Comparison of Metabolomic Profiles of Organs in Mice of Different Strains Based on SPME-LC-HRMS
Abstract
1. Introduction
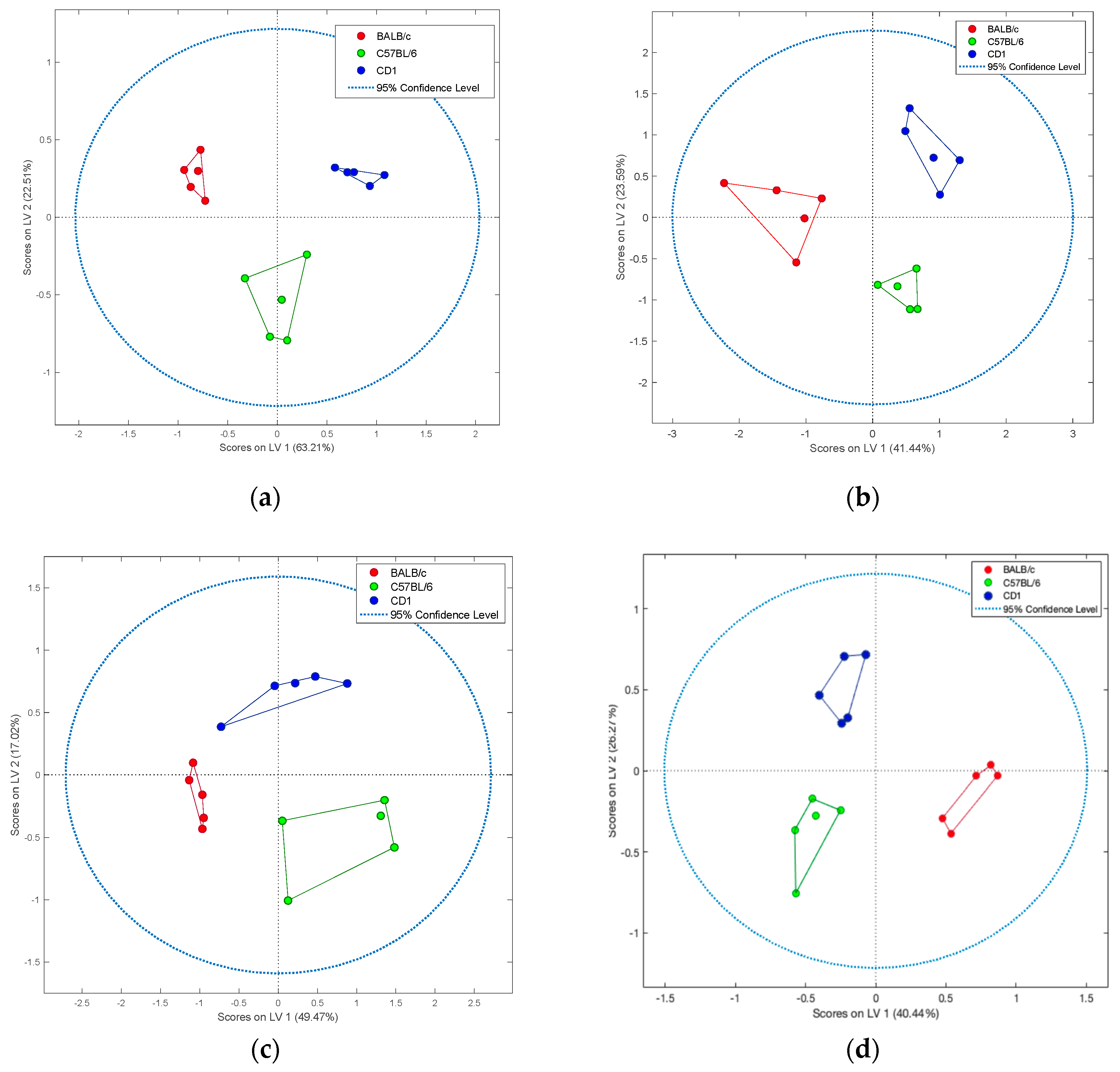
2. Results
3. Discussion
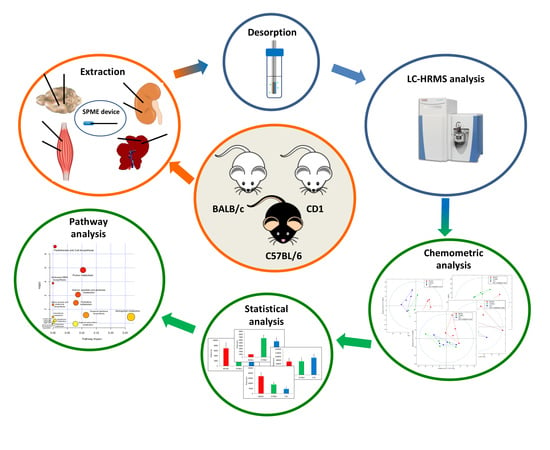
4. Materials and Methods
4.1. Chemicals
4.2. Materials
4.3. Animal Handling and Tissue Collection
4.4. Solid Phase Procedure and Sample Preparation
4.5. Liquid Chromatography–High Resolution Mass Spectrometry Analysis (LC–HRMS)
4.6. Data Processing and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bryda, E.C. The mighty mouse: The impact of rodents on advances in biomedical research. Mo. Med. 2013, 110, 207–211. [Google Scholar] [PubMed]
- Perlman, R.L. Mouse models of human disease: An evolutionary perspective. Evol. Med. Public Health 2016, 1, 170–176. [Google Scholar] [CrossRef] [PubMed]
- 2019 Report on the Statistics on the Use of Animals for Scientific Purposes in the Member States of the European Union in 2015–2017; The Commission to the European Parliament and the Council: Brussels, Belgium, 2020.
- The Importance of Genetic Background in Mouse Models. Available online: https://www.biocompare.com/Bench-Tips/341470-The-Importance-of-Genetic-Background-in-Mouse-Models (accessed on 4 May 2020).
- Hedrich, H.J. Strains, stocks and mutant mice. In The Laboratory Mouse, 2nd ed.; Elsevier Ltd.: Amsterdam, The Netherlands, 2012; pp. 37–56. [Google Scholar]
- Potter, M. History of the BALB/c family. In The BALB/c Mouse. Current Topics in Microbiology and Immunology; Potter, M., Ed.; Springer: Berlin/Heidelberg, Germany, 1985; Volume 122, pp. 1–5. [Google Scholar]
- Tuttle, A.H.; Philip, V.M.; Chesler, E.J.; Mogil, J.S. Comparing phenotypic variation between inbred and outbred mice. Nat. Methods 2018, 15, 994–996. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.L. Understanding mouse models of disease through metabolomics. Curr. Opin. Chem. Biol. 2006, 10, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Kosmides, A.K.; Kamisoglu, K.; Calvano, S.E.; Corbett, S.A.; Androulakis, I.P. Metabolomic fingerprinting: Challenges and opportunities. Crit. Rev. Biomed. Eng. 2013, 41, 205–221. [Google Scholar] [CrossRef]
- Kim, S.J.; Kim, S.H.; Kim, J.H.; Hwang, S.; Yoo, H.J. Understanding metabolomics in biomedical research. Endocrinol. Metab. 2016, 31, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted metabolomics strategies—challenges and emerging directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef]
- Wishart, D.S. Metabolomics for investigating physiological and pathophysiological processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef]
- Klupczyńska, A.; Dereziński, P.; Kokot, Z.J. Metabolomics in medical sciences—Trends, challenges and perspectives. Acta Pol. Pharm. 2015, 72, 629–641. [Google Scholar]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Nash, W.J.; Dunn, W.B. From mass to metabolite in human untargeted metabolomics: Recent advances in annotation of metabolites applying liquid chromatography-mass spectrometry data. TrAC Trends Anal. Chem. 2019, 120. [Google Scholar] [CrossRef]
- Vinayavekhin, N.; Saghatelian, A. Untargeted metabolomics. Curr. Protoc. Mol. Biol. 2010, 90. [Google Scholar] [CrossRef]
- Naz, S.; Moreira Dos Santos, D.C.; García, A.; Barbas, C. Analytical protocols based on LC-MS, GC-MS and CE-MS for nontargeted metabolomics of biological tissues. Bioanalysis 2014, 6, 1657–1677. [Google Scholar] [CrossRef]
- Zukunft, S.; Prehn, C.; Röhring, C.; Möller, G.; Hraběde Angelis, M.; Adamski, J.; Tokarz, J. High-throughput extraction and quantification method for targeted metabolomics in murine tissues. Metabolomics 2018, 14. [Google Scholar] [CrossRef] [PubMed]
- Stryjak, I.; Warmuzińska, N.; Bogusiewicz, J.; Łuczykowski, K.; Bojko, B. Monitoring of the influence of long-term oxidative stress and ischemia on the condition of kidneys using solid-phase microextraction chemical biopsy coupled with liquid chromatography–high–resolution mass spectrometry. J. Sep. Sci. 2020, 43, 1867–1878. [Google Scholar] [CrossRef] [PubMed]
- Bojko, B.; Gorynski, K.; Gomez-Rios, G.A.; Knaak, J.M.; Machuca, T.; Spetzler, V.N.; Cudjoe, E.; Hsin, M.; Cypel, M.; Selzner, M.; et al. Solid phase microextraction fills the gap in tissue sampling protocols. Anal. Chim. Acta 2013, 803, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Bojko, B.; Pawliszyn, J. In vivo and ex vivo SPME: A low invasive sampling and sample preparation tool in clinical bioanalysis. Bioanalysis 2014, 6, 1227–1239. [Google Scholar] [CrossRef] [PubMed]
- Vuckovic, D.; de Lannoy, I.; Gien, B.; Shirey, E.; Sidisky, L.M.; Dutta, S.; Pawliszyn, J. In vivo solid-phase microextraction: Capturing the elusive portion of metabolome. Angew. Chem. 2011, 50, 5344–5348. [Google Scholar] [CrossRef] [PubMed]
- Vuckovic, D.; Risticevic, S.; Pawliszyn, J. In vivo solid-phase microextraction in metabolomics: Opportunities for the direct investigation of biological systems. Angew. Chem. 2011, 50, 5618–5628. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhou, L.; Chen, H.; Wang, C.Z.; Xia, Z.; Yuan, C.S. Solid-phase microextraction technology for in vitro and in vivo metabolite analysis. TrAC Trends Anal. Chem. 2016, 80, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Overmyer, K.A.; Thonusin, C.; Qi, N.R.; Burant, C.F.; Evans, C.R. Impact of anesthesia and euthanasia on metabolomics of mammalian tissues: Studies in a C57BL/6J mouse model. PLoS ONE 2015, 10, e0117232. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Yim, S.H.; Lee, S.-G.; Kim, E.B.; Lee, S.R.; Chang, K.T.; Buffenstein, R.; Lewis, K.N.; Park, T.J.; Miller, R.A.; et al. Organization of the mammalian metabolome according to organ function, lineage specialization, and longevity. Cell Metab. 2015, 22, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.M.; Greenaway, S.; White, J.K.; Fuchs, H.; Gailus-Durner, V.; Wells, S.; Sorg, T.; Wong, K.; Bedu, E.; Cartwright, E.J.; et al. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol. 2013, 14, R82. [Google Scholar] [CrossRef] [PubMed]
- Wells, A.E. Discovering the impact of diet, genetic, and sex effects on adipose, skeletal muscle, and liver metabolomes in mice using untargeted liquid-chromatography mass-spectrometry. Ph.D. Dissertation, University of Tennessee, Knoxville, TN, USA, December 2017. [Google Scholar]
- Waagepetersen, H.S.; Sonnewald, U.; Schousboe, A. 1 Glutamine, Glutamate, and GABA: Metabolic Aspects. In Handbook of Neurochemistry and Molecular Neurobiology; Lajtha, A., Oja, S.S., Schousboe, A., Saransaari, P., Eds.; Springer: Boston, MA, USA, 2007; pp. 1–21. [Google Scholar]
- Wyse, A.T.S.; Netto, C.A. Behavioral and neurochemical effects of proline. Metab. Brain Dis. 2011, 26, 159–172. [Google Scholar] [CrossRef]
- Nadler, J.V.; Wang, A.; Hakim, A. Toxicity of L-proline toward rat hippocampal neurons. Brain Res. 1988, 456, 168–172. [Google Scholar] [CrossRef]
- Moffett, J.R.; Ross, B.; Arun, P.; Madhavarao, C.N.; Namboodiri, A.M.A. N-Acetylaspartate in the CNS: From neurodiagnostics to neurobiology. Prog. Neurobiol. 2007, 81, 89–131. [Google Scholar] [CrossRef]
- Birken, D.L.; Oldendorf, W.H. N-Acetyl-L-Aspartic acid: A literature review of a compound prominent in 1H-NMR spectroscopic studies of brain. Neurosci. Biobehav. Rev. 1989, 13, 23–31. [Google Scholar] [CrossRef]
- Weatherly, C.A.; Du, S.; Parpia, C.; Santos, P.T.; Hartman, A.L.; Armstrong, D.W. D-amino acid levels in perfused mouse brain tissue and blood: A comparative study. ACS Chem. Neurosci. 2017, 8, 1251–1261. [Google Scholar] [CrossRef]
- Kumar, A.; Bachhawat, A.K. Pyroglutamic acid: Throwing light on a lightly studied metabolite. Curr. Sci. 2012, 102, 288–297. [Google Scholar]
- Marinello, E.; Arezzini, L.; Pizzichini, M.; Frosi, B.; Porcelli, B.; Terzuoli, L. Purine nucleotide catabolism in rat liver: Labelling of uric acid and allantoin after administration of various labelled precursors. Life Sci. 2002, 70, 2931–2941. [Google Scholar] [CrossRef]
- Pani, A.K.; Jiao, Y.; Sample, K.J.; Smeyne, R.J. Neurochemical measurement of adenosine in discrete brain regions of five strains of inbred mice. PLoS ONE 2014, 9, e92422. [Google Scholar] [CrossRef] [PubMed]
- Stein, E.D.; Diamond, J.M. Do dietary levels of pantothenic acid regulate its intestinal uptake in mice? J. Nutr. 1989, 119, 1973–1983. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Ríos, G.A.; Tascon, M.; Reyes-Garcés, N.; Boyaci, E.; Poole, J.; Pawliszyn, J. Quantitative analysis of biofluid spots by coated blade spray mass spectrometry, a new approach to rapid screening. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Vuckovic, D.; Pawliszyn, J. Systematic evaluation of solid-phase microextraction coatings for untargeted metabolomic profiling of biological fluids by liquid chromatography-mass spectrometry. Anal. Chem. 2011, 83, 1944–1954. [Google Scholar] [CrossRef] [PubMed]


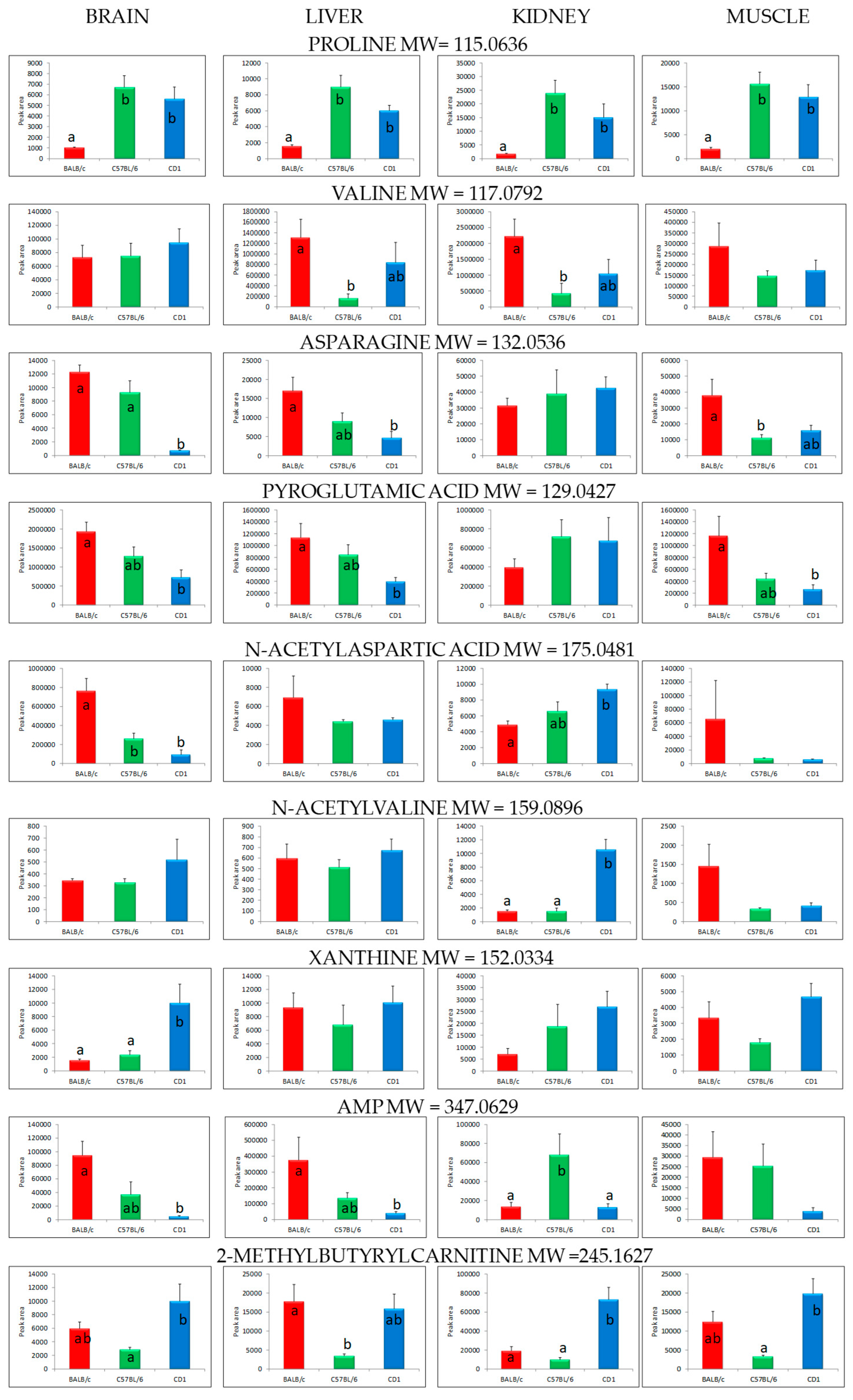

| Organ | Class | Compound | Molecular Weight | Retention Time (min) | p-Value (ANOVA) |
|---|---|---|---|---|---|
| Brain | Alpha-amino acids and derivatives | Proline | 115.0636 | 1.80 | 0.0019 |
| Valine | 117.0792 | 1.28 | 0.6876 | ||
| Asparagine | 132.0536 | 1.19 | 0.0000 | ||
| Pyroglutamic acid | 129.0427 | 1.22 | 0.0104 | ||
| Cystine | 240.0237 | 1.17 | 0.0441 | ||
| N-acetylaspartic acid | 175.0481 | 2.12 | 0.0005 | ||
| Tetrahydrodipicolinate | 171.0532 | 1.36 | 0.0145 | ||
| N-acyl-alpha-amino acids | N-acetylvaline | 159.0896 | 7.69 | 0.3731 | |
| Purine derivatives | Xanthine | 152.0334 | 3.23 | 0.0065 | |
| Purine nucleotides | Adenosine monophosphate (AMP) | 347.0629 | 1.34 | 0.0063 | |
| Purine nucleosides | Inosine | 268.0807 | 6.68 | 0.0070 | |
| 8-hydroxydeoxyguanosine | 283.0916 | 6.93 | 0.0005 | ||
| Pyrimidine derivatives | Uracil | 112.0276 | 1.34 | 0.0091 | |
| Pyrimidine nucleosides | 2’-deoxycytidine | 227.0905 | 7.05 | 0.0002 | |
| Fatty acid esters | 2-methylbutyrylcarnitine | 245.1627 | 17.87 | 0.0277 | |
| Hydroxy fatty acids | Mevalonic acid | 148.0737 | 1.33 | 0.0483 | |
| Fatty amides | Oleamide | 281.2718 | 20.64 | 0.0079 | |
| Alcohols and polyols | Pantothenic acid | 219.1107 | 7.08 | 0.0273 | |
| Liver | Alpha-amino acids and derivatives | Proline | 115.0636 | 1.80 | 0.0005 |
| Valine | 117.0792 | 1.28 | 0.0466 | ||
| Asparagine | 132.0536 | 1.19 | 0.0190 | ||
| Pyroglutamic acid | 129.0427 | 1.22 | 0.0083 | ||
| Tetrahydrodipicolinate | 171.0532 | 1.36 | 0.0185 | ||
| N-acetylaspartic acid | 175.0481 | 2.12 | 0.5917 | ||
| N-acyl-alpha-amino acids | N-acetylvaline | 159.0896 | 7.69 | 0.3605 | |
| Purine derivatives | Xanthine | 152.0334 | 3.23 | 0.6444 | |
| 5-hydroxyisourate | 184.0233 | 1.34 | 0.0004 | ||
| Purine nucleotides | Adenosine monophosphate (AMP) | 347.0629 | 1.34 | 0.0490 | |
| Purine nucleosides | Inosine | 268.0807 | 6.68 | 0.0120 | |
| 8-hydroxydeoxyguanosine | 283.0916 | 6.93 | 0.0033 | ||
| Fatty acid esters | 2-methylbutyrylcarnitine | 245.1627 | 17.87 | 0.0235 | |
| Ethyl eicosapentaenoic acid | 330.2557 | 21.65 | 0.0148 | ||
| Ceramides | Ceramide (d40:1) | 621.6063 | 26.30 | 0.0175 | |
| Benzoic acids and derivatives | 2-aminobenzoic acid | 137.0476 | 1.33 | 0.0048 | |
| Imidazoles | Allantoin | 158.0439 | 1.15 | 0.0442 | |
| Kidney | Alpha-amino acids and derivatives | Proline | 115.0636 | 1.80 | 0.0069 |
| Valine | 117.0792 | 1.28 | 0.0446 | ||
| Asparagine | 132.0536 | 1.19 | 0.7313 | ||
| Pyroglutamic acid | 129.0427 | 1.22 | 0.4256 | ||
| N-acetylaspartic acid | 175.0481 | 2.12 | 0.0080 | ||
| N-acyl-alpha-amino acids | N-acetylvaline | 159.0896 | 7.69 | 0.0000 | |
| Purine derivatives | Xanthine | 152.0334 | 3.23 | 0.1554 | |
| Purine nucleotides | Adenosine monophosphate (AMP) | 347.0629 | 1.34 | 0.0176 | |
| Fatty acid esters | 2-methylbutyrylcarnitine | 245.1627 | 17.87 | 0.0003 | |
| Ethyl eicosapentaenoic acid | 330.2557 | 21.65 | 0.0066 | ||
| Benzoic acids and derivatives | 2-aminobenzoic acid | 137.0476 | 1.33 | 0.0067 | |
| Muscle | Alpha-amino acids and derivatives | Proline | 115.0636 | 1.80 | 0.0015 |
| Valine | 117.0792 | 1.28 | 0.3540 | ||
| Asparagine | 132.0536 | 1.19 | 0.0236 | ||
| Pyroglutamic acid | 129.0427 | 1.22 | 0.0206 | ||
| N-acetylaspartic acid | 175.0481 | 2.12 | 0.3746 | ||
| N-acyl-alpha-amino acids | N-acetylvaline | 159.0896 | 7.69 | 0.6755 | |
| N-tridecanoylglycine | 271.2147 | 22.52 | 0.0407 | ||
| Purine derivatives | Xanthine | 152.0334 | 3.23 | 0.0635 | |
| 5-hydroxyisourate | 184.0233 | 1.34 | 0.0038 | ||
| Purine nucleotides | Adenosine monophosphate (AMP) | 347.0629 | 1.34 | 0.1549 | |
| Fatty acid esters | 2-methylbutyrylcarnitine | 245.1627 | 17.87 | 0.0051 | |
| Alcohols and polyols | Pantothenic acid | 219.1107 | 7.08 | 0.0305 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burlikowska, K.; Stryjak, I.; Bogusiewicz, J.; Kupcewicz, B.; Jaroch, K.; Bojko, B. Comparison of Metabolomic Profiles of Organs in Mice of Different Strains Based on SPME-LC-HRMS. Metabolites 2020, 10, 255. https://doi.org/10.3390/metabo10060255
Burlikowska K, Stryjak I, Bogusiewicz J, Kupcewicz B, Jaroch K, Bojko B. Comparison of Metabolomic Profiles of Organs in Mice of Different Strains Based on SPME-LC-HRMS. Metabolites. 2020; 10(6):255. https://doi.org/10.3390/metabo10060255
Chicago/Turabian StyleBurlikowska, Katarzyna, Iga Stryjak, Joanna Bogusiewicz, Bogumiła Kupcewicz, Karol Jaroch, and Barbara Bojko. 2020. "Comparison of Metabolomic Profiles of Organs in Mice of Different Strains Based on SPME-LC-HRMS" Metabolites 10, no. 6: 255. https://doi.org/10.3390/metabo10060255
APA StyleBurlikowska, K., Stryjak, I., Bogusiewicz, J., Kupcewicz, B., Jaroch, K., & Bojko, B. (2020). Comparison of Metabolomic Profiles of Organs in Mice of Different Strains Based on SPME-LC-HRMS. Metabolites, 10(6), 255. https://doi.org/10.3390/metabo10060255