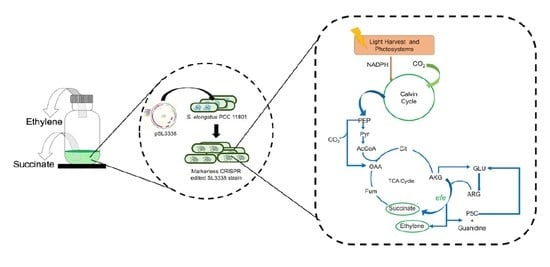
Photosynthetic Co-production of Succinate and Ethylene in a Fast-Growing Cyanobacterium, Synechococcus elongatus PCC 11801
Abstract
:1. Introduction
2. Results and Discussion
2.1. Optimising Growth Condition for Improved Succinate Production in the Recombinant Strain
2.2. Nitrate Limitation Positively Affected Succinate Production
2.3. Efficient Carbon Partitioning Between Product Formation and Biomass
2.4. Co-Production of Ethylene in S. elongatus PCC 11801-efe Strain
2.5. Metabolic Profiling and 13C Labelling
2.5.1. Partitioning of Carbon in the Recombinant Strain
2.5.2. Lower Part of Glycolysis and the TCA Cycle
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Strain Development Strategy
3.2.1. Plasmid Construction
3.2.2. Recombinant Strain Construction via Conjugation
3.3. Strain Cultivation and Determination of Cell Biomass
3.4. Optimisation of Conditions for Succinate Production
3.5. Effect of Nitrate Limitation on Succinate Production
3.6. Estimation of Ethylene Production Using GC-FID
3.7. Isolation of Total RNA and Reverse Transcriptase (RT)-PCR of Selected Genes
3.8. Metabolite Pool Size Estimation of SL3338 Strain
3.9. 13C Enrichment Analysis of SL3338 Strain
3.10. Determination of Photosynthetic Efficiency and Capacity
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nozzi, N.E.; Oliver, J.W.K.; Atsumi, S. Cyanobacteria as a Platform for Biofuel Production. Front. Bioeng. Biotechnol. 2013, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, B.; Madhu, S.; Wangikar, P.P. The role of systems biology in developing non-model cyanobacteria as hosts for chemical production. Curr. Opin. Biotechnol. 2020, 64, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, A.; Pakrasi B., H.; Wangikar P., P. Recent advances in synthetic biology of cyanobacteria. Appl. Microbiol. Biotechnol. 2018, 102, 5457–5471. [Google Scholar] [CrossRef] [PubMed]
- Santos-Merino, M.; Singh, A.K.; Ducat, D.C. New Applications of Synthetic Biology Tools for Cyanobacterial Metabolic Engineering. Front. Bioeng. Biotechnol. 2019, 7. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Liberton, M.; Cliften, P.F.; Head, R.D.; Jacobs, J.M.; Smith, R.D.; Koppenaal, D.W.; Brand, J.J.; Pakrasi, H.B. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO₂. Sci. Rep. 2015, 5, 8132. [Google Scholar] [CrossRef]
- Jaiswal, D.; Sengupta, A.; Sohoni, S.; Sengupta, S.; Phadnavis, A.G.; Pakrasi, H.B.; Wangikar, P.P. Genome Features and Biochemical Characteristics of a Robust, Fast Growing and Naturally Transformable Cyanobacterium Synechococcus elongatus PCC 11801 Isolated from India. Sci. Rep. 2018, 8, 16632. [Google Scholar] [CrossRef]
- Jaiswal, D.; Sengupta, A.; Sengupta, S.; Madhu, S.; Pakrasi, H.B.; Wangikar, P.P. A Novel Cyanobacterium Synechococcus elongatus PCC 11802 has Distinct Genomic and Metabolomic Characteristics Compared to its Neighbor PCC 11801. Sci. Rep. 2020, 10, 191. [Google Scholar] [CrossRef] [Green Version]
- Włodarczyk, A.; Selão, T.T.; Norling, B.; Nixon, P.J. Newly discovered Synechococcus sp. PCC 11901 is a robust cyanobacterial strain for high biomass production. Commun. Biol. 2020, 3, 215. [Google Scholar] [CrossRef]
- Vasudevan, R.; Gale, G.A.R.; Schiavon, A.A.; Puzorjov, A.; Malin, J.; Gillespie, M.D.; Vavitsas, K.; Zulkower, V.; Wang, B.; Howe, C.J.; et al. CyanoGate: A Modular Cloning Suite for Engineering Cyanobacteria Based on the Plant MoClo Syntax. Plant Physiol. 2019, 180, 39–55. [Google Scholar] [CrossRef] [Green Version]
- Knoot, C.J.; Khatri, Y.; Hohlman, R.M.; Sherman, D.H.; Pakrasi, H.B. Engineered Production of Hapalindole Alkaloids in the Cyanobacterium Synechococcus sp. UTEX 2973. ACS Synth. Biol. 2019, 8, 1941–1951. [Google Scholar] [CrossRef]
- Li, S.; Sun, T.; Xu, C.; Chen, L.; Zhang, W. Development and optimization of genetic toolboxes for a fast-growing cyanobacterium Synechococcus elongatus UTEX 2973. Metab. Eng. 2018, 48, 163–174. [Google Scholar] [CrossRef]
- Ungerer, J.; Pakrasi, H.B. Cpf1 Is A Versatile Tool for CRISPR Genome Editing Across Diverse Species of Cyanobacteria. Sci. Rep. 2016, 6, 39681. [Google Scholar] [CrossRef]
- Wendt, K.E.; Ungerer, J.; Cobb, R.E.; Zhao, H.; Pakrasi, H.B. CRISPR/Cas9 mediated targeted mutagenesis of the fast growing cyanobacterium Synechococcus elongatus UTEX 2973. Microb. Cell Fact. 2016, 15, 115. [Google Scholar] [CrossRef] [Green Version]
- Jaiswal, D.; Prasannan, C.B.; Hendry, J.I.; Wangikar, P.P. SWATH Tandem Mass Spectrometry Workflow for Quantification of Mass Isotopologue Distribution of Intracellular Metabolites and Fragments Labeled with Isotopic 13C Carbon. Anal. Chem. 2018, 90, 6486–6493. [Google Scholar] [CrossRef]
- Dange, M.C.; Mishra, V.; Mukherjee, B.; Jaiswal, D.; Merchant, M.S.; Prasannan, C.B.; Wangikar, P.P. Evaluation of freely available software tools for untargeted quantification of 13C isotopic enrichment in cellular metabolome from HR-LC/MS data. Metab. Eng. Commun. 2020, 10, e00120. [Google Scholar] [CrossRef]
- Abernathy, M.H.; Yu, J.; Ma, F.; Liberton, M.; Ungerer, J.; Hollinshead, W.D.; Gopalakrishnan, S.; He, L.; Maranas, C.D.; Pakrasi, H.B.; et al. Deciphering cyanobacterial phenotypes for fast photoautotrophic growth via isotopically nonstationary metabolic flux analysis. Biotechnol. Biofuels 2017, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knoot, C.J.; Ungerer, J.; Wangikar, P.P.; Pakrasi, H.B. Cyanobacteria: Promising biocatalysts for sustainable chemical production. J. Biol. Chem. 2018, 293, 5044–5052. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Bryant, D.A. The tricarboxylic acid cycle in cyanobacteria. Science 2011, 334, 1551–1553. [Google Scholar] [CrossRef] [PubMed]
- Lan, E.I.; Wei, C.T. Metabolic engineering of cyanobacteria for the photosynthetic production of succinate. Metab. Eng. 2016, 38, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Shen, C.R.; Huang, C.; Sung, L.; Wu, M.; Hu, Y. CRISPR-Cas9 for the genome engineering of cyanobacteria and succinate production. Metab. Eng. 2016, 38, 293–302. [Google Scholar] [CrossRef]
- Huang, C.H.; Shen, C.R.; Li, H.; Sung, L.Y.; Wu, M.Y.; Hu, Y.C. CRISPR interference (CRISPRi) for gene regulation and succinate production in cyanobacterium S. elongatus PCC 7942. Microb. Cell Fact. 2016, 15, 196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasunuma, T.; Matsuda, M.; Kato, Y.; Vavricka, C.J.; Kondo, A. Temperature enhanced succinate production concurrent with increased central metabolism turnover in the cyanobacterium Synechocystis sp. PCC 6803. Metab. Eng. 2018, 48, 109–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shabestary, K.; Anfelt, J.; Ljungqvist, E.; Jahn, M.; Yao, L.; Hudson, E.P. Targeted Repression of Essential Genes to Arrest Growth and Increase Carbon Partitioning and Biofuel Titers in Cyanobacteria. ACS Synth. Biol. 2018, 7, 1669–1675. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, V.; Vuorio, E.; Aro, E.M.; Kallio, P. Enhanced stable production of ethylene in photosynthetic cyanobacterium Synechococcus elongatus PCC 7942. World J. Microbiol. Biotechnol. 2019, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ungerer, J.; Tao, L.; Davis, M.; Ghirardi, M.; Maness, P.C.; Yu, J. Sustained photosynthetic conversion of CO2 to ethylene in recombinant cyanobacterium Synechocystis 6803. Energy Environ. Sci. 2012, 5, 8998–9006. [Google Scholar] [CrossRef]
- Mo, H.; Xie, X.; Zhu, T.; Lu, X. Effects of global transcription factor NtcA on photosynthetic production of ethylene in recombinant Synechocystis sp. PCC 6803. Biotechnol. Biofuels 2017, 10. [Google Scholar] [CrossRef]
- Eckert, C.; Xu, W.; Xiong, W.; Lynch, S.; Ungerer, J.; Tao, L.; Gill, R.; Maness, P.-C.; Yu, J. Ethylene-forming enzyme and bioethylene production. Biotechnol. Biofuels 2014, 7, 33. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Dong, T.; Myrlie, A.; Gu, L.; Zhu, H.; Xiong, W.; Maness, P.; Zhou, R.; Yu, J. Photosynthetic production of the nitrogen-rich compound guanidine. Green Chem. 2019, 21, 2928–2937. [Google Scholar] [CrossRef] [Green Version]
- Durall, C.; Lindberg, P.; Yu, J.; Lindblad, P. Increased ethylene production by overexpressing phosphoenolpyruvate carboxylase in the cyanobacterium Synechocystis PCC 6803. Biotechnol. Biofuels 2020, 13, 16. [Google Scholar] [CrossRef]
- Xiong, W.; Morgan, J.A.; Ungerer, J.; Wang, B.; Maness, P.-C.; Yu, J. The plasticity of cyanobacterial metabolism supports direct CO2 conversion to ethylene. Nat. Plants 2015, 1, 15053. [Google Scholar] [CrossRef]
- Zhu, T.; Xie, X.; Li, Z.; Tan, X.; Lu, X. Enhancing photosynthetic production of ethylene in genetically engineered Synechocystis sp. PCC 6803. Green Chem. 2015, 17, 421–434. [Google Scholar] [CrossRef]
- Osanai, T.; Shirai, T.; Iijima, H.; Nakaya, Y.; Okamoto, M.; Kondo, A.; Hirai, M.Y. Genetic manipulation of a metabolic enzyme and a transcriptional regulator increasing succinate excretion from unicellular cyanobacterium. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veetil, V.P.; Angermayr, S.A.; Hellingwerf, K.J. Ethylene production with engineered Synechocystis sp PCC 6803 strains. Microb. Cell Fact. 2017, 16. [Google Scholar] [CrossRef] [Green Version]
- Hagemann, M.; Hess, W.R. Systems and synthetic biology for the biotechnological application of cyanobacteria. Curr. Opin. Biotechnol. 2018, 49, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Fell, D.A. Increasing the flux in metabolic pathways: A metabolic control analysis perspective. Biotechnol. Bioeng. 1998, 58, 121–124. [Google Scholar] [CrossRef]
- Cheah, Y.E.; Xu, Y.; Sacco, S.A.; Babele, P.K.; Zheng, A.O.; Johnson, C.H.; Young, J.D. Systematic identification and elimination of flux bottlenecks in the aldehyde production pathway of Synechococcus elongatus PCC 7942. Metab. Eng. 2020, 60, 56–65. [Google Scholar] [CrossRef] [PubMed]
- McAtee, A.G.; Jazmin, L.J.; Young, J.D. Application of isotope labeling experiments and 13C flux analysis to enable rational pathway engineering. Curr. Opin. Biotechnol. 2015, 36, 50–56. [Google Scholar] [CrossRef] [Green Version]
- Nagahama, K.; Ogawa, T.; Fujii, T.; Tazaki, M.; Tanase, S.; Morino, Y.; Fukuuda, H. Purification and Properties of an ethylene-forming enzyme from Pseudomonas syringae pv. phaseolicola PK2. J. Gen. Microbiol. 1991, 137, 2281–2286. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, H.; Takahira, O.; Masato, T.; Kazuhiro, N.; Takao, F.; Sumio, T.; Yoshimasa, M. Two reactions are simultaneously catalyzed by a single enzyme: The arginine-dependent simultaneous formation of two products, ethylene and succinate, from 2-oxoglutarate by an enzyme from Pseudomonas syringae. Biochem. Biophys. Res. Commun. 1992, 188, 483–489. [Google Scholar] [CrossRef]
- Knoop, H.; Steuer, R. A Computational Analysis of Stoichiometric Constraints and Trade-Offs in Cyanobacterial Biofuel Production. Front. Bioeng. Biotechnol. 2015, 3. [Google Scholar] [CrossRef]
- Sengupta, A.; Sunder, A.V.; Sohoni, S.V.; Wangikar, P.P. Fine-Tuning Native Promoters of Synechococcus elongatus PCC 7942 to Develop a Synthetic Toolbox for Heterologous Protein Expression. ACS Synth. Biol. 2019, 8, 1219–1223. [Google Scholar] [CrossRef] [PubMed]
- Asplund-Samuelsson, J.; Janasch, M.; Hudson, E.P. Thermodynamic analysis of computed pathways integrated into the metabolic networks of E. coli and Synechocystis reveals contrasting expansion potential. Metab. Eng. 2018, 45, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Brune, D.; Vermaas, W.F.J. The γ-aminobutyric acid shunt contributes to closing the tricarboxylic acid cycle in Synechocystis sp. PCC 6803. Mol. Microbiol. 2014, 93, 786–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stockel, J.; Welsh, E.A.; Liberton, M.; Kunnvakkam, R.; Aurora, R.; Pakrasi, H.B. Global transcriptomic analysis of Cyanothece 51142 reveals robust diurnal oscillation of central metabolic processes. Proc. Natl. Acad. Sci. USA 2008, 105, 6156–6161. [Google Scholar] [CrossRef] [Green Version]
- Hewett, J.P.; Wolfe, A.K.; Bergmann, R.A.; Stelling, S.C.; Davis, K.L. Human health and environmental risks posed by synthetic biology R D for energy applications: A literature analysis. J. ABSA Int. 2016, 21, 177–184. [Google Scholar] [CrossRef] [Green Version]
- Yen, U.C.; Huang, T.C.; Yen, T.C. Observation of the circadian photosynthetic rhythm in cyanobacteria with a dissolved-oxygen meter. Plant. Sci. 2004, 166, 949–952. [Google Scholar] [CrossRef]
- Sengupta, A.; Wangikar, P.P. A method to compute instantaneous oxygen evolution rates in cyanobacterial cultures grown in shake flasks. Eng. Reports 2020, 2. [Google Scholar] [CrossRef]
- Gründel, M.; Scheunemann, R.; Lockau, W.; Zilliges, Y. Impaired glycogen synthesis causes metabolic overflow reactions and affects stress responses in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 2012, 158, 3032–3043. [Google Scholar] [CrossRef]
- Salar-García, M.J.; Bernal, V.; Pastor, J.M.; Salvador, M.; Argandoña, M.; Nieto, J.J.; Vargas, C.; Cánovas, M. Understanding the interplay of carbon and nitrogen supply for ectoines production and metabolic overflow in high density cultures of Chromohalobacter salexigens. Microb. Cell Fact. 2017, 16. [Google Scholar] [CrossRef] [Green Version]
- Yao, L.; Cengic, I.; Anfelt, J.; Hudson, E.P. Multiple Gene Repression in Cyanobacteria Using CRISPRi. ACS Synth. Biol. 2016, 5, 207–212. [Google Scholar] [CrossRef]
- Díaz-Troya, S.; Roldán, M.; Mallén-Ponce, M.J.; Ortega-Martínez, P.; Florencio, F.J. Lethality caused by ADP-glucose accumulation is suppressed by salt-induced carbon flux redirection in cyanobacteria. J. Exp. Bot. 2019, 71, 2005–2017. [Google Scholar] [CrossRef]
- Cano, M.; Holland, S.C.; Artier, J.; Burnap, R.L.; Ghirardi, M.; Morgan, J.A.; Yu, J. Glycogen Synthesis and Metabolite Overflow Contribute to Energy Balancing in Cyanobacteria. Cell Rep. 2018, 23, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Visser, D.; Heijnen, J.J. The mathematics of Metabolic Control Analysis revisited. Metab. Eng. 2002, 4, 114–123. [Google Scholar] [CrossRef]
- Hendry, J.I.; Prasannan, C.; Ma, F.; Möllers, K.B.; Jaiswal, D.; Digmurti, M.; Allen, D.K.; Frigaard, N.U.; Dasgupta, S.; Wangikar, P.P. Rerouting of carbon flux in a glycogen mutant of cyanobacteria assessed via isotopically non-stationary 13C metabolic flux analysis. Biotechnol. Bioeng. 2017, 114, 2298–2308. [Google Scholar] [CrossRef] [PubMed]
- Prasannan, C.B.; Jaiswal, D.; Davis, R.; Wangikar, P.P. An improved method for extraction of polar and charged metabolites from cyanobacteria. PLoS ONE 2018, 13. [Google Scholar] [CrossRef] [PubMed]
- Prasannan, C.B.; Mishra, V.; Jaiswal, D.; Wangikar, P.P. Mass Isotopologue Distribution of dimer ion adducts of intracellular metabolites for potential applications in 13C Metabolic Flux Analysis. PLoS ONE 2019, 14. [Google Scholar] [CrossRef] [PubMed]
- Nöh, K.; Grönke, K.; Luo, B.; Takors, R.; Oldiges, M.; Wiechert, W. Metabolic flux analysis at ultra short time scale: Isotopically non-stationary 13C labeling experiments. J. Biotechnol. 2007, 129, 249–267. [Google Scholar] [CrossRef] [PubMed]
- Abernathy, M.H.; Czajka, J.J.; Allen, D.K.; Hill, N.C.; Jeffrey, C.; Tang, Y.J. Cyanobacterial carboxysome mutant analysis reveals the influence of enzyme compartmentalization on cellular metabolism and metabolic network rigidity. Metab. Eng. 2019, 54, 222–231. [Google Scholar] [CrossRef]
- Janasch, M.; Asplund-Samuelsson, J.; Steuer, R.; Hudson, E.P. Kinetic modeling of the Calvin cycle identifies flux control and stable metabolomes in Synechocystis carbon fixation. J. Exp. Bot. 2019, 70, 1017–1031. [Google Scholar]
- Liang, F.; Lindberg, P.; Lindblad, P. Engineering photoautotrophic carbon fixation for enhanced growth and productivity. Sustain. Energy Fuels 2018, 2, 2583–2600. [Google Scholar] [CrossRef] [Green Version]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Cai, Y.; Guo, Y.; Chen, F.; Zhu, Z.J. MetDIA: Targeted Metabolite Extraction of Multiplexed MS/MS Spectra Generated by Data-Independent Acquisition. Anal. Chem. 2016, 88, 8757–8764. [Google Scholar] [CrossRef] [PubMed]
- Ungerer, J.; Wendt, K.E.; Hendry, J.I.; Maranas, C.D.; Pakrasi, H.B. Comparative genomics reveals the molecular determinants of rapid growth of the cyanobacterium Synechococcus elongatus UTEX 2973. Proc. Natl. Acad. Sci. USA 2018, 115, E11761–E11770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison III, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Bandyopadhyay, A.; Stöckel, J.; Min, H.; Sherman, L.A.; Pakrasi, H.B. High rates of photobiological H2 production by a cyanobacterium under aerobic conditions. Nat. Commun. 2010, 1, 139. [Google Scholar] [CrossRef]
- Millard, P.; Delépine, B.; Guionnet, M.; Heuillet, M.; Bellvert, F.; Létisse, F. IsoCor: Isotope correction for high-resolution MS labeling experiments. Bioinformatics 2019, 35, 4484–4487. [Google Scholar] [CrossRef]
- Millard, P.; Letisse, F.; Sokol, S.; Portais, J.C. IsoCor: Correcting MS data in isotope labeling experiments. Bioinformatics 2012, 28, 1294–1296. [Google Scholar] [CrossRef] [Green Version]
- Varshney, P.; Beardall, J.; Bhattacharya, S.; Wangikar, P.P. Effect of elevated carbon dioxide and nitric oxide on the physiological responses of two green algae, Asterarcys quadricellulare and Chlorella sorokiniana. J. Appl. Phycol. 2020, 32, 189–204. [Google Scholar] [CrossRef]
- Pierangelini, M.; Stojkovic, S.; Orr, P.T.; Beardall, J. Photosynthetic characteristics of two Cylindrospermopsis raciborskii strains differing in their toxicity. J. Phycol. 2014, 50, 292–302. [Google Scholar] [CrossRef]
- Wolter, K.M. Taylor Series Methods. In Introduction to Variance Estimation; Springer: New York, NY, USA, 2007; pp. 226–271. [Google Scholar]








| SL. No. | Year | Organisms | Genetic Modifications | Peak Ethylene Productivity (µmole/g/h) | Peak Succinate Productivity (mg/g/h) | Ref. |
|---|---|---|---|---|---|---|
| 1 | 2013 | Synechocystis sp. PCC 6803 | 2X efe | 44 | --- | [25] |
| 2 | 2015 | Synechocystis sp. PCC 6803 | 1X efe, modified RBS | 128.21 | --- | [30] |
| 3 | 2015 | Synechocystis sp. PCC 6803 | * | 86.61 | --- | [31] |
| 4 | 2015 | Synechocystis sp. PCC 6803 | ∆ackA and O.E.(SigE) | --- | 0.2 | [32] |
| 5 | 2016 | S. elongatus PCC 7942 | O.E.(gabD/kgd/ppc/gltA) | --- | 10.67 | [19] |
| 6 | 2017 | Synechocystis sp. PCC 6803 | # | 439 | --- | [26] |
| 7 | 2017 | Synechocystis sp. PCC 6803 | slr0168:: Ptrc-Rbs30-efe-Kmr | 15.82 | --- | [33] |
| 8 | 2018 | Synechocystis sp. PCC 6803 | O.E.(ppc) and ∆ackA | --- | 1.0 | [22] |
| 9 | 2019 | S. elongatus PCC 7942 | NSI:: Ptrc- efe-TrrnB | 25 | --- | [24] |
| 10 | 2020 | Synechocystis sp. PCC 6803 | PnrsB-efe, O.E.(2Xppc, ppsA7002) | 118.84 | --- | [29] |
| 11 | S. elongatus PCC 11801 | NSI::PcpcB300-efe-TrrnB | 338.26 | 121.1 | This study |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sengupta, A.; Pritam, P.; Jaiswal, D.; Bandyopadhyay, A.; Pakrasi, H.B.; Wangikar, P.P. Photosynthetic Co-production of Succinate and Ethylene in a Fast-Growing Cyanobacterium, Synechococcus elongatus PCC 11801. Metabolites 2020, 10, 250. https://doi.org/10.3390/metabo10060250
Sengupta A, Pritam P, Jaiswal D, Bandyopadhyay A, Pakrasi HB, Wangikar PP. Photosynthetic Co-production of Succinate and Ethylene in a Fast-Growing Cyanobacterium, Synechococcus elongatus PCC 11801. Metabolites. 2020; 10(6):250. https://doi.org/10.3390/metabo10060250
Chicago/Turabian StyleSengupta, Annesha, Prem Pritam, Damini Jaiswal, Anindita Bandyopadhyay, Himadri B. Pakrasi, and Pramod P. Wangikar. 2020. "Photosynthetic Co-production of Succinate and Ethylene in a Fast-Growing Cyanobacterium, Synechococcus elongatus PCC 11801" Metabolites 10, no. 6: 250. https://doi.org/10.3390/metabo10060250
APA StyleSengupta, A., Pritam, P., Jaiswal, D., Bandyopadhyay, A., Pakrasi, H. B., & Wangikar, P. P. (2020). Photosynthetic Co-production of Succinate and Ethylene in a Fast-Growing Cyanobacterium, Synechococcus elongatus PCC 11801. Metabolites, 10(6), 250. https://doi.org/10.3390/metabo10060250