Identification of Plasma Lipidome Changes Associated with Low Dose Space-Type Radiation Exposure in a Murine Model
Abstract
1. Introduction
2. Results
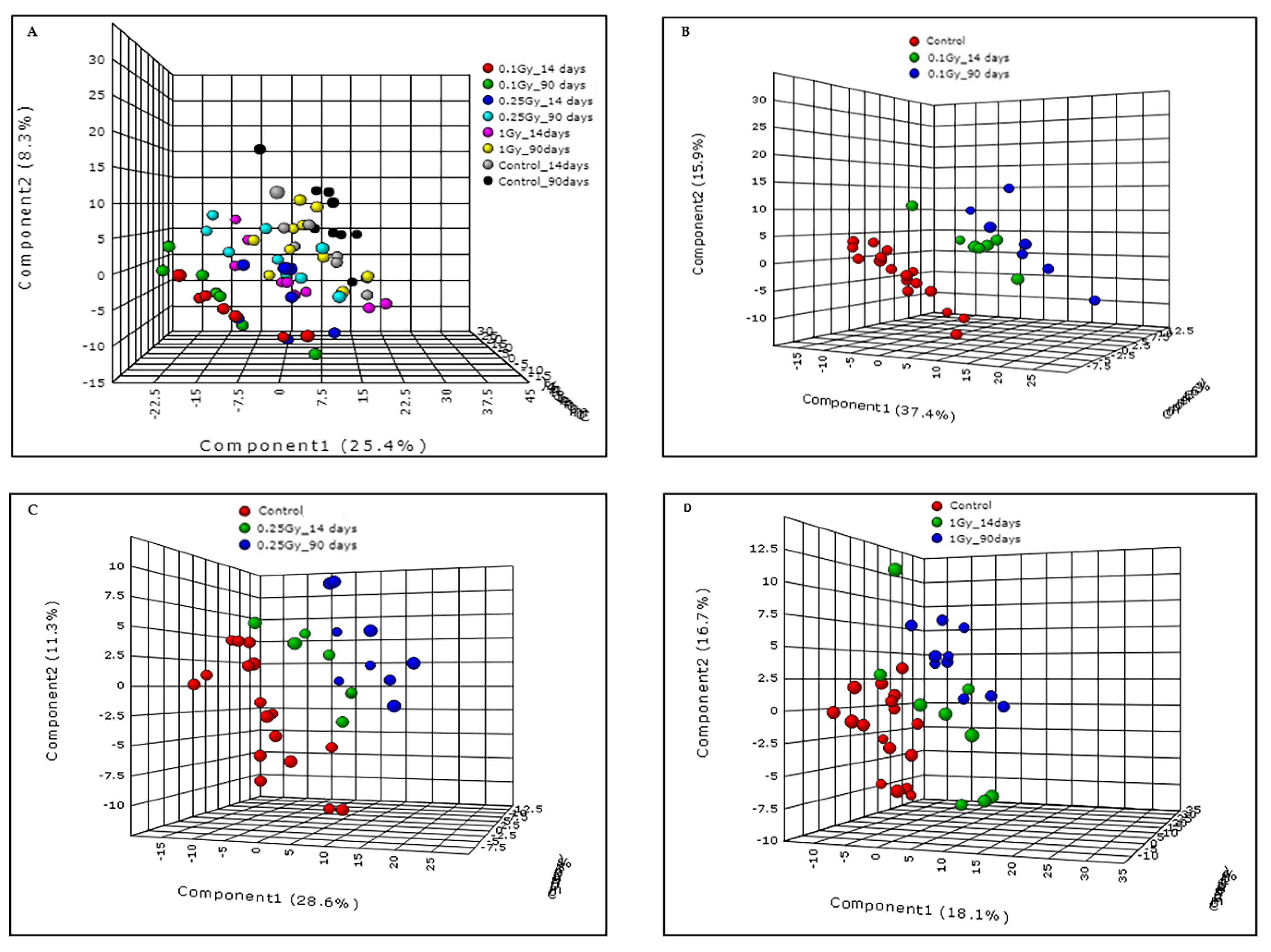
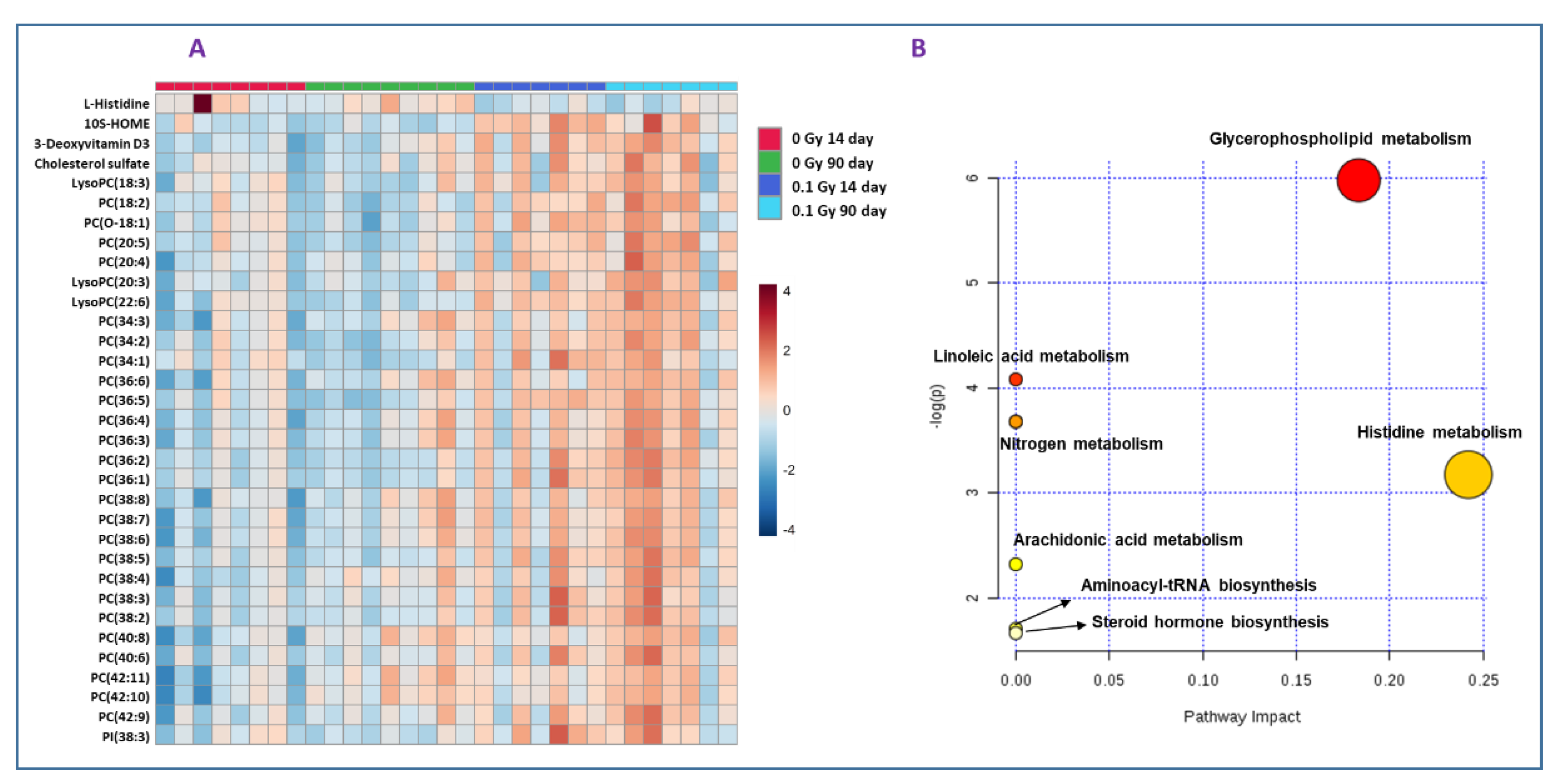
2.1. Exposure to 16O Radiation Elicits Robust Changes in Plasma Lipidome
2.2. Metabolic Changes in Mouse Plasma Following Exposure to 1H
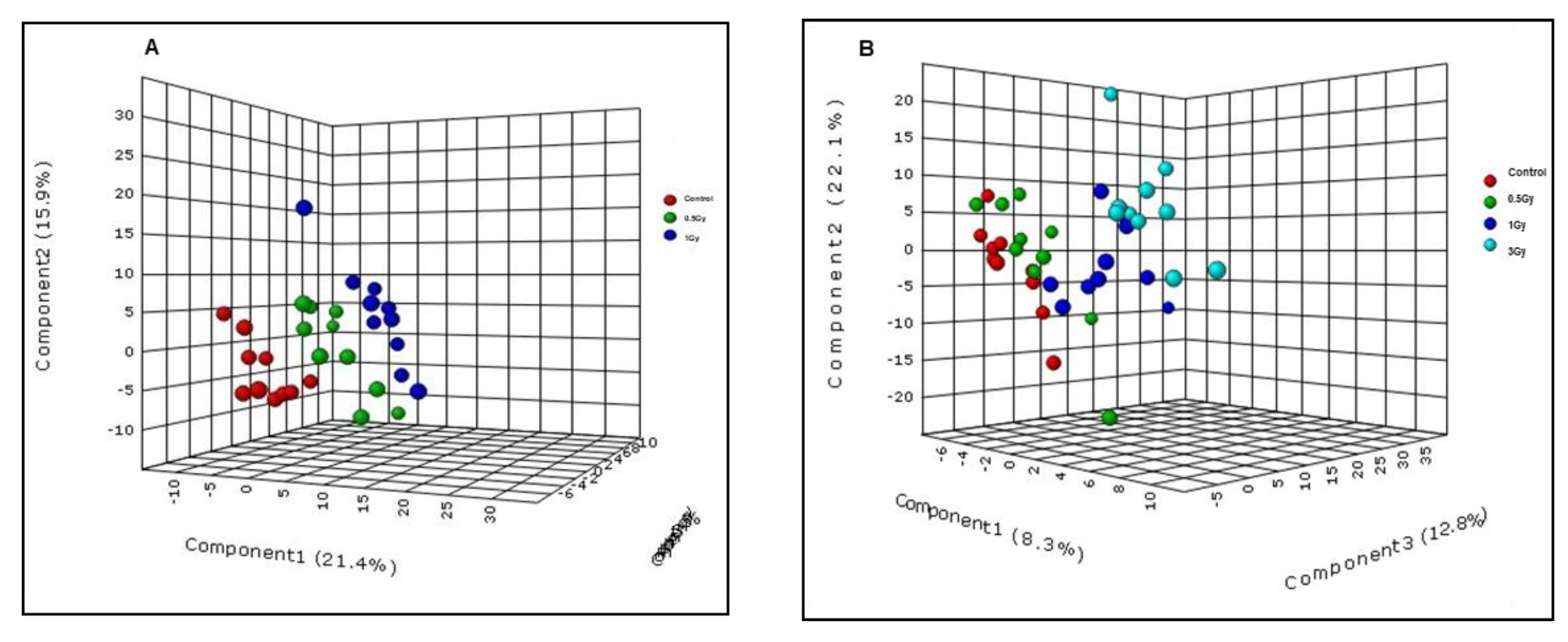
2.3. Low LET γ-Radiation Induced Changes in Plasma of Male C57BL/6J Mice
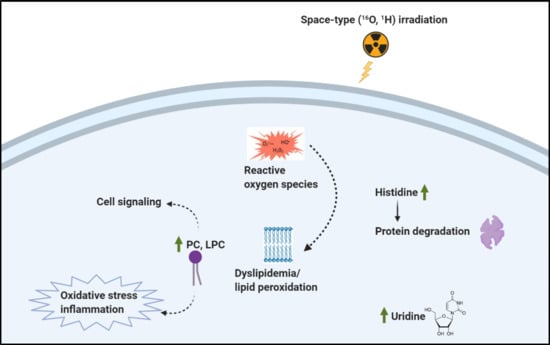
3. Discussion
4. Materials and Methods
4.1. Animals and Radiation Exposure
4.2. Plasma Collection
4.3. Plasma Metabolomics/Lipidomics Using UPLC-QToF
4.4. Statistical Analyses of Mass Spectrometry Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Durante, M.; Cucinotta, F.A. Heavy ion carcinogenesis and human space exploration. Nat. Rev. Cancer 2008, 8, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Blakely, E.A. Biological effects of cosmic radiation: Deterministic and stochastic. Health Phys. 2000, 79, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Badhwar, G.D.; O’Neill, P.M. Long-term modulation of galactic cosmic radiation and its model for space exploration. Adv. Space Res. 1994, 14, 749–757. [Google Scholar] [CrossRef]
- Cucinotta, F.A.; Durante, M. Cancer risk from exposure to galactic cosmic rays: Implications for space exploration by human beings. Lancet Oncol. 2006, 7, 431–435. [Google Scholar] [CrossRef]
- Zeitlin, C. Physical interactions of charged particles for radiotherapy and space applications. Health Phys. 2012, 103, 540–546. [Google Scholar] [CrossRef]
- Hu, S.; Kim, M.-H.Y.; McClellan, G.E.; Cucinotta, F.A. Modeling the acute health effects of astronauts from exposure to large solar particle events. Health Phys. 2009, 96, 465–476. [Google Scholar] [CrossRef]
- Datta, K.; Suman, S.; Trani, D.; Doiron, K.; Rotolo, J.A.; Kallakury, B.V.S.; Kolesnick, R.; Cole, M.F.; Fornace, A.J., Jr. Accelerated hematopoietic toxicity by high energy 56Fe radiation. Int. J. Radiat. Biol. 2011, 88, 213–222. [Google Scholar] [CrossRef]
- Suman, S.; Datta, K.; Trani, D.; Laiakis, E.C.; Strawn, S.J.; Fornace, A.J., Jr. Relative biological effectiveness of 12C and 28Si radiation in C57BL/6J mice. Radiat. Environ. Biophys. 2012, 51, 303–309. [Google Scholar] [CrossRef][Green Version]
- Reisz, J.A.; Bansal, N.; Qian, J.; Zhao, W.; Furdui, C.M. Effects of ionizing radiation on biological molecules—Mechanisms of damage and emerging methods of detection. Antioxid. Redox Signal. 2014, 21, 260–292. [Google Scholar] [CrossRef]
- Jones, J.A.; Riggs, P.K.; Yang, T.C.; Pedemonte, C.H.; Clarke, M.S.F.; Feeback, D.L.; Au, W.W. Ionizing radiation-induced bioeffects in space and strategies to reduce cellular injury and carcinogenesis. Aviat. Space Environ. Med. 2007, 78, A67–A78. [Google Scholar]
- Romano, E.; Ferrucci, L.; Nicolai, F.; Derme, V.; De Stefano, G.F. Increase of chromosomal aberrations induced by ionising radiation in peripheral blood lymphocytes of civil aviation pilots and crew members. Mutat. Res. 1997, 377, 89–93. [Google Scholar] [CrossRef]
- Burlaka, A.P.; Druzhyna, M.O.; Vovk, A.V.; Lukin, S.M. Disordered redox metabolism of brain cells in rats exposed to low doses of ionizing radiation or UHF electromagnetic radiation. Exp. Oncol. 2016, 38, 238–241. [Google Scholar] [CrossRef]
- Sasi, S.P.; Yan, X.; Zuriaga-Herrero, M.; Gee, H.; Lee, J.; Mehrzad, R.; Song, J.; Onufrak, J.; Morgan, J.; Enderling, H.; et al. Different sequences of fractionated low-dose proton and single iron-radiation-induced divergent biological responses in the heart. Radiat. Res. 2017, 188, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Howe, A.; Kiffer, F.; Alexander, T.C.; Sridharan, V.; Wang, J.; Ntagwabira, F.; Rodriguez, A.; Boerma, M.; Allen, A.R. Long-term changes in cognition and physiology after low-dose 16O irradiation. Int. J. Mol. Sci. 2019, 20, 188. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, Y.; Wang, S.; Guan, H.; Hu, S.; Huang, R.; Zhou, P. Impact of low-dose ionising radiation on the composition of the gut microbiota of mice. Toxicol. Sci. 2019, 258–268. [Google Scholar] [CrossRef]
- Kwon, Y.-K.; Ha, I.J.; Bae, H.-W.; Jang, W.G.; Yun, H.J.; Kim, S.R.; Lee, E.K.; Kang, C.-M.; Hwang, G.-S. Dose-dependent metabolic alterations in human cells exposed to gamma irradiation. PLoS ONE 2014, 9, e113573. [Google Scholar] [CrossRef]
- Cheema, A.K.; Suman, S.; Kaur, P.; Singh, R.; Fornace, A.J., Jr.; Datta, K. Long-term differential changes in mouse intestinal metabolomics after gamma and heavy ion radiation exposure. PLoS ONE 2014, 9, e87079. [Google Scholar] [CrossRef]
- Lin, H.-R.; Liao, C.-C.; Lin, T.-C. Improved identification of multiple drugs of abuse and relative metabolites in urine samples using liquid chromatography/triple quadrupole mass spectrometry coupled with a library search. Rapid Commun. Mass Spectrom. 2014, 28, 2043–2053. [Google Scholar] [CrossRef]
- De Leoz, M.L.A.; Simon-Manso, Y.; Woods, R.J.; Stein, S.E. Cross-ring fragmentation patterns in the tandem mass spectra of underivatized sialylated oligosaccharides and their special suitability for spectrum library searching. J. Am. Soc. Mass Spectrom. 2019, 30, 426–438. [Google Scholar] [CrossRef]
- Cooper, B.T.; Yan, X.; Simon-Manso, Y.; Tchekhovskoi, D.V.; Mirokhin, Y.A.; Stein, S.E. Hybrid search: A method for identifying metabolites absent from tandem mass spectrometry libraries. Anal. Chem. 2019, 91, 13924–13932. [Google Scholar] [CrossRef]
- Hanson, W.R.; Fry, R.J.M.; Sallese, A.R.; Frischer, H.; Ahmad, T.; Ainsworth, E.J. Comparison of intestine and bone marrow radiosensitivity of the BALB/c and the C57BL/6 mouse strains and their B6CF1 Offspring. Radiat. Res. 1987, 110, 340–352. [Google Scholar] [CrossRef] [PubMed]
- Virmani, R.; Farb, A.; Carter, A.J.; Jones, R.M. Pathology of radiation-induced coronary artery disease in human and pig. Cardiovasc. Radiat. Med. 1999, 1, 98–101. [Google Scholar] [CrossRef]
- Wang, C.; Yang, J.; Nie, J. Plasma phospholipid metabolic profiling and biomarkers of rats following radiation exposure based on liquid chromatography-mass spectrometry technique. Biomed. Chromatogr. 2009, 23, 1079–1085. [Google Scholar] [CrossRef] [PubMed]
- Seawright, J.W.; Sridharan, V.; Landes, R.D.; Cao, M.; Singh, P.; Koturbash, I.; Mao, X.-W.; Miousse, I.R.; Singh, S.P.; Nelson, G.A.; et al. Effects of low-dose oxygen ions and protons on cardiac function and structure in male C57BL/6J mice. Life Sci. Space Res. (Amst) 2019, 20, 72–84. [Google Scholar] [CrossRef]
- Tapio, S. Pathology and biology of radiation-induced cardiac disease. J. Radiat. Res. 2016, 57, 439–448. [Google Scholar] [CrossRef]
- Sutherland, B.M.; Bennett, P.V.; Sidorkina, O.; Laval, J. Clustered damages and total lesions induced in DNA by ionizing radiation: Oxidized bases and strand breaks. Biochemistry 2000, 39, 8026–8031. [Google Scholar] [CrossRef]
- Goodhead, D.T. Initial events in the cellular effects of ionizing radiations: Clustered damage in DNA. Int. J. Radiat. Biol. 1994, 65, 7–17. [Google Scholar] [CrossRef]
- Mishra, B.; Ripperdan, R.; Ortiz, L.; Luderer, U. Very low doses of heavy oxygen ion radiation induce premature ovarian failure. Reproduction 2017, 154, 123–133. [Google Scholar] [CrossRef]
- Choudhary, D.; Srivastava, M.; Sarma, A.; Kale, R.K. Effect of high linear energy transfer radiation on biological membranes. Radiat. Environ. Biophys. 1998, 37, 177–185. [Google Scholar] [CrossRef]
- Zhang, W.; Sun, G.; Aitken, D.; Likhodii, S.; Liu, M.; Martin, G.; Furey, A.; Randell, E.; Rahman, P.; Jones, G.; et al. Lysophosphatidylcholines to phosphatidylcholines ratio predicts advanced knee osteoarthritis. Rheumatology (Oxford) 2016, 55, 1566–1574. [Google Scholar] [CrossRef]
- Casero, D.; Gill, K.; Sridharan, V.; Koturbash, I.; Nelson, G.; Hauer-Jensen, M.; Boerma, M.; Braun, J.; Cheema, A.K. Space-type radiation induces multimodal responses in the mouse gut microbiome and metabolome. Microbiome 2017, 5, 105. [Google Scholar] [CrossRef] [PubMed]
- York, J.M.; Blevins, N.A.; Meling, D.D.; Peterlin, M.B.; Gridley, D.S.; Cengel, K.A.; Freund, G.G. The biobehavioral and neuroimmune impact of low-dose ionizing radiation. Brain Behav. Immun. 2012, 26, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Leyko, W.; Bartosz, G. Membrane effects of ionizing radiation and hyperthermia. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1986, 49, 743–770. [Google Scholar] [CrossRef] [PubMed]
- Okunieff, P.; Chen, Y.; Maguire, D.J.; Huser, A.K. Molecular markers of radiation-related normal tissue toxicity. Cancer Metastasis Rev. 2008, 27, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Azzam, E.I.; Jay-Gerin, J.-P.; Pain, D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett. 2012, 327, 48–60. [Google Scholar] [CrossRef]
- Cheema, A.K.; Pathak, R.; Zandkarimi, F.; Kaur, P.; Alkhalil, L.; Singh, R.; Zhong, X.; Ghosh, S.; Aykin-Burns, N.; Hauer-Jensen, M. Liver metabolomics reveals increased oxidative stress and fibrogenic potential in gfrp transgenic mice in response to ionizing radiation. J. Proteome Res. 2014, 13, 3065–3074. [Google Scholar] [CrossRef]
- Koundouros, N.; Poulogiannis, G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef]
- Liu, H.; Wang, Z.; Zhang, X.; Qiao, Y.; Wu, S.; Dong, F.; Chen, Y. Selection of candidate radiation biomarkers in the serum of rats exposed to gamma-rays by GC/TOFMS-based metabolomics. Radiat. Prot. Dosim. 2013, 154, 9–17. [Google Scholar] [CrossRef]
- Lee, D.Y.; Bowen, B.P.; Nguyen, D.H.; Parsa, S.; Huang, Y.; Mao, J.-H.; Northen, T.R. Low-dose ionizing radiation-induced blood plasma metabolic response in a diverse genetic mouse population. Radiat. Res. 2012, 178, 551–555. [Google Scholar] [CrossRef]
- Wu, R.; Zeng, Y. Does angiotensin II-aldosterone have a role in radiation-induced heart disease? Med. Hypotheses 2009, 72, 263–266. [Google Scholar] [CrossRef]
- LaRosa, J.C.; Hunninghake, D.; Bush, D.; Criqui, M.H.; Getz, G.S.; Gotto, A.M., Jr.; Grundy, S.M.; Rakita, L.; Robertson, R.M.; Weisfeldt, M.L.; et al. The cholesterol facts. A summary of the evidence relating dietary fats, serum cholesterol, and coronary heart disease. A joint statement by the American Heart Association and the National Heart, Lung, and Blood Institute. Commissioned by the task force on cholesterol issues, American Heart Association. Circulation 1990, 81, 1721–1733. [Google Scholar] [CrossRef]
- Circulating cholesterol level and risk of death from cancer in men aged 40 to 69 years. Experience of an international collaborative group. JAMA 1982, 248, 2853–2859. [Google Scholar] [CrossRef] [PubMed]
- Sherwin, R.W.; Wentworth, D.N.; Cutler, J.A.; Hulley, S.B.; Kuller, L.H.; Stamler, J. Serum cholesterol levels and cancer mortality in 361,662 men screened for the multiple risk factor intervention trial. JAMA 1987, 257, 943–948. [Google Scholar] [CrossRef] [PubMed]
- Mathews, C.K.; Sinha, N.K. Are DNA precursors concentrated at replication sites? Proc. Natl. Acad. Sci. USA 1982, 79, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Datta, K.; Suman, S.; Kallakury, B.V.S.; Fornace, A.J., Jr. Exposure to heavy ion radiation induces persistent oxidative stress in mouse intestine. PLoS ONE 2012, 7, e42224. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Upadhyay, M.; Rajagopal, M.; Gill, K.; Li, Y.; Bansal, S.; Sridharan, V.; Tyburski, J.B.; Boerma, M.; Cheema, A.K. Identification of Plasma Lipidome Changes Associated with Low Dose Space-Type Radiation Exposure in a Murine Model. Metabolites 2020, 10, 252. https://doi.org/10.3390/metabo10060252
Upadhyay M, Rajagopal M, Gill K, Li Y, Bansal S, Sridharan V, Tyburski JB, Boerma M, Cheema AK. Identification of Plasma Lipidome Changes Associated with Low Dose Space-Type Radiation Exposure in a Murine Model. Metabolites. 2020; 10(6):252. https://doi.org/10.3390/metabo10060252
Chicago/Turabian StyleUpadhyay, Maarisha, Meena Rajagopal, Kirandeep Gill, Yaoxiang Li, Shivani Bansal, Vijayalakshmi Sridharan, John B. Tyburski, Marjan Boerma, and Amrita K. Cheema. 2020. "Identification of Plasma Lipidome Changes Associated with Low Dose Space-Type Radiation Exposure in a Murine Model" Metabolites 10, no. 6: 252. https://doi.org/10.3390/metabo10060252
APA StyleUpadhyay, M., Rajagopal, M., Gill, K., Li, Y., Bansal, S., Sridharan, V., Tyburski, J. B., Boerma, M., & Cheema, A. K. (2020). Identification of Plasma Lipidome Changes Associated with Low Dose Space-Type Radiation Exposure in a Murine Model. Metabolites, 10(6), 252. https://doi.org/10.3390/metabo10060252