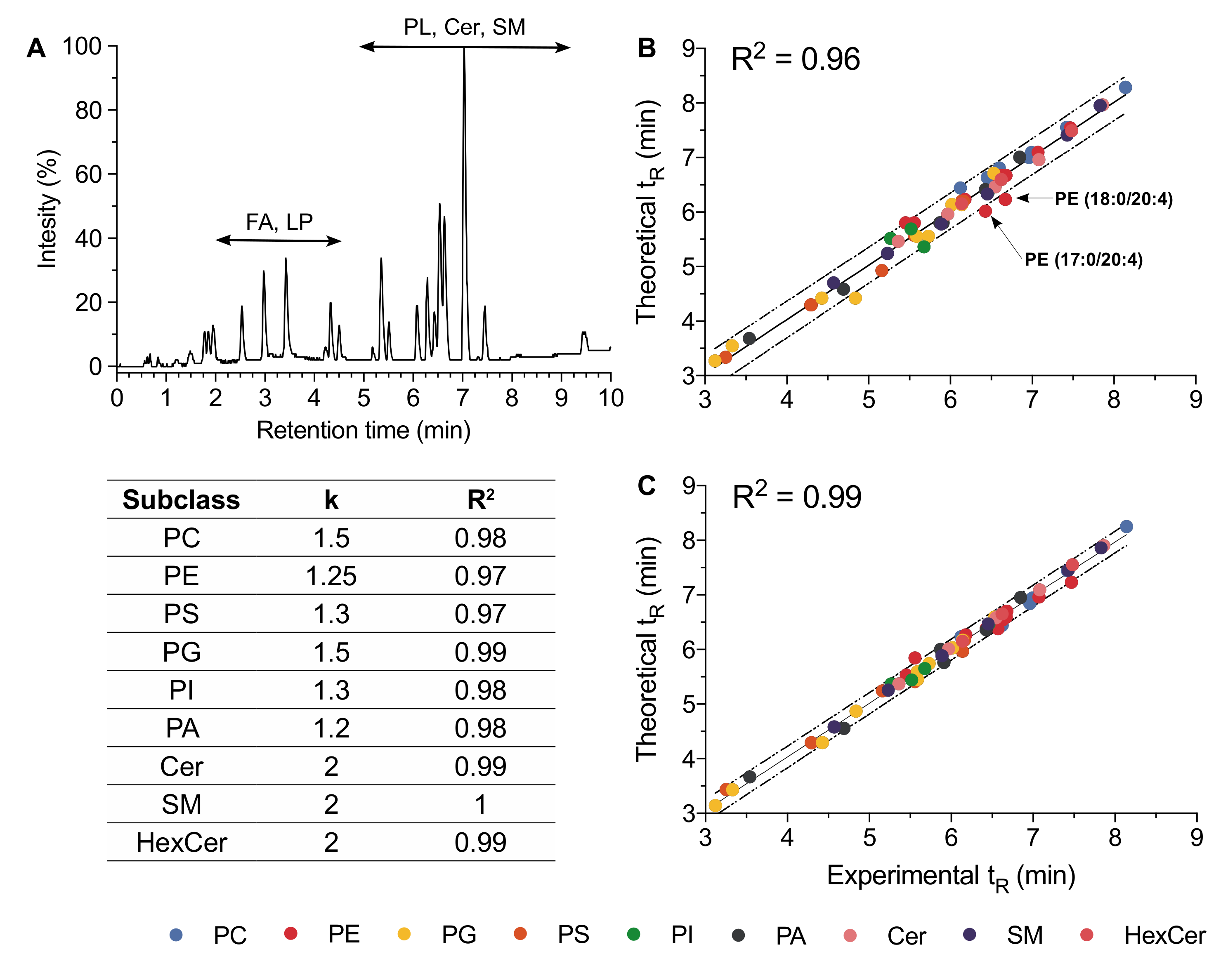
2.3. Retention Time Prediction
2.4. Instrument Stability
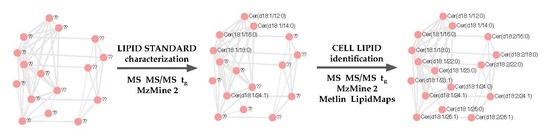
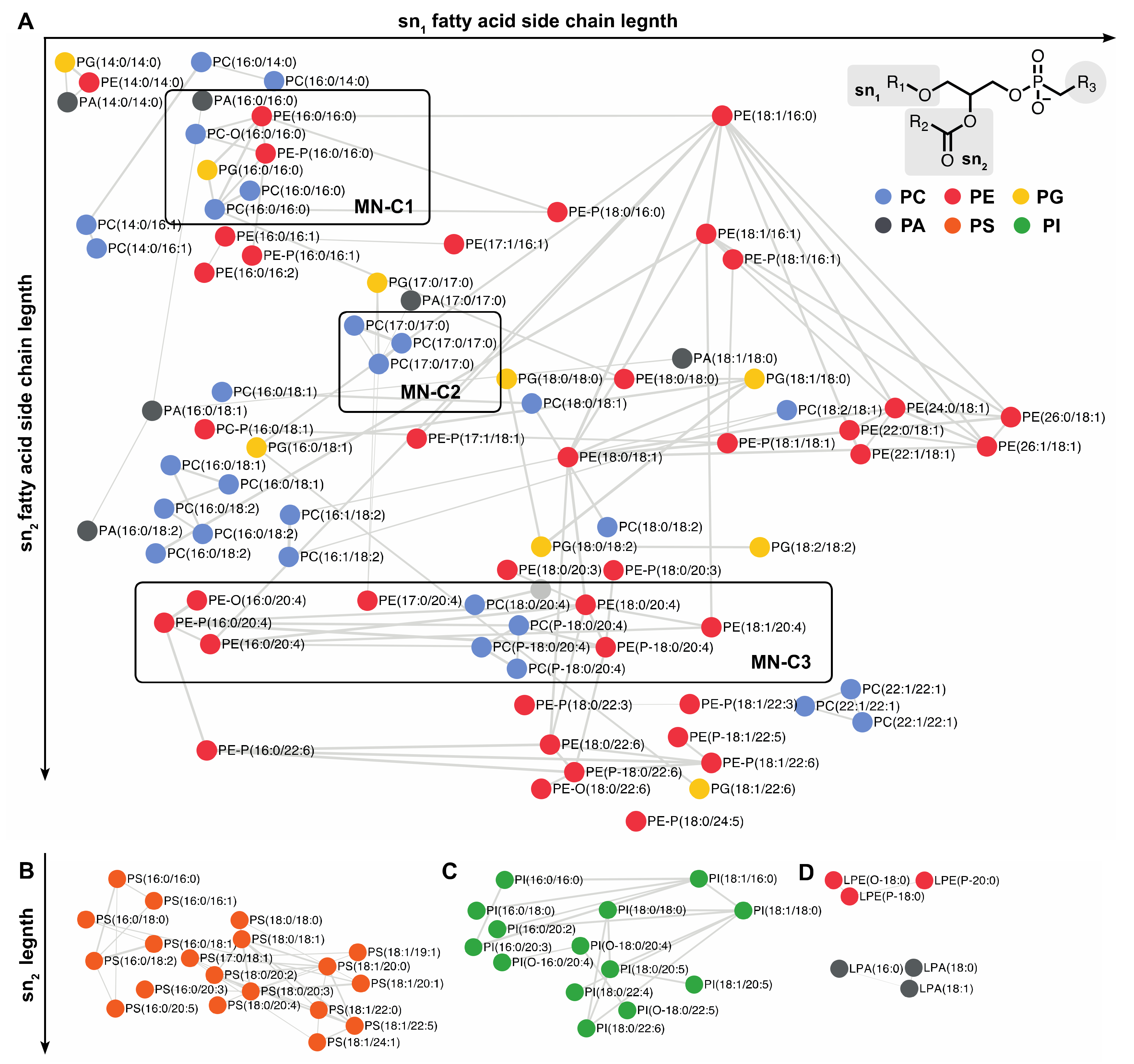
2.5. Lipidic Networking of Human Corneal Epithelial Cells
2.6. Use of Retention Time Prediction for Lipid Annotation
2.7. Use of Existing Lipid Library Database
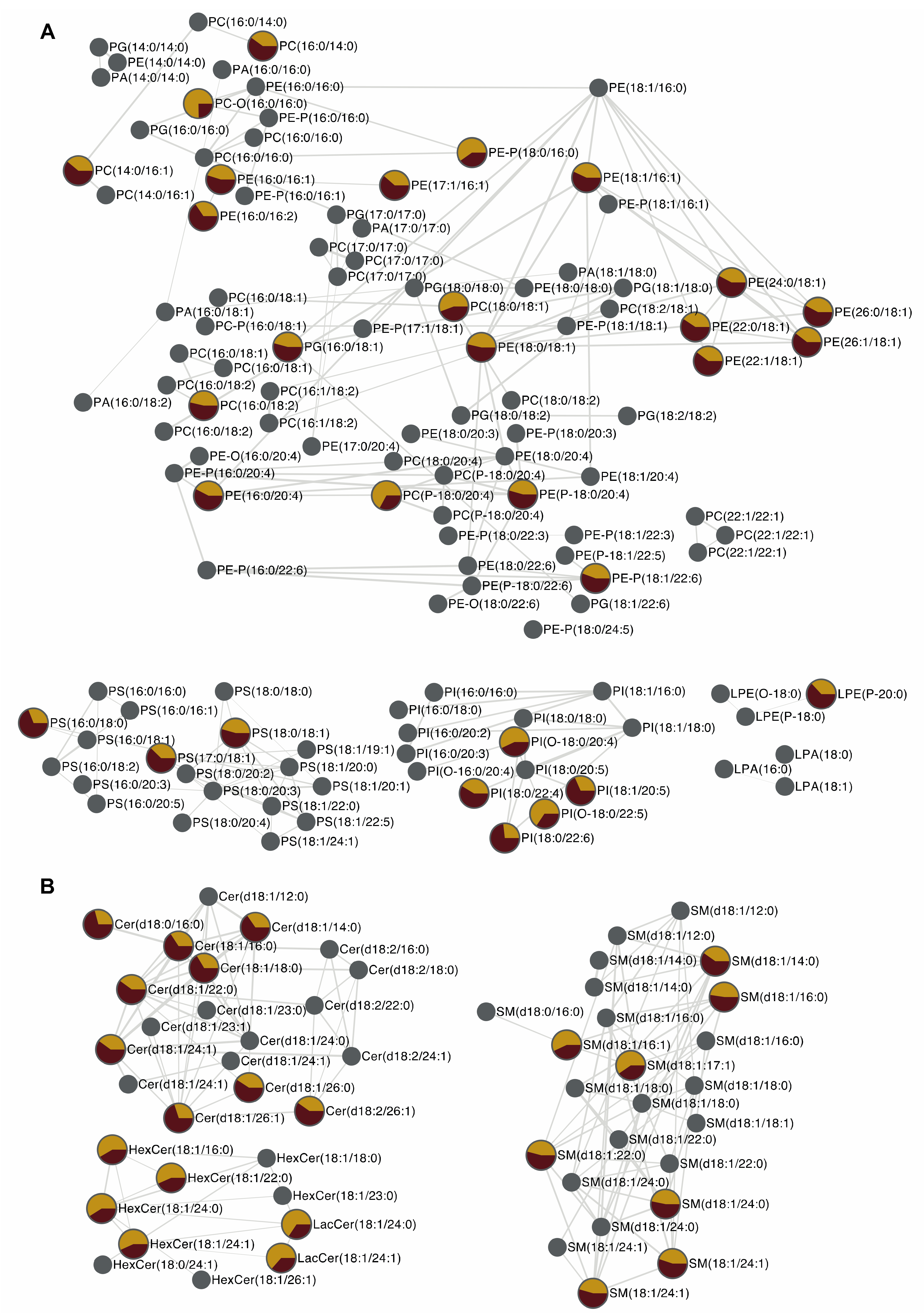
2.8. Effect of HO on HCE Cells
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Sample Preparation
3.3. Data-Dependent LC-ESI-HRMS/MS Analysis
3.4. Data-Preprocessing Parameters
3.5. Molecular Network Analysis
3.6. Lipid Structure Assignment
3.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhang, C.; Wang, K.; Yang, L.; Liu, R.; Chu, Y.; Qin, X.; Yang, P.; Yu, H. Lipid metabolism in inflammation-related diseases. Analyst 2018, 143, 4526–4536. [Google Scholar] [CrossRef] [PubMed]
- Stith, J.L.; Velazquez, F.N.; Obeid, L.M. Advances in determining signaling mechanisms of ceramide and role in disease. J. Lipid Res. 2019, 60, 913–918. [Google Scholar] [CrossRef] [PubMed]
- Meikle, P.J.; Summers, S.A. Sphingolipids and phospholipids in insulin resistance and related metabolic disorders. Nat. Rev. Endocrinol. 2017, 13, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Magtanong, L.; Ko, P.J.; Dixon, S.J. Emerging roles for lipids in non-apoptotic cell death. Cell Death Differ. 2016, 23, 1099–1109. [Google Scholar] [CrossRef]
- Agmon, E.; Stockwell, B.R. Lipid homeostasis and regulated cell death. Curr. Opin. Chem. Biol. 2017, 39, 83–89. [Google Scholar] [CrossRef]
- Serhan, C.N.; Savill, J. Resolution of inflammation: The beginning programs the end. Nat. Immunol. 2005, 6, 1191–1197. [Google Scholar] [CrossRef]
- Maceyka, M.; Spiegel, S. Sphingolipid metabolites in inflammatory disease. Nature 2014, 510, 58–67. [Google Scholar] [CrossRef]
- Holčapek, M.; Liebisch, G.; Ekroos, K. Lipidomic Analysis. Anal. Chem. 2018, 90, 4249–4257. [Google Scholar] [CrossRef]
- Ivanisevic, J.; Want, E.J. From samples to insights into metabolism: Uncovering biologically relevant information in LC- HRMS metabolomics data. Metabolites 2019, 9, 308. [Google Scholar] [CrossRef]
- Hsu, F.F. Mass spectrometry-based shotgun lipidomics – a critical review from the technical point of view. Anal. Bioanal. Chem. 2018, 410, 6387–6409. [Google Scholar] [CrossRef]
- Wang, J.; Wang, C.; Han, X. Tutorial on lipidomics. Anal. Chim. Acta 2019, 1061, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Yang, K.; Gross, R.W. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom. Rev. 2012, 31, 134–178. [Google Scholar] [PubMed]
- Züllig, T.; Trötzmüller, M.; Köfeler, H.C. Lipidomics from sample preparation to data analysis: A primer. Anal. Bioanal. Chem. 2019, 412, 2191–2209. [Google Scholar] [PubMed]
- Hinz, C.; Liggi, S.; Griffin, J.L. The potential of Ion Mobility Mass Spectrometry for high-throughput and high-resolution lipidomics. Curr. Opin. Chem. Biol. 2018, 42, 42–50. [Google Scholar]
- Leaptrot, K.L.; May, J.C.; Dodds, J.N.; McLean, J.A. Ion mobility conformational lipid atlas for high confidence lipidomics. Nat. Commun. 2019, 10, 985. [Google Scholar]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar]
- Allard, P.-M.; Genta-Jouve, G.; Wolfender, J.-L. Deep metabolome annotation in natural products research: Towards a virtuous cycle in metabolite identification. Curr. Opin. Chem. Biol. 2017, 36, 40–49. [Google Scholar]
- Allard, P.M.; Péresse, T.; Bisson, J.; Gindro, K.; Marcourt, L.; Pham, V.C.; Roussi, F.; Litaudon, M.; Wolfender, J.L. Integration of Molecular Networking and In-Silico MS/MS Fragmentation for Natural Products Dereplication. Anal. Chem. 2016, 88, 3317–3323. [Google Scholar] [CrossRef]
- Quinn, R.A.; Nothias, L.F.; Vining, O.; Meehan, M.; Esquenazi, E.; Dorrestein, P.C. Molecular Networking As a Drug Discovery, Drug Metabolism, and Precision Medicine Strategy. Trends Pharmacol. Sci. 2017, 38, 143–154. [Google Scholar] [CrossRef]
- Bauvais, C.; Bonneau, N.; Blond, A.; Pérez, T.; Bourguet-Kondracki, M.-L.; Zirah, S. Furanoterpene Diversity and Variability in the Marine Sponge Spongia officinalis, from Untargeted LC–MS/MS Metabolomic Profiling to Furanolactam Derivatives. Metabolites 2017, 7, 27. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [PubMed]
- Olivon, F.; Elie, N.; Grelier, G.; Roussi, F.; Litaudon, M.; Touboul, D. MetGem Software for the Generation of Molecular Networks Based on the t-SNE Algorithm. Anal. Chem. 2018, 90, 13900–13908. [Google Scholar] [PubMed]
- Tada, I.; Tsugawa, H.; Meister, I.; Zhang, P.; Shu, R.; Katsumi, R.; Wheelock, C.E.; Arita, M.; Chaleckis, R. Creating a Reliable Mass Spectral–Retention Time Library for All Ion Fragmentation-Based Metabolomics. Metabolites 2019, 9, 251. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Aicheler, F.; Li, J.; Hoene, M.; Lehmann, R.; Xu, G.; Kohlbacher, O. Retention Time Prediction Improves Identification in Nontargeted Lipidomics Approaches. Anal. Chem. 2015, 87, 7698–7704. [Google Scholar]
- Randazzo, G.M.; Tonoli, D.; Hambye, S.; Guillarme, D.; Jeanneret, F.; Nurisso, A.; Goracci, L.; Boccard, J.; Rudaz, S. Prediction of retention time in reversed-phase liquid chromatography as a tool for steroid identification. Anal. Chim. Acta 2016, 916, 8–16. [Google Scholar]
- Clouzeau, C.; Godefroy, D.; Riancho, L.; Rostène, W.; Baudouin, C.; Brignole-Baudouin, F. Hyperosmolarity potentiates toxic effects of benzalkonium chloride on conjunctival epithelial cells in vitro. Mol. Vis. 2012, 18, 851–863. [Google Scholar]
- Warcoin, E.; Clouzeau, C.; Roubeix, C.; Raveu, A.L.; Godefroy, D.; Riancho, L.; Baudouin, C.; Brignole-Baudouin, F. Hyperosmolarity and Benzalkonium Chloride Differently Stimulate Inflammatory Markers in Conjunctiva-Derived Epithelial Cells in vitro. Ophthalmic Res. 2017, 58, 40–48. [Google Scholar]
- Craig, J.P.; Nichols, K.K.; Akpek, E.K.; Caffery, B.; Dua, H.S.; Joo, C.; Liu, Z.; Nelson, J.D.; Nichols, J.J.; Tsubota, K.; et al. The Ocular Surface TFOS DEWS II Definition and Classification Report. Ocul. Surf. 2017, 15, 276–283. [Google Scholar]
- Pflugfelder, S.C.; de Paiva, C.S. The Pathophysiology of Dry Eye Disease: What We Know and Future Directions for Research. Ophthalmology 2017, 124, S4–S13. [Google Scholar]
- Baudouin, C.; Aragona, P.; Messmer, E.M.; Tomlinson, A.; Calonge, M.; Boboridis, K.G.; Akova, Y.A.; Geerling, G.; Labetoulle, M.; Rolando, M. Role of hyperosmolarity in the pathogenesis and management of dry eye disease: Proceedings of the ocean group meeting. Ocul. Surf. 2013, 11, 246–258. [Google Scholar] [PubMed]
- Robciuc, A.; Hyo, T. Ceramides in the Pathophysiology of the Anterior Segment of the Eye. Curr. Eye Res. 2013, 38, 1006–1016. [Google Scholar] [PubMed]
- Olivon, F.; Roussi, F.; Litaudon, M.; Touboul, D. Optimized experimental workflow for tandem mass spectrometry molecular networking in metabolomics. Anal. Bioanal. Chem. 2017, 409, 5767–5778. [Google Scholar] [CrossRef] [PubMed]
- Godzien, J.; Ciborowski, M.; Martínez-Alcázar, M.P.; Samczuk, P.; Kretowski, A.; Barbas, C. Rapid and Reliable Identification of Phospholipids for Untargeted Metabolomics with LC-ESI-QTOF-MS/MS. J. Proteome Res. 2015, 14, 3204–3216. [Google Scholar]
- Hsu, F.F.; Turk, J. Electrospray ionization with low-energy collisionally activated dissociation tandem mass spectrometry of glycerophospholipids: Mechanisms of fragmentation and structural characterization. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2009, 877, 2673–2695. [Google Scholar] [CrossRef]
- Maccarone, A.T.; Duldig, J.; Mitchell, T.W.; Blanksby, S.J.; Duchoslav, E.; Campbell, J.L. Characterization of acyl chain position in unsaturated phosphatidylcholines using differential mobility-mass spectrometry. J. Lipid Res. 2014, 55, 1668–1677. [Google Scholar] [CrossRef]
- Ann, Q.; Adams, J. Structure-Specific Collision-Induced Fragmentations of Ceramides Cationized with Alkali-Metal Ions. Anal. Chem. 1993, 65, 7–13. [Google Scholar] [CrossRef]
- Lanzini, J.; Dargère, D.; Regazzetti, A.; Tebani, A.; Laprévote, O.; Auzeil, N. Changing in lipid profile induced by the mutation of Foxn1 gene: A lipidomic analysis of Nude mice skin. Biochimie 2015, 118, 234–243. [Google Scholar]
- Petit, J.; Wakx, A.; Gil, S.; Fournier, T.; Auzeil, N.; Rat, P.; Laprévote, O. Lipidome-wide disturbances of human placental JEG-3 cells by the presence of MEHP. Biochimie 2018, 149, 1–8. [Google Scholar] [CrossRef]
- Ovčačíková, M.; Lísa, M.; Cífková, E.; Holčapek, M. Retention behavior of lipids in reversed-phase ultrahigh-performance liquid chromatography-electrospray ionization mass spectrometry. J. Chromatogr. A 2016, 1450, 76–85. [Google Scholar] [CrossRef]
- Kind, T.; Liu, K.H.; Lee, D.Y.; Defelice, B.; Meissen, J.K.; Fiehn, O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods 2013, 10, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Cajka, T.; Fiehn, O. LC–MS-based lipidomics and automated identification of lipids using the LipidBlast in-silico MS/MS library. Methods Mol. Biol. 2017, 1609, 149–170. [Google Scholar] [PubMed]
- Hutchins, P.D.; Russell, J.D.; Coon, J.J. LipiDex: An Integrated Software Package for High-Confidence Lipid Identification. Cell Syst. 2018, 6, 621–625.e5. [Google Scholar] [CrossRef] [PubMed]
- Bron, A.J.; de Paiva, C.S.; Chauhan, S.K.; Bonini, S.; Gabison, E.E.; Jain, S.; Knop, E.; Markoulli, M.; Ogawa, Y.; Perez, V.; et al. TFOS DEWS II pathophysiology report. Ocul. Surf. 2017, 15, 438–510. [Google Scholar] [CrossRef]
- Baudouin, C.; Messmer, E.M.; Aragona, P.; Geerling, G.; Akova, Y.A.; Benítez-Del-Castillo, J.; Boboridis, K.G.; Merayo-Lloves, J.; Rolando, M.; Labetoulle, M. Revisiting the vicious circle of dry eye disease: A focus on the pathophysiology of meibomian gland dysfunction. Br. J. Ophthalmol. 2016, 100, 300–306. [Google Scholar] [CrossRef]
- Baudouin, C.; Irkeç, M.; Messmer, E.M.; Benítez-del-Castillo, J.M.; Bonini, S.; Figueiredo, F.C.; Geerling, G.; Labetoulle, M.; Lemp, M.; Rolando, M.; et al. Clinical impact of inflammation in dry eye disease: Proceedings of the ODISSEY group meeting. Acta Ophthalmol. 2018, 96, 111–119. [Google Scholar] [CrossRef]
- Robciuc, A.; Hyötyläinen, T.; Jauhiainen, M.; Holopainen, J.M. Hyperosmolarity-induced lipid droplet formation depends on ceramide production by neutral sphingomyelinase 2. J. Lipid Res. 2012, 53, 2286–2295. [Google Scholar] [CrossRef]
- Warcoin, E.; Clouzeau, C.; Brignole-Baudouin, F.; Baudouin, C. Hyperosmolarité: Effets intracellulaires et implication dans la sécheresse oculaire. J. Fr. Ophtalmol. 2016, 39, 641–651. [Google Scholar] [CrossRef]
- Warcoin, E.; Baudouin, C.; Gard, C.; Brignole-Baudouin, F. In Vitro Inhibition of NFAT5-Mediated Induction of CCL2 in Hyperosmotic Conditions by Cyclosporine and Dexamethasone on Human HeLa-Modified Conjunctiva-Derived Cells. PLoS ONE 2016, 11, e0159983. [Google Scholar] [CrossRef]
- Cavet, M.E.; Harrington, K.L.; Vollmer, T.R.; Ward, K.W.; Zhang, J.-Z. Anti-inflammatory and anti-oxidative effects of the green tea polyphenol epigallocatechin gallate in human corneal epithelial cells. Mol. Vis. 2011, 17, 533–542. [Google Scholar]
- Ren, Y.; Lu, H.; Reinach, P.S.; Zheng, Q.; Li, J.; Tan, Q.; Zhu, H.; Chen, W. Hyperosmolarity-induced AQP5 upregulation promotes inflammation and cell death via JNK1/2 Activation in human corneal epithelial cells. Sci. Rep. 2017, 7, 4727. [Google Scholar] [PubMed]
- Hua, X.; Deng, R.; Li, J.; Chi, W.; Su, Z.; Lin, J.; Pflugfelder, S.; Li, D. Protective Effects of L-Carnitine against Oxidative Injury by Hyperosmolarity in Human Corneal Epithelial Cells. Investig. Ophthalmol. Vis. Sci. 2015, 56, 5503–5511. [Google Scholar] [CrossRef]
- Deng, R.; Hua, X.; Li, J.; Chi, W.; Zhang, Z.; Lu, F.; Zhang, L.; Pflugfelder, S.C.; Li, D.-Q. Oxidative stress markers induced by hyperosmolarity in primary human corneal epithelial cells. PLoS ONE 2015, 10, e0126561. [Google Scholar] [CrossRef]
- Wang, P.; Sheng, M.; Li, B.; Jiang, Y.; Chen, Y. High osmotic pressure increases reactive oxygen species generation in rabbit corneal epithelial cells by endoplasmic reticulum. Am. J. Transl. Res. 2016, 8, 860. [Google Scholar]
- Braverman, N.E.; Moser, A.B. Functions of plasmalogen lipids in health and disease. Biochim. Biophys. Acta Mol. Basis Dis. 2012, 1822, 1442–1452. [Google Scholar]
- Dogru, M.; Kojima, T.; Simsek, C.; Tsubotav, K. Potential role of oxidative stress in ocular surface inflammation and dry eye disease. Investig. Ophthalmol. Vis. Sci. 2018, 59, DES163–DES168. [Google Scholar]
- Choi, W.; Lian, C.; Ying, L.; Kim, G.E.; You, I.C.; Park, S.H.; Yoon, K.C. Expression of Lipid Peroxidation Markers in the Tear Film and Ocular Surface of Patients with Non-Sjogren Syndrome: Potential Biomarkers for Dry Eye Disease. Curr. Eye Res. 2016, 41, 1143–1149. [Google Scholar]
- Chaurasia, B.; Summers, S.A. Ceramides—Lipotoxic Inducers of Metabolic Disorders. Trends Endocrinol. Metab. 2015, 26, 538–550. [Google Scholar] [CrossRef]
- Magny, R.; Kessal, K.; Regazzetti, A.; Yedder, A. Ben; Baudouin, C.; Parsadaniantz, S.M.; Brignole-Baudouin, F.; Laprévote, O.; Auzeil, N. Lipidomic analysis of epithelial corneal cells following hyperosmolarity and benzalkonium chloride exposure: New insights in dry eye disease. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids 2020, 158728. [Google Scholar]
