Allelopathic Potential of Rice and Identification of Published Allelochemicals by Cloud-Based Metabolomics Platform
Abstract
1. Introduction
2. Results and Discussion
2.1. Allelopathy of Rice on Barnyardgrass
2.2. Identification of Previously-Published Allelochemicals
2.3. Confirmation of Allelochemicals in OM 5930 Rice Cultivar
3. Materials and Methods
3.1. Extraction and Allelopathy Bioassays
3.1.1. Plant Material
3.1.2. Extraction of Nine OM Rice Cultivars by Methanol (MeOH) Separation Method
3.1.3. Bioassays
3.1.4. Bioassay Data Analysis
3.2. Identification of Previously Published Allelochemicals Using Cloud-Based Metabolomics Platform
3.2.1. Rice Materials
3.2.2. UHPLC/MS Analytical Method
3.2.3. Data Analysis Platform
3.2.4. Statistical Analysis
3.3. Confirmation of Allelochemicals in OM 5930 Rice Cultivar
3.3.1. Sample Preparation
3.3.2. Chromatographic Conditions of HPLC/UV-Vis and Its Optimization
3.3.3. Molecular Bioassay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cosslett, T.; Cosslett, P. Rice Trade of the Mainland Southeast Asian Countries: Cambodia, Laos, Thailand, and Vietnam. In Sustainable Development of Rice and Water Resources in Mainland Southeast Asia and Mekong River Basin; Springer: Singapore, 2018; pp. 55–83. ISBN 978-981-10-5613-0. [Google Scholar]
- Khai, N.; Tinh, T.; Tin, H.; NV, S. Reducing Greenhouse Gas Emissions in Rice Grown in the Mekong Delta of Vietnam. Environ. Pollut. Clim. Chang. 2018, 2, 158. [Google Scholar] [CrossRef]
- Chauhan, B. Strategies to manage weedy rice in Asia. Crop. Prot. 2013, 48, 51–56. [Google Scholar] [CrossRef]
- Smith, R. Weed Thresholds in Southern U.S. rice, Oryza sativa. Weed. Technol. 1988, 2, 232–241. [Google Scholar] [CrossRef]
- Chin, D. Biology and management of Barnyardgrass, red Sprangletop and weedy rice. Weed Biol. Manag. 2001, 1, 37–41. [Google Scholar] [CrossRef]
- Rimando, A.; Olofsdotter, M.; Dayan, F.; Duke, S. Searching for Rice Allelochemicals. Agron. J. 2001, 93, 16–20. [Google Scholar] [CrossRef]
- Khanh, T.; Xuan, T.; Chin, D.; Chung, I.; Elzaawely, A.; Tawata, S. Current status of biological control of paddy weeds in Vietnam. Weed Biol. Manag. 2006, 6, 1–9. [Google Scholar] [CrossRef]
- Khanh, T.; Xuan, T.; Chung, I. Rice allelopathy and the possibility for weed management. Ann. Appl. Biol. 2007, 151, 325–339. [Google Scholar] [CrossRef]
- Amb, M.; Ahluwalia, A. Allelopathy: Potential Role to Achieve New Milestones in Rice Cultivation. Rice Sci. 2016, 23, 165–183. [Google Scholar] [CrossRef]
- Thi, H.; Lin, C.; Smeda, R.; Fritschi, F. Isolation and purification of growth-inhibitors from Vietnamese rice cultivars: Growth-inhibitors from Vietnamese rice cultivars. Weed Biol. Manag. 2014, 14, 221–231. [Google Scholar] [CrossRef]
- Thi, H.; Lin, C.; Smeda, R.; Leigh, N.; Wycoff, W.; Fritschi, F. Isolation and identification of an allelopathic phenylethylamine in rice. Phytochemistry 2014, 108, 109–121. [Google Scholar] [CrossRef]
- Thi, H.L.; Zhou, H.; Lin, C.-H.; Liu, S.; Berezin, M.Y.; Smeda, R.J.; Fritschi, F.B. Synthesis and plant growth inhibitory activity of N-trans-cinnamoyltyramine: Its possible inhibition mechanisms and biosynthesis pathway. J. Plant Interact. 2017, 12, 51–57. [Google Scholar] [CrossRef]
- Olofsdotter, M. Allelopathy in Rice; International Rice Research Institute: Los Baños, Philippines, 1998; ISBN 978-971-22-0101-1. [Google Scholar]
- Salam, M.; Kato-Noguchi, H. Screening of Allelopathic Potential Bangladesh Rice Cultivars by Donor-Receiver Bioassay. Asian J. Plant Sci. 2009, 8, 761–795. [Google Scholar] [CrossRef][Green Version]
- Olofsdotter, M.; Rebulanan, M.; Madrid, A.; Dali, W.; Navarez, D.; Olf, D.C. Why phenolic acids are unlikely primary allelochemicals in rice. J. Chem. Ecol. 2002, 28, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Moore, W.; Meyers, D.A.; Wenzel, S.; Teague, W.; Li, H.; Li, X.; D’Agostino, R.; Castro, M.; Curran-Everett, D.; Fitzpatrick, A.M.; et al. Identification of asthma phenotypes using cluster analysis in the severe asthma research program. Am. J. Respir. Crit. Care Med. 2010, 181, 315–323. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Shindo, H.; Ohta, S.; Kuwatsuka, S. Behavior of phenolic substances in the decaying process of plants. Soil Sci. Plant Nutr. 1978, 24, 233–243. [Google Scholar] [CrossRef]
- Macías, F.; Chinchilla, N.; Varela, R.; Molinillo, J. Bioactive steroids from Oryza sativa L. Steroids 2006, 71, 603–608. [Google Scholar] [CrossRef]
- Bouillant, M.; Jacoud, C.; Zanella, I.; Favre-Bonvin, J.; Bally, R. Identification of 5-(12-heptadecenyl)-resorcinol in rice root exudates. Phytochemistry 1994, 35, 768–771. [Google Scholar] [CrossRef]
- Seal, A.; Haig, T.; Pratley, J. Evaluation of putative allelochemicals in rice root exudates for their role in the suppression of arrowhead root growth. J. Chem. Ecol. 2004, 30, 1663–1678. [Google Scholar] [CrossRef]
- Chou, C.; Lin, H. Autointoxication mechanism of Oryza sativa I. Phytotoxic effects of decomposing rice residues in soil. J. Chem. Ecol. 1976, 2, 353–367. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Q.; Ruan, X.; Pan, C.; Jiang, D. Phenolics and plant allelopathy. Molecules 2010, 15, 8933–8952. [Google Scholar] [CrossRef]
- Chou, C.; Chang, F.; HI, O. Allelopathic potentials of a wild rice, Oryza Perennis. Taiwania 1991, 36, 201–210. [Google Scholar]
- Chung, I.; Kim, K.; Ahn, J.; Chun, S.; Kim, C.; Kim, J.; Kim, S. Screening of allelochemicals on barnyardgrass (Echinochloa crus-galli) and identification of potentially allelopathic compounds from rice (Oryza sativa) variety hull extracts. Crop. Prot. 2002, 21, 913–920. [Google Scholar] [CrossRef]
- Mattice, J.; Lavy, T.; Skulman, B.; Dilday, R. Searching for allelochemicals in rice that control ducksalad. In Allelopathy in Rice; International Rice Research Institute: Los Baños, Philippines, 1998; pp. 81–98. [Google Scholar]
- Kato-Noguchi, H. Allelochemicals released from rice plants. Jpn. J. Plant Sci. 2008, 2, 18–25. [Google Scholar]
- Chou, C.; Patrick, Z. Identification and phytotoxic activity of compounds produced during decomposition of corn and rye residues in soil. J. Chem. Ecol. 1976, 2, 369–387. [Google Scholar] [CrossRef]
- Fernandez, C.; Lelong, B.; Vila, B.; Mévy, J.; Robles, C.; Greff, S.; Dupouyet, S.; Bousquet-Mélou, A. Potential allelopathic effect of Pinus halepensis in the secondary succession: An experimental approach. Chemoecology 2006, 16, 97–105. [Google Scholar] [CrossRef]
- Zhou, B.; Kong, C.; Li, Y.; Wang, P.; Xu, X. Crabgrass (Digitaria sanguinalis) allelochemicals that interfere with crop growth and the soil microbial community. J. Agric. Food Chem. 2013, 61, 5310–5317. [Google Scholar] [CrossRef]
- Dornbos, D.; Spencer, G. Natural products phytotoxicity A bioassay suitable for small quantities of slightly water-soluble compounds. J. Chem. Ecol. 1990, 16, 339–352. [Google Scholar] [CrossRef]
- Yamamoto, Y. Movement of allelopathic compound coumarin from plant residue of sweet vernalgrass (Anthoxanthum odoratum L.) to soil. Grassl. Sci. 2009, 55, 36–40. [Google Scholar] [CrossRef]
- Chon, S.; Kim, Y.; Lee, J. Herbicidal potential and quantification of causative allelochemicals from several compositae weeds. Weed Res. 2003, 43, 444–450. [Google Scholar] [CrossRef]
- Chon, S.; Kim, Y. Herbicidal potential and quantification of suspected allelochemicals from four grass crop extracts. J. Agron. Crop. Sci. 2004, 190, 145–150. [Google Scholar] [CrossRef]
- Takemura, T.; Kamo, T.; Sakuno, E.; Hiradate, S. Discovery of coumarin as the predominant allelochemical in Guricidia sepium. J. Trop. Sci. 2013, 25, 268–272. [Google Scholar]
- Choi, G.-H.; Ro, J.-H.; Park, B.-J.; Lee, D.-Y.; Cheong, M.-S.; Lee, D.-Y.; Seo, W.-D.; Kim, J.H. Benzaldehyde as a new class plant growth regulator on Brassica campestris. J. Appl. Biol. Chem. 2016, 59, 159–164. [Google Scholar] [CrossRef]
- Wang, P.; Kong, C.-H.; Hu, F.; Xu, X. Allantoin involved in species interactions with rice and other organisms in paddy soil. Plant Soil 2007, 296, 43–51. [Google Scholar] [CrossRef]
- Hegab, M.; Khodary, S.; Hammouda, O.; Abdelgawad, H. Autotoxicity of chard and its allelopathic potentiality on germination and some metabolic activities associated with growth of wheat seedlings. Afr. J. Biotechnol. 2008, 7, 884–892. [Google Scholar]
- Yu, J.; Matsui, Y. Effects of root exudates of cucumber (Cucumis sativus) and allelochemicals on ion uptake by cucumber seedlings. J. Chem. Ecol. 1997, 23, 817–827. [Google Scholar] [CrossRef]
- Einhellig, F.; Rasmussen, J. Effects of three phenolic acids on chlorophyll content and growth of soybean and grain sorghum seedlings. J. Chem. Ecol. 1979, 5, 815–824. [Google Scholar] [CrossRef]
- Reigosa, M.; Sánchez-Moreiras, A.; González, L. Ecophysiological approach in allelopathy. Crit. Rev. Plant Sci. 1999, 26, 577–608. [Google Scholar] [CrossRef]
- Duke, S.I. Ecophysiological aspects of allelopathy. Planta 2003, 217, 529–539. [Google Scholar] [CrossRef]
- Weir, T.; Park, S.; Vivanco, J. Biochemical and physiological mechanisms mediated by allelochemicals. Curr. Opin. Plant Biol. 2004, 7, 472–479. [Google Scholar] [CrossRef]
- Wagner, A.; Donaldson, L.; Ralph, J. Chapter 2—Lignification and lignin manipulations in conifers. In Advances in Botanical Research; Jouanin, L., Lapierre, C., Eds.; Academic Press: Cambridge, MA, USA, 2012; Volume 61, pp. 37–76. [Google Scholar]
- Baleroni, C.; Ferrarese, M.; Souza, N.; Ferrarese-Filho, O. Lipid accumulation during canola seed germination in response to cinnamic acid derivatives. Biol. Plant 2000, 43, 313–316. [Google Scholar] [CrossRef]
- Ding, J.; Sun, Y.; Xiao, C.; Shi, K.; Zhou, Y.; Yu, J. Physiological basis of different allelopathic reactions of cucumber and figleaf gourd plants to cinnamic acid. J. Exp. Bot. 2007, 58, 3765–73. [Google Scholar] [CrossRef]
- Politycka, B.; Mielcarz, B. Involvement of ethylene in growth inhibition of cucumber roots by ferulic and p-coumaric acids. Allelopath. J. 2007, 19, 451–460. [Google Scholar]
- Roger, M.; Malvido-Pazos, E. Phytotoxic effects of 21 plant secondary metabolites on arabidopsis thaliana germination and root growth. J. Chem. Ecol. 2007, 33, 1456–1466. [Google Scholar] [CrossRef]
- Salvador, A.; Cavaleiro, A.; Sousa, D.; Alves, M.; Pereira, M. Endurance of methanogenic Archaea in anaerobic bioreactors treating oleate-based wastewater. Appl. Microbiol. Biotechnol. 2013, 97, 2211–2218. [Google Scholar] [CrossRef][Green Version]
- Bacilio-Jimenez, M.; Aguilar-Flores, S.; Ventura-Zapata, E.; Perez, E.; Bouquelet, S.; Zenteno, E. Chemical characterization of root exudates from rice (Oryza sativa) and their effects on the chemotactic response of endophytic bacteria. Plant Soil 2003, 249, 7. [Google Scholar] [CrossRef]
- Kato-Noguchi, H.; Ino, T.; Sata, N.; Yamamura, S. Isolation and identification of a potent allelopathic substance in rice root exudates. Physiol. Plant. 2002, 115, 401–405. [Google Scholar] [CrossRef]
- Kato-Noguchi, H.; Ino, T. Rice seedlings release momilactone B into the environment. Phytochemistry 2003, 63, 551–554. [Google Scholar] [CrossRef]
- Kong, J.; Liang, N.H.; Xu, F.K.; Hu, F.; Wang, P.; Jiang, Y. Release and Activity of Allelochemicals from Allelopathic Rice Seedlings. J. Agric. Food Chem. 2004, 52, 2861–2865. [Google Scholar] [CrossRef]




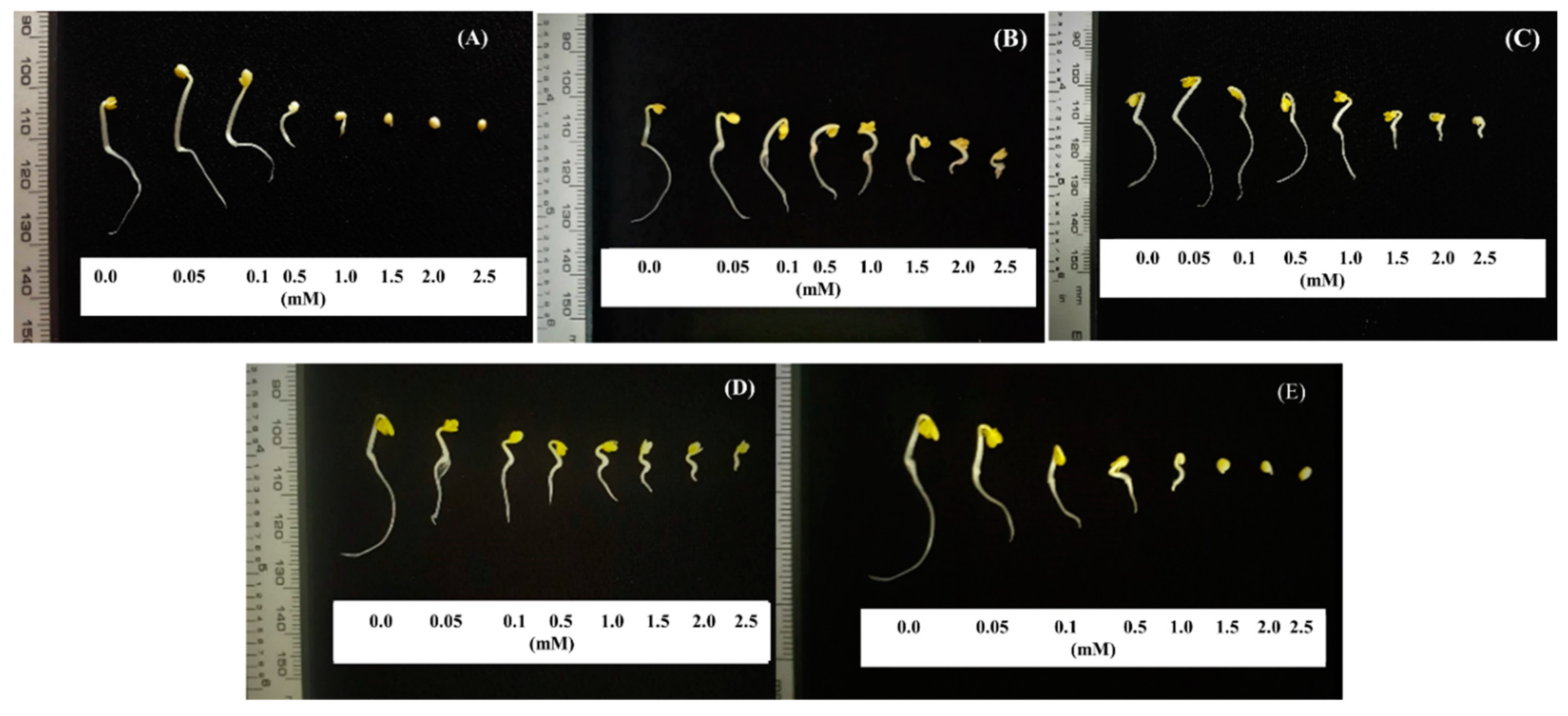
| Concentrationg mL−1 | Inhibition (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| OM 2395 | OM 3536 | OM 4498 | OM 5451 | OM 5930 | OM 6976 | OM 7347 | OM 380 | OM N406 | |
| Control | 0.00b ± 0.73 | 0.00b ± 0.50 | 0.00a ± 0.40 | 0.00a ± 0.60 | 0.00a ± 0.20 | 0.00a ± 0.40 | 0.00a ± 0.30 | 0.00a ± 0.28 | 0.00a ± 0.70 |
| 0.01 | −4.50a ± 0.85 | −4.23a ± 0.62 | 0.16a ± 0.67 | 9.54b ± 0.67 | 5.60b ± 0.51 | 7.42b ± 0.46 | 2.44a ± 0.48 | 12.2ab ± 0.55 | 0.96a ± 0.54 |
| 0.03 | 0.29b ± 0.78 | 2.45b ± 0.77 | 6.36b ± 0.70 | 12.30c ± 0.56 | 12.42c ± 0.50 | 19.17c ± 0.56 | 8.99b ± 0.31 | 17.3b ± 0.63 | 15.85b ± 1.33 |
| 0.1 | 21.00c ± 0.79 | 22.14c ± 1.02 | 8.65b ± 0.95 | 17.13d ± 0.59 | 20.83d ± 0.83 | 24.66d ± 0.73 | 10.42b ± 0.43 | 18.2b ± 0.44 | 18.98c ± 0.73 |
| 0.3 | 57.11d ± 0.79 | 38.36d ± 1.21 | 32.98c ± 1.02 | 18.71d ± 0.65 | 57.39e ± 0.40 | 27.98e ± 0.52 | 12.13c ± 0.50 | 38.5c ± 0.81 | 25.15d ± 0.49 |
| 0.5 | 64.58e ± 0.27 | 55.01e ± 0.90 | 54.01d ± 1.05 | 26.03e ± 0.49 | 64.96f ± 0.53 | 50.58f ± 0.57 | 23.08d ± 0.57 | 45.3c ± 0.56 | 37.59e ± 0.56 |
| 1.0 | 76.22f ± 0.42 | 61.02f ± 1.16 | 90.75e ± 0.15 | 72.12f ± 0.37 | 98.77g ± 0.09 | 87.17g ± 0.23 | 30.77e ± 0.54 | 60.3d ± 0.13 | 54.83f ± 1.05 |
| F | ** | ** | ** | ** | ** | ** | ** | ** | ** |
| C.V. (%) | 1.73 | 1.92 | 1.95 | 1.86 | 0.82 | 1.86 | 1.06 | 0.79 | 0.81 |
| Concentrationg mL−1 | Inhibition (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| OM 2395 | OM 3536 | OM 4498 | OM 5451 | OM 5930 | OM 6976 | OM 7347 | OM 380 | OM N406 | |
| Control | 0.00a ± 0.79 | 0.00b ± 0.80 | 0.00a ± 0.80 | 0.00a ± 0.60 | 0.00a ± 0.40 | 0.00b ± 0.80 | 0.00a ± 0.70 | 0.00a ± 0.93 | 0.00a ± 0.80 |
| 0.01 | 21.27b ± 0.63 | −10.44a ± 0.83 | 6.74b ± 1.00 | 0.58a ± 0.67 | 11.76b ± 0.83 | −19.62a ± 0.48 | 12.70b ± 0.68 | 12.8b ± 0.81 | 4.63b ± 0.75 |
| 0.03 | 30.48c ± 0.48 | 1.50b ± 0.85 | 22.63c ± 0.82 | 16.26b ± 0.81 | 29.29c ± 0.90 | −17.93a ± 0.54 | 28.24c ± 0.63 | 28.5bc ± 0.11 | 12.20c ± 0.63 |
| 0.1 | 32.46d ± 0.60 | 27.70c ± 1.36 | 44.05d ± 0.85 | 22.87c ± 0.60 | 49.71d ± 1.1 | 16.52c ± 0.89 | 30.64d ± 0.64 | 38.1c ± 0.96 | 18.74d ± 1.11 |
| 0.3 | 46.09e ± 0.84 | 46.67d ± 0.94 | 74.58e ± 0.86 | 42.48d ± 0.49 | 66.93e ± 0.80 | 24.13d ± 0.37 | 32.71e ± 0.51 | 45.2d ± 0.75 | 19.53d ± 0.68 |
| 0.5 | 57.65f ± 0.62 | 56.09e ± 0.46 | 88.12f ± 0.42 | 51.95e ± 0.56 | 85.67f ± 0.64 | 35.41e ± 0.72 | 34.40f ± 0.65 | 49.4d ± 0.39 | 30.19e ± 0.96 |
| 1.0 | 78.03g ± 0.40 | 68.05f ± 0.43 | 92.83g ± 0.12 | 78.05f ± 0.60 | 99.39g ± 0.05 | 86.56f ± 0.12 | 50.30g ± 0.84 | 56.6e ± 0.14 | 69.63f ± 1.06 |
| F | ** | ** | ** | ** | ** | ** | ** | ** | ** |
| CV (%) | 1.39 | 1.82 | 1.18 | 1.93 | 0.76 | 1.81 | 1.02 | 1.25 | 1.03 |
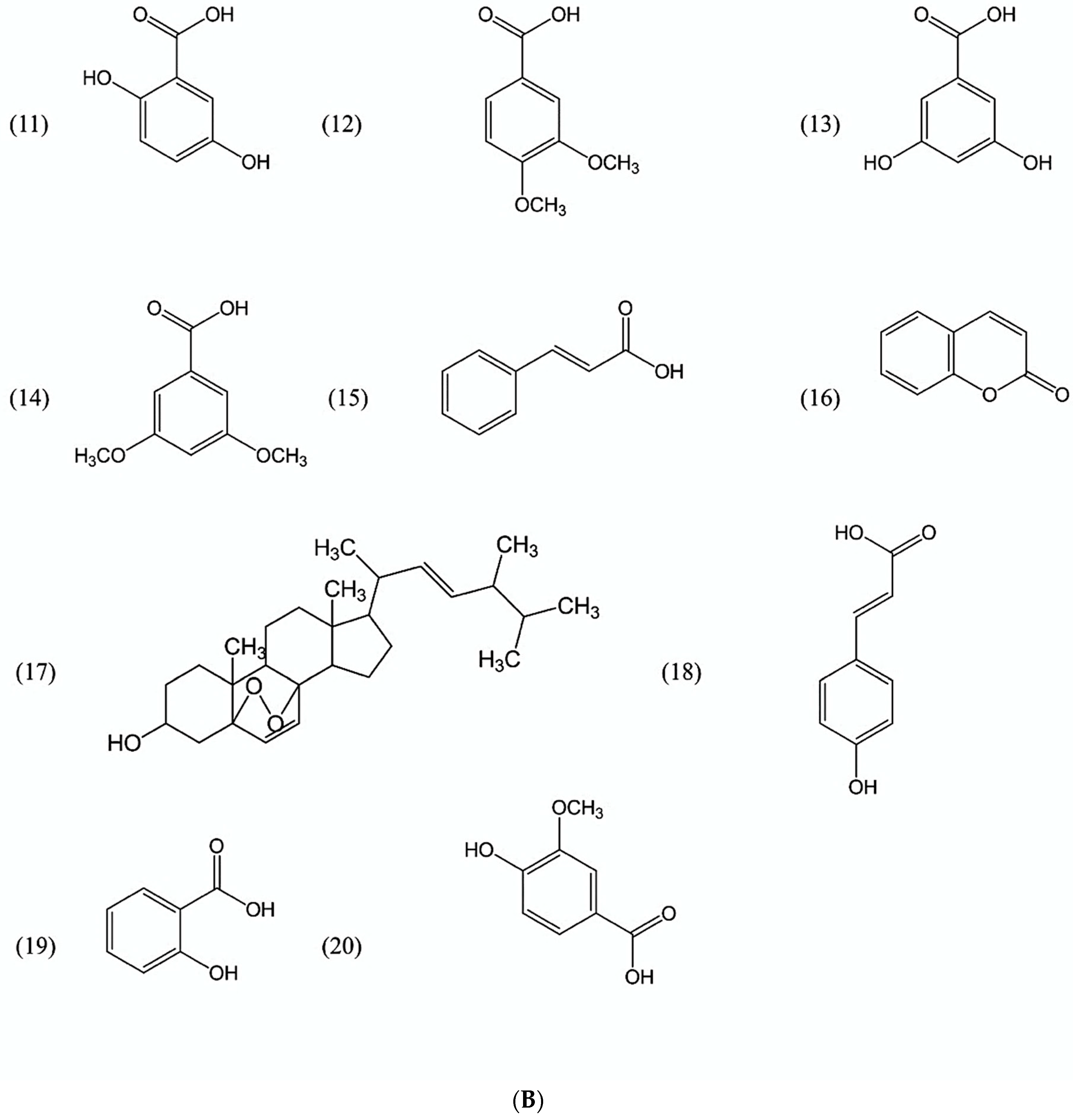
| No. | Allelochemicals * | Cultivars (Oryza sativa L.) ** | Detection | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | Frequency (%) | ||
| 1 | 2,4-dihydroxybenzaldehyde | √ | 11.1 | ||||||||
| 2 | 2,6-dimethoxybenzoic acid | √ | √ | 22.2 | |||||||
| 3 | 3,4-dihydroxybenzoic acid | √ | √ | √ | √ | √ | 55.6 | ||||
| 4 | 3,4-dihydroxyphenylacetic acid | √ | √ | √ | 33.3 | ||||||
| 5 | 3-hydroxybenzoic acid | √ | √ | √ | √ | √ | √ | 66.7 | |||
| 6 | 4-hydroxybenzoic acid | √ | √ | √ | √ | √ | √ | 66.7 | |||
| 7 | 5-methoxysalicylic acid | √ | 11.1 | ||||||||
| 8 | 7-oxostigmasterol | √ | √ | √ | √ | √ | √ | √ | 77.8 | ||
| 9 | Benzoic acid | √ | 11.1 | ||||||||
| 10 | 2,4-dimethoxybenzoic acid | √ | √ | √ | √ | √ | √ | 66.7 | |||
| 11 | 2,5-dihydroxybenzoic acid | √ | √ | √ | √ | √ | 55.6 | ||||
| 12 | 3,4-dimethoxybenzoic acid | √ | √ | √ | √ | √ | 55.6 | ||||
| 13 | 3,5-dihydroxybenzoic acid | √ | √ | √ | √ | √ | 55.6 | ||||
| 14 | 3,5-dimethoxybenzoic acid | √ | √ | √ | √ | √ | 55.6 | ||||
| 15 | Cinnamic acid | √ | √ | √ | √ | √ | √ | √ | √ | 88.9 | |
| 16 | Coumarin | √ | 11.1 | ||||||||
| 17 | Ergosterol peroxide | √ | 11.1 | ||||||||
| 18 | p-hydroxycinnamic acid | √ | √ | √ | √ | √ | √ | √ | √ | √ | 100.0 |
| 19 | Salicylic acid | √ | √ | √ | √ | √ | √ | 66.7 | |||
| 20 | Vanillic acid | √ | √ | √ | 33.3 | ||||||
| Total | 9 | 7 | 10 | 9 | 14 | 9 | 13 | 13 | 2 | ||
| Percentage (%) | 45 | 35 | 50 | 45 | 70 | 45 | 65 | 65 | 10 | ||
| No. | Allelochemicals ** | Formula | Molecular Ion | Theoretical Mass | Observed Mass | Mass Error |
|---|---|---|---|---|---|---|
| 1 | Allantoin | C4H6N4O3 | [M + H]+ | 159.0512 | 159.0513 | 0.63 |
| 2 | Benzoic acid | C7H6O2 | [M + H]+ | 123.0441 | 123.0441 | 0.81 |
| 3 | Cinnamic acid | C9H8O2 | [M + H]+ | 149.0597 | 149.0596 | −0.67 |
| 4 | Vanillic acid | C8H8O4 | [M + H]+ | 169.0495 | 169.0496 | 0.59 |
| 5 | Coumarin | C9H6O2 | [M + H]+ | 147.0441 | 147.0440 | −0.68 |
| 6 | Salicylic acid | C7H6O3 | [M-H]- | 137.0245 | 137.0244 | 0.73 |
| 7 | 7-oxostigmasterol | C29H46O2 | [M + H-H2O]+ | 409.3465 | 409.3471 | 1.46 |
| 8 | Ergosterol peroxide | C28H44O3 | [M + H-H2O]+ | 411.3258 | 411.3269 | 2.67 |
| OM Cultivar | Predominant Allelochemicals in 9 Cultivars * | |||||||
|---|---|---|---|---|---|---|---|---|
| Cinnamic Acid | Vanillic Acid | Coumarin | Benzoic Acid | 7-Oxostigmasterol | Ergosterol Peroxide | Allantoin | Salicylic Acid | |
| 5451 | 13.94 ab | 1.07 c | 8.44 a | 7.35 ab | 2.04 cd | - | 9.16 a | 17.56 a |
| 380 | 11.24 cd | 1.32 c | 6.23 bc | 5.41 c | 1.62 d | - | 5.39 b | 6.67 e |
| 3536 | 10.83 d | 1.33 b | 6.98 b | 4.75 d | 2.25 c | - | 4.27 b | 12.07 bcd |
| 6976 | 14.19 a | - | 5.93 bc | 8.26 a | 1.31 e | - | - | 10.92 cde |
| 4498 | 12.55 bc | 1.64 a | 7.75 b | - | 2.00 cd | - | 4.96 b | 19.54 a |
| 2395 | 11.89 cd | - | 7.25b | 6.36 bc | 3.06 b | - | 4.68 b | 14.76 abc |
| N406 | 12.99 abc | - | 7.25 b | 5.98 c | 3.32 b | - | 2.84 c | 16.94 ab |
| 5930 | 14.11 ab | 1.33 b | 8.23 a | 5.89 c | 4.85 a | 2.36 | - | 17.06 ab |
| 7347 | - | - | 4.65 c | - | 1.51 de | - | 3.81bc | 8.20 de |
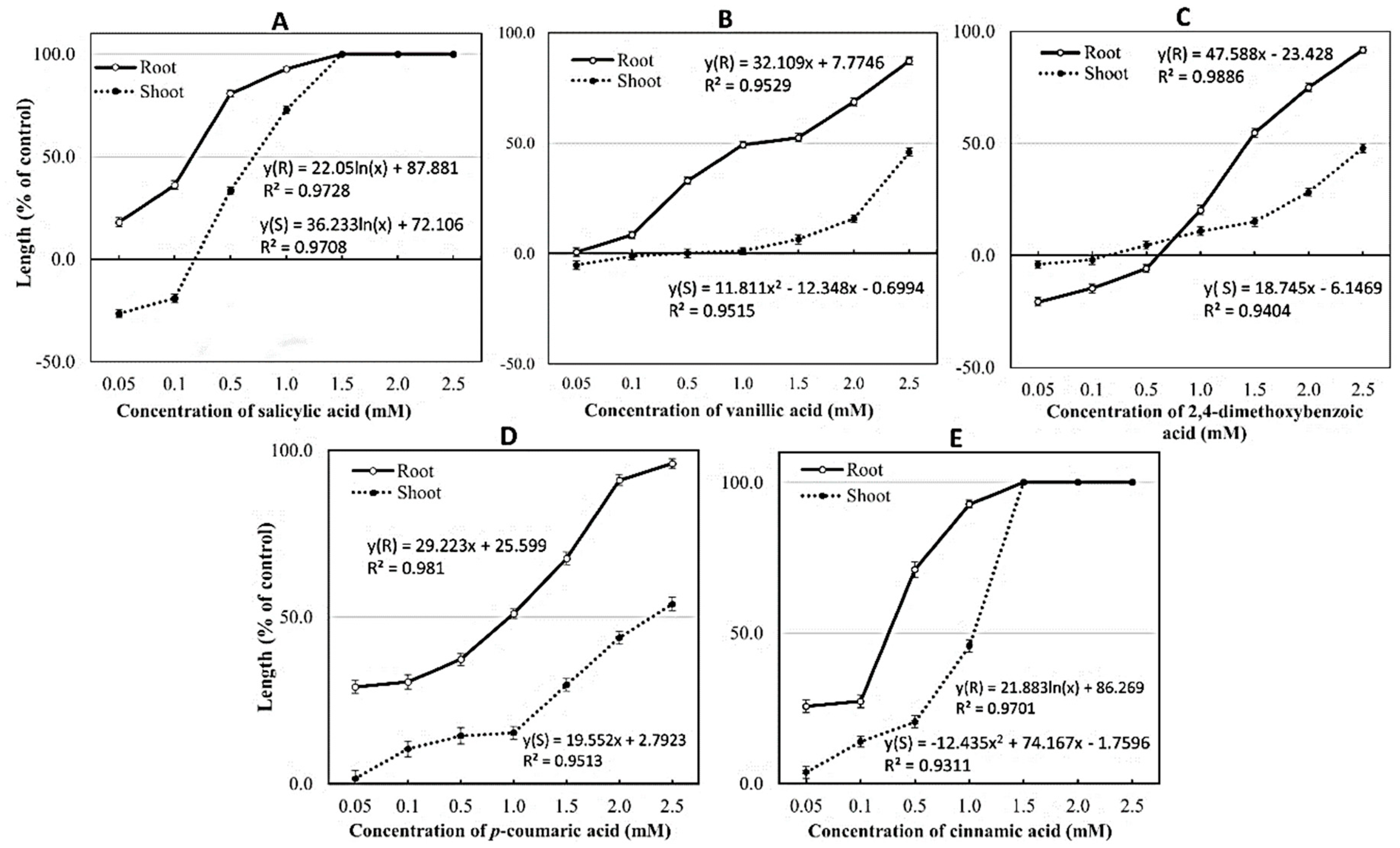
| No. | Allelochemicals ** | Retention | Purity | Allelochemical Content | |
|---|---|---|---|---|---|
| Time (min.) | (%) | In Rice Extract (mg mL−1) | In 1g Fresh Rice (mg g−1) | ||
| 1. | Salicylic acid | 11.469 | 98.9 | 0.7715 | 0.0501 |
| 2. | Vanillic acid | 11.126 | 99.7 | 0.0192 | 0.0012 |
| 3. | p-Coumaric acid | 20.269 | 99.7 | 0.0245 | 0.0016 |
| 4. | 2,4-dimethoxybenzoic acid | 30.058 | 99.7 | 0.0161 | 0.0010 |
| 5. | Cinnamic acid | 29.902 | 98.7 | 0.5210 | 0.0333 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ho, T.L.; Nguyen, T.T.C.; Vu, D.C.; Nguyen, N.Y.; Nguyen, T.T.T.; Phong, T.N.H.; Nguyen, C.T.; Lin, C.-H.; Lei, Z.; Sumner, L.W.; et al. Allelopathic Potential of Rice and Identification of Published Allelochemicals by Cloud-Based Metabolomics Platform. Metabolites 2020, 10, 244. https://doi.org/10.3390/metabo10060244
Ho TL, Nguyen TTC, Vu DC, Nguyen NY, Nguyen TTT, Phong TNH, Nguyen CT, Lin C-H, Lei Z, Sumner LW, et al. Allelopathic Potential of Rice and Identification of Published Allelochemicals by Cloud-Based Metabolomics Platform. Metabolites. 2020; 10(6):244. https://doi.org/10.3390/metabo10060244
Chicago/Turabian StyleHo, Thi L., Tu T. C. Nguyen, Danh C. Vu, Nhu Y. Nguyen, Trang T. T. Nguyen, Trieu N. H. Phong, Cuong T. Nguyen, Chung-Ho Lin, Zhentian Lei, Lloyd W. Sumner, and et al. 2020. "Allelopathic Potential of Rice and Identification of Published Allelochemicals by Cloud-Based Metabolomics Platform" Metabolites 10, no. 6: 244. https://doi.org/10.3390/metabo10060244
APA StyleHo, T. L., Nguyen, T. T. C., Vu, D. C., Nguyen, N. Y., Nguyen, T. T. T., Phong, T. N. H., Nguyen, C. T., Lin, C.-H., Lei, Z., Sumner, L. W., & Le, V. V. (2020). Allelopathic Potential of Rice and Identification of Published Allelochemicals by Cloud-Based Metabolomics Platform. Metabolites, 10(6), 244. https://doi.org/10.3390/metabo10060244







