Review of Oxepine-Pyrimidinone-Ketopiperazine Type Nonribosomal Peptides
Abstract
:1. Introduction
2. Results
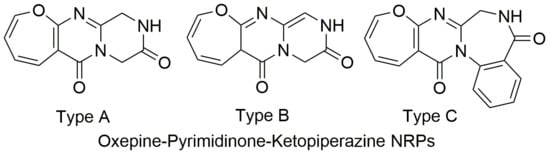
2.1. Biological Sources and Chemical Structures
2.2. Bioactivities
2.2.1. Plant Growth Regulation
2.2.2. Anti-Inflammatory Activity
2.2.3. Antifungal Activity
2.2.4. Cytotoxicity
2.2.5. Antibacterial Activity
2.2.6. Anti-Plasmodial Activity
2.2.7. Transcriptional Activation
2.3. Biosynthesis
3. Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schwarzer, D.; Finking, R.; Marahiel, M.A. Nonribosomal peptides: From genes to products. Nat. Prod. Rep. 2003, 20, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Cohen, D.J.; Loertscher, R.; Rubin, M.F.; Tilney, N.L.; Carpenter, C.B.; Strom, T.B. Cyclosporine: A new immunosuppressive agent for organ transplantation. Ann. Intern. Med. 1984, 101, 667–682. [Google Scholar] [CrossRef] [PubMed]
- Raja, A.; LaBonte, J.; Lebbos, J.; Kirkpatrick, P. Fresh from the pipeline: Daptomycin. Nat. Rev. Drug Discov. 2003, 2, 943–944. [Google Scholar] [CrossRef] [PubMed]
- Winn, M.; Fyans, J.K.; Zhuo, Y.; Micklefield, J. Recent advances in engineering nonribosomal peptide assembly lines. Nat. Prod. Rep. 2016, 33, 317–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niquille, D.L.; Hansen, D.A.; Mori, T.; Fercher, D.; Kries, H.; Hilvert, D. Nonribosomal biosynthesis of backbone-modified peptides. Nat. Chem. 2018, 10, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Cutler, H.G.; Springer, J.P.; Arrendale, R.F.; Arison, B.H.; Cole, P.D.; Roberts, R.G. Cinereain: A novel metabolite with plant growth regulating properties from Botrytis cinerea. Agric. Biol. Chem. 1988, 52, 1725–1733. [Google Scholar] [CrossRef]
- He, D.; Wang, M.; Zhao, S.; Shu, Y.; Zeng, H.; Xiao, C.; Lu, C.; Liu, Y. Pharmaceutical prospects of naturally occurring quinazolinone and its derivatives. Fitoterapia 2017, 119, 136–149. [Google Scholar] [CrossRef]
- Li, G.Y.; Yang, T.; Luo, Y.G.; Chen, X.Z.; Fang, D.M.; Zhang, G.L. Brevianamide J, a New indole alkaloid dimer from fungus Aspergillus versicolor. Org. Lett. 2009, 11, 3714–3717. [Google Scholar] [CrossRef]
- Li, G.; Li, L.; Yang, T.; Chen, X.; Fang, D. Four New Alkaloids, Brevianamides O – R, from the fungus Aspergillus versicolor. Helv. Chim. Acta 2010, 93, 2075–2080. [Google Scholar] [CrossRef]
- Zhang, P.; Li, X.M.; Wang, J.N.; Wang, B.G. Oxepine-containing diketopiperazine alkaloids from the algal-derived endophytic fungus Paecilomyces variotii EN-291. Helv. Chim. Acta 2015, 98, 800–804. [Google Scholar] [CrossRef]
- Kshirsagar, U.A. Recent developments in the chemistry of quinazolinone alkaloids. Org. Biomol. Chem. 2015, 13, 9336–9352. [Google Scholar] [CrossRef]
- Hameed, A.; Al-Rashida, M.; Uroos, M.; Ali, S.A.; Arshia; Ishtiaq, M.; Khan, K.M. Quinazoline and quinazolinone as important medicinal scaffolds: A comparative patent review (2011–2016). Expert Opin. Ther. Pat. 2018, 28, 281–297. [Google Scholar] [CrossRef]
- Resende, D.I.S.P.; Boonpothong, P.; Sousa, E.; Kijjoa, A.; Pinto, M.M.M. Chemistry of the fumiquinazolines and structurally related alkaloids. Nat. Prod. Rep. 2019, 36, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.M.; Yi, X.X.; Zhou, Y.; Su, X.; Peng, Y.; Gao, C.H. An update on 2,5-Diketopiperazines from marine organisms. Mar. Drugs 2014, 12, 6213–6235. [Google Scholar] [CrossRef] [PubMed]
- Doveston, R.G.; Steendam, R.; Jones, S.; Taylor, R.J.K. Total synthesis of an oxepine natural product, (±)-janoxepin. Org. Lett. 2012, 14, 1122–1125. [Google Scholar] [CrossRef]
- Doveston, R.G.; Taylor, R.J.K. An expedient synthesis of the proposed biosynthetic precursor of the oxepine natural product, janoxepin. Tetrahedron Lett. 2012, 53, 2533–2536. [Google Scholar] [CrossRef]
- Fuchser, J. Beeinflussung der Sekundarstoffbildung bei Aspergillus ochraceus durch Variation der Kulturbedingungen sowie Isolierung, Strukturaufklarung und Biosynthese der neuen Naturstoffe. Ph. D. Thesis, Universität Göttingen, Göttingen, Germany, 1996. [Google Scholar]
- Michael, B. Pyrralsäuren, eine neue Klasse pyrrolmaskierter Aminosäuren aus Nocardia sp. und neue Sekundärmetabolite aus Aspergillus ochraceus. Ph. D. Thesis, Universität Göttingen, Göttingen, Germany, 1999. [Google Scholar]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Belofsky, G.N.; Anguera, M.; Jensen, P.R.; Fenical, W.; Köck, M. Oxepinamides A-C and fumiquinazolines H-I: Bioactive metabolites from a marine isolate of a fungus of the genus Acremonium. Chem. Eur. J. 2000, 6, 1355–1360. [Google Scholar] [CrossRef]
- Sprogøe, K.; Manniche, S.; Larsen, T.O.; Christophersen, C. Janoxepin and brevicompanine B: Antiplasmodial metabolites from the fungus Aspergillus janus. Tetrahedron 2005, 61, 8718–8721. [Google Scholar] [CrossRef]
- Rahbæk, L.; Breinholt, J.; Frisvad, J.C.; Christophersen, C. Circumdatin A, B, and C: Three new benzodiazepine alkaloids isolated from a culture of the fungus Aspergillus ochraceus. J. Org. Chem. 1999, 64, 1689–1692. [Google Scholar]
- Ookura, R.; Kito, K.; Ooi, T.; Namikoshi, M.; Kusumi, T. Structure revision of circumdatins A and B, benzodiazepine alkaloids produced by marine fungus Aspergillus ostianus, by X-ray crystallography. J. Org. Chem. 2008, 73, 4245–4247. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.H.; Shi, Q.W.; Zheng, Z.H.; Ke, A.B.; Zhang, H.; Huo, C.H.; Ma, Y.; Ren, X.; Li, Y.Y.; Lin, J.; et al. Oxepinamides: Novel liver X receptor agonists from aspergillus puniceus. Eur. J. Org. Chem. 2011, 802–807. [Google Scholar] [CrossRef]
- Lee, S.U.; Asami, Y.; Lee, D.; Jang, J.H.; Ahn, J.S.; Oh, H. Protuboxepins A and B and protubonines A and B from the marine-derived fungus Aspergillus sp. SF-5044. J. Nat. Prod. 2011, 74, 1284–1287. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Zhang, X.Y.; Tu, Z.C.; Xu, X.Y.; Qi, S.H. Alkaloids from the deep-sea-derived fungus Aspergillus westerdijkiae DFFSCS013. J. Nat. Prod. 2013, 76, 983–987. [Google Scholar] [CrossRef]
- Zhuravleva, O.I.; Afiyatullov, S.S.; Yurchenko, E.A.; Denisenko, V.A.; Kirichuk, N.N.; Dmitrenok, P.S. New metabolites from the algal associated marine-derived fungus Aspergillus carneus. Nat. Prod. Commun. 2013, 8, 1071–1074. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Mandi, A.; Li, X.M.; Du, F.Y.; Wang, J.N.; Li, X.; Kurtan, T.; Wang, B.G. Varioxepine a, a 3H-oxepine-containing alkaloid with a new oxa-cage from the marine algal-derived endophytic fungus Paecilomyces variotii. Org. Lett. 2014, 16, 4834–4837. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; He, W.; Huang, X.; Tian, X.; Liao, S.; Yang, B.; Wang, F.; Zhou, X.; Liu, Y. Antifungal New oxepine-containing alkaloids and xanthones from the deep-sea-derived fungus Aspergillus versicolor SCSIO 05879. J. Agric. Food Chem. 2016, 64, 2910–2916. [Google Scholar] [CrossRef]
- Pan, C.; Shi, Y.; Chen, X.; Chen, C.T.A.; Tao, X.; Wu, B. New compounds from a hydrothermal vent crab-associated fungus Aspergillus versicolor XZ-4. Org. Biomol. Chem. 2017, 15, 1155–1163. [Google Scholar] [CrossRef]
- Tian, Y.Q.; Lin, S.N.; Zhou, H.; Lin, S.T.; Wang, S.Y.; Liu, Y.H. Protuboxepin C and protuboxepin D from the sponge-derived fungus Aspergillus sp. SCSIO XWS02F40. Nat. Prod. Res. 2018, 32, 2510–2515. [Google Scholar] [CrossRef]
- Tian, Y.Q.; Lin, X.P.; Wang, Z.; Zhou, X.F.; Qin, X.C.; Kaliyaperumal, K.; Zhang, T.Y.; Tu, Z.C.; Liu, Y. Asteltoxins with antiviral activities from the marine sponge-derived fungus Aspergillus 308 sp. SCSIO XWS02F40. Molecules 2016, 21, 34. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.F.; Mao, N.; Xue, X.J.; Qi, Y.X.; Wei, M.Y.; Wang, C.Y.; Shao, C.L. Structures and absolute configurations of diketopiperazine alkaloids chrysopiperazines A-C from the gorgonian-derived Penicillium chrysogenum fungus. Mar. Drugs 2019, 17, 250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, X.; Chen, C.; Tao, H.; Lin, X.; Yang, B.; Zhou, X.; Liu, Y. Structurally diverse diketopiperazine alkaloids from the marine-derived fungus: Aspergillus versicolor SCSIO 41016. Org. Chem. Front. 2019, 6, 736–740. [Google Scholar] [CrossRef]
- Liang, X.; Zhang, X.; Lu, X.; Zheng, Z.; Ma, X.; Qi, S. Diketopiperazine-type alkaloids from a deep-sea-derived Aspergillus puniceus fungus and their effects on Liver X Receptor α. J. Nat. Prod. 2019, 82, 1558–1564. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.Q.; Li, P.H.; Chao, Y.X.; Chen, H.; Du, N.; He, Q.X.; Liu, K.C. Alkaloids with cardiovascular effects from the marine-derived fungus Penicillium expansum Y32. Mar. Drugs 2015, 13, 6489–6504. [Google Scholar] [CrossRef]
- Asami, Y.; Jang, J.H.; Soung, N.K.; He, L.; Moon, D.O.; Kim, J.W.; Oh, H.; Muroi, M.; Osada, H.; Kim, B.Y.; et al. Protuboxepin A, a marine fungal metabolite, inducing metaphase arrest and chromosomal misalignment in tumor cells. Bioorganic Med. Chem. 2012, 20, 3799–3806. [Google Scholar] [CrossRef] [PubMed]
- Ames, B.D.; Walsh, C.T. Anthranilate-activating modules from fungal nonribosomal peptide assembly lines. Biochemistry 2010, 49, 3351–3365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ames, B.D.; Liu, X.; Walsh, C.T. Enzymatic processing of fumiquinazoline F: A tandem oxidative-acylation strategy for the generation of multicyclic scaffolds in fungal indole alkaloid biosynthesis. Biochemistry 2010, 49, 8564–8576. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Chooi, Y.H.; Ames, B.D.; Wang, P.; Walsh, C.T.; Tang, Y. Fungal indole alkaloid biosynthesis: Genetic and biochemical investigation of the tryptoquialanine pathway in penicillium aethiopicum. J. Am. Chem. Soc. 2011, 133, 2729–2741. [Google Scholar] [CrossRef] [Green Version]
- Yan, D.; Chen, Q.; Gao, J.; Bai, J.; Liu, B.; Zhang, Y.; Zhang, L.; Zhang, C.; Zou, Y.; Hu, Y. Complexity and diversity generation in the biosynthesis of fumiquinazoline-related peptidyl alkaloids. Org. Lett. 2019, 21, 1475–1479. [Google Scholar] [CrossRef]
- Henderson, A.P.; Mutlu, E.; Leclercq, A.; Bleasdale, C.; Clegg, W.; Henderson, R.A.; Golding, B.T. Trapping of benzene oxide-oxepin and methyl-substituted derivatives with 4-phenyl- and 4-pentafluorophenyl-1,2,4-triazoline-3,5-dione. Chem. Commun. 2002, 8, 1956–1957. [Google Scholar] [CrossRef]
- Samson, R.A.; Houbraken, J.A.M.P.; Kuijpers, A.F.A.; Frank, J.M.; Frisvad, J.C. New ochratoxin A or sclerotium producing species in Aspergillus section Circumdati. Stud. Mycol. 2004, 50, 45–61. [Google Scholar]
- Chen, A.J.; Frisvad, J.C.; Sun, B.D.; Varga, J.; Kocsubé, S.; Dijksterhuis, J.; Kim, D.H.; Hong, S.B.; Houbraken, J.; Samson, R.A. Aspergillus section Nidulantes (formerly Emericella): Polyphasic taxonomy, chemistry and biology. Stud. Mycol. 2016, 84. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, C.; Matsushita, T.; Doi, M.; Minoura, K.; Shingu, T.; Kumeda, Y.; Numata, A. Fumiquinazolines A-G, novel metabolites of a fungus separated from a Pseudolabrus marine fish. J. Chem. Soc. Perkin Trans. 1 1995, 2345–2353. [Google Scholar] [CrossRef]
- Vesth, T.C.; Nybo, J.L.; Theobald, S.; Frisvad, J.C.; Larsen, T.O.; Nielsen, K.F.; Hoof, J.B.; Brandl, J.; Salamov, A.; Riley, R.; et al. Investigation of inter- and intraspecies variation through genome sequencing of Aspergillus section Nigri. Nat. Genet. 2018, 50, 1688–1695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kjærbølling, I.; Vesth, T.; Frisvad, J.C.; Nybo, J.L.; Theobald, S.; Kildgaard, S.; Petersen, T.I.; Kuo, A.; Sato, A.; Lyhne, E.K.; et al. A comparative genomics study of 23 Aspergillus species from section Flavi. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| No. | Name | Substitution groups | Sources | ||||
|---|---|---|---|---|---|---|---|
| R1 | R2 | R3 | R4 | R5 | |||
| 1 | Cinereain | =CHCH(CH3)2, Z | n/a | H | CH(CH3)2 | H | Botrytis cinerea ATCC 64157 [6] Aspergillus carneus KMM 4638 [27] |
| 4 | Oxepinamide A | CH(CH3)CH2CH3 | OH | OCH3 | H | CH3 | Acremonium sp. [20] |
| 5 | Oxepinamide B | OH | CH(CH3)CH2CH3 | OCH3 | H | CH3 | Acremonium sp. [20] |
| 6 | Oxepinamide C | CH2CH(CH3)2 | OCH3 | OCH3 | H | CH3 | Acremonium sp. [20] |
| 7 | Janoxepin | =CHCH(CH3)2, Z | n/a | H | H | CH2CH(CH3)2 | Aspergillus janus IBT 22274 [21] |
| 10 | Brevianamide O | OH | CH(CH3)CH2CH3 | H | Benzyl | H | Aspergillus versicolor (AS 3.4186) [9] |
| 11 | Brevianamide P | H | CH(CH3)CH2CH3 | H | Benzyl | H | Aspergillus versicolor (AS 3.4186) [9] |
| 12 | Oxepinamide D | OH | Benzyl | H | H | CH3 | Aspergillus puniceus F02Z-1744 [24] |
| 16 | Protuboxepin A | CH(CH3)CH2CH3 | H | H | H | Benzyl | Aspergillus sp. SF-5044 [25] Penicillium expansum Y32 [36] |
| 17 | Protuboxepin B | CH(CH3)2 | H | H | H | Benzyl | Aspergillus sp. SF-5044 [25] Paecilomyces variotii EN-291 [10] Penicillium expansum Y32 [36] |
| 19 | Dihydrocinereain | H | CH2CH(CH3)2 | H | CH(CH3)2 | H | Aspergillus carneus KMM 4638 [27] |
| 22 | Varioloid B | OCH3 | CH(CH3)2 | H | Benzyl | H | Paecilomyces variotii EN-291 [10] |
| 23 | Versicoloid A | H | CH(CH3)CH2CH3 | OCH3 | CH(CH3)2 | H | Aspergillus versicolor SCSIO 05879 [29] Penicillium chrysogenum CHNSCLM-0019 [33] |
| 24 | Versicoloid B | OH | CH(CH3)CH2CH3 | OCH3 | CH(CH3)2 | H | Aspergillus versicolor SCSIO 05879 [29] Penicillium chrysogenum CHNSCLM-0019 [33] |
| 25 | Versicomide D | CH(CH3)CH2CH3, 18S | H | OCH3 | CH(CH3)2 | H | Aspergillus versicolor XZ-4 [30] |
| 26 | Protuboxepin C | CH(CH3)CH2CH3, 16S | OCH3 | H | H | Benzyl | Aspergillus sp. SCSIO XWS02F40 [31] |
| 27 | Protuboxepin D | CH(CH3)CH2CH3, 16S | OH | H | H | Benzyl | Aspergillus sp. SCSIO XWS02F40 [31] |
| 28 | Chryzopiperazine A | CH(CH3)CH2CH3, 19S | OCH3 | OCH3 | H | CH(CH3)2 | Penicillium chrysogenum CHNSCLM-0019 [33] |
| 29 | Chrysopiperazine B | OCH3 | CH(CH3)CH2CH3, 19S | OCH3 | H | CH(CH3)2 | Penicillium chrysogenum CHNSCLM-0019 [33] |
| 30 | Protuboxepin F | =CHCH(CH3)2, Z | n/a | H | H | Benzyl | Aspergillus versicolor SCSIO 41016 [34] |
| 31 | Protuboxepin G | =CHCH(CH3)2, E | n/a | H | H | Benzyl | Aspergillus versicolor SCSIO 41016 [34] |
| 32 | Oxepinamide H | OCH3 | Benzyl | H | H | CH3 | Aspergillus puniceus SCSIO z021 [35] |
| 33 | Oxepinamide I | Benzyl | OCH3 | H | H | CH3 | Aspergillus puniceus SCSIO z021 [35] |
| 34 | Oxepinamide J | Benzyl | OH | H | H | CH3 | Aspergillus puniceus SCSIO z021 [35] |
| 35 | Oxepinamide K | =CH-Phenyl, Z | H | H | H | CH3 | Aspergillus puniceus SCSIO z021 [35] |
| No. | Name | Substitution Groups | Sources | |
|---|---|---|---|---|
| R6 | R7 | |||
| 9 | Brevianamide L | CH(CH3)CH2CH3 | OH, 12S | Aspergillus versicolor (AS 3.4186) [8] |
| 13 | Oxepinamide E | CH(CH3)CH2CH3, 17S | OH, 12R | Aspergillus puniceus F02Z-1744 [24] |
| 14 | Oxepinamide F | CH(CH3)CH2CH3, 17S | OCH3, 12R | Aspergillus puniceus F02Z-1744 [24] |
| 15 | Oxepinamide G | CH(CH3)2 | OCH3, 12R | Aspergillus puniceus F02Z-1744 [24] |
| 20 | Varioxepine A | CH(CH3)2 | See Figure 1 | Paecilomyces variotii EN-291 [28] |
| 21 | Varioloid A | CH(CH3)2 | O(CH2)COCH(CH3)2, 12R | Paecilomyces variotii EN-291 [10] |
| No. | Name | Scaffold | Sources |
|---|---|---|---|
| 2 | Asperloxin A (Circumdatin A) | 7/6/7/6/5, Figure 1 | Aspergillus ochraceus DSM 7428 [17] Aspergillus ochraceus IBT 12704 [22] Aspergillus ostianus 01F313 [23] |
| 3 | Asperloxin B | 7/6/7/6/5, Figure 1 | Aspergillus ochraceus DSM 7428 [18] |
| 8 | Circumdatin B | 7/6/7/6/5, Figure 1 | Aspergillus ochraceus IBT 12704 [22] Aspergillus ostianus 01F313 [23] |
| 18 | Circumdatin L | 7/6/7/6, Figure 1 | Aspergillus westerdijkiae DFFSC S013 [26] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Frisvad, J.C.; Larsen, T.O. Review of Oxepine-Pyrimidinone-Ketopiperazine Type Nonribosomal Peptides. Metabolites 2020, 10, 246. https://doi.org/10.3390/metabo10060246
Guo Y, Frisvad JC, Larsen TO. Review of Oxepine-Pyrimidinone-Ketopiperazine Type Nonribosomal Peptides. Metabolites. 2020; 10(6):246. https://doi.org/10.3390/metabo10060246
Chicago/Turabian StyleGuo, Yaojie, Jens C. Frisvad, and Thomas O. Larsen. 2020. "Review of Oxepine-Pyrimidinone-Ketopiperazine Type Nonribosomal Peptides" Metabolites 10, no. 6: 246. https://doi.org/10.3390/metabo10060246
APA StyleGuo, Y., Frisvad, J. C., & Larsen, T. O. (2020). Review of Oxepine-Pyrimidinone-Ketopiperazine Type Nonribosomal Peptides. Metabolites, 10(6), 246. https://doi.org/10.3390/metabo10060246