- Myers, O.D.; Sumner, S.J.; Li, S.; Barnes, S.; Du, X. One Step Forward for Reducing False Positive and False Negative Compound Identifications from Mass Spectrometry Metabolomics Data: New Algorithms for Constructing Extracted Ion Chromatograms and Detecting Chromatographic Peaks. Anal. Chem. 2017, 89, 8696–8703. [Google Scholar]
- GNPS Home Page. Available online: http://gnps.ucsd.edu (accessed on 26 May 2020).
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magny, R.; Regazzetti, A.; Kessal, K.; Genta-Jouve, G.; Baudouin, C.; Mélik-Parsadaniantz, S.; Brignole-Baudouin, F.; Laprévote, O.; Auzeil, N. Lipid Annotation by Combination of UHPLC-HRMS (MS), Molecular Networking, and Retention Time Prediction: Application to a Lipidomic Study of In Vitro Models of Dry Eye Disease. Metabolites 2020, 10, 225. https://doi.org/10.3390/metabo10060225
Magny R, Regazzetti A, Kessal K, Genta-Jouve G, Baudouin C, Mélik-Parsadaniantz S, Brignole-Baudouin F, Laprévote O, Auzeil N. Lipid Annotation by Combination of UHPLC-HRMS (MS), Molecular Networking, and Retention Time Prediction: Application to a Lipidomic Study of In Vitro Models of Dry Eye Disease. Metabolites. 2020; 10(6):225. https://doi.org/10.3390/metabo10060225
Chicago/Turabian StyleMagny, Romain, Anne Regazzetti, Karima Kessal, Gregory Genta-Jouve, Christophe Baudouin, Stéphane Mélik-Parsadaniantz, Françoise Brignole-Baudouin, Olivier Laprévote, and Nicolas Auzeil. 2020. "Lipid Annotation by Combination of UHPLC-HRMS (MS), Molecular Networking, and Retention Time Prediction: Application to a Lipidomic Study of In Vitro Models of Dry Eye Disease" Metabolites 10, no. 6: 225. https://doi.org/10.3390/metabo10060225
APA StyleMagny, R., Regazzetti, A., Kessal, K., Genta-Jouve, G., Baudouin, C., Mélik-Parsadaniantz, S., Brignole-Baudouin, F., Laprévote, O., & Auzeil, N. (2020). Lipid Annotation by Combination of UHPLC-HRMS (MS), Molecular Networking, and Retention Time Prediction: Application to a Lipidomic Study of In Vitro Models of Dry Eye Disease. Metabolites, 10(6), 225. https://doi.org/10.3390/metabo10060225