Metabolomic Profiling in Neuromyelitis Optica Spectrum Disorder Biomarker Discovery
Abstract
1. Introduction
1.1. A Role for AQPA-IgG1 and MOG-IgG1 Autoantibodies in NMOSD Pathology
1.2. The Urgency for Novel NMOSD Biomarker Discovery
1.3. Metabolomic Profiling Applied to Autoimmune Disease
1.4. Immunometabolism in Autoimmune Disease
1.5. A Role for Metabolomics in Multiple Sclerosis
2. Metabolomic Profiling of NMOSD
2.1. Metabolomic Studies to Discriminate NMOSD
2.2. NMOSD Metabolomics Profiling of SCFA in Fatty Acid Metabolism and Glycolysis
2.3. NMOSD Metabolomics Profiling of Lactate/Lactic Acid in Fatty Acid Metabolism and Glycolysis
2.4. NMOSD Metabolomics Profiling of Lipids and Lipoproteins in Energy Metabolism and Glycolysis
2.5. NMOSD Metabolomic Profiling of Amino Acids in NMOSD
2.6. Magnetic Resonance Spectroscopy (MRS) in NMOSD Metabolomic Studies
3. Proteomics in NMOSD
3.1. Proteomic Studies to Discriminate NMOSD
3.2. Proteomic-Based Acute Phase Protein Perturbations
3.3. Proteomic-Based Perturbations in Humoral Immunity
3.4. Targeted Immunoassay-Based Studies to Discriminate NMOSD
4. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Jarius, S.; Wildemann, B. The history of neuromyelitis optica. J. Neuroinflamm. 2013, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Weinshenker, B.G.; Wingerchuk, D.M. Neuromyelitis Spectrum Disorders. Mayo Clin. Proc. 2017, 92, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.; Wildemann, B.; Jarius, S.; Orellano, B.; Sasidharan, S.; Weber, M.S.; Stuve, O. Immunopathogenesis of neuromyelitis optica. Adv. Immunol. 2014, 121, 213–242. [Google Scholar] [CrossRef] [PubMed]
- Wingerchuk, D.M.; Lennon, V.A.; Lucchinetti, C.F.; Pittock, S.J.; Weinshenker, B.G. The spectrum of neuromyelitis optica. Lancet Neurol. 2007, 6, 805–815. [Google Scholar] [CrossRef]
- Pereira, W.L.; Reiche, E.M.; Kallaur, A.P.; Kaimen-Maciel, D.R. Epidemiological, clinical, and immunological characteristics of neuromyelitis optica: A review. J. Neurol. Sci. 2015, 355, 7–17. [Google Scholar] [CrossRef]
- Lennon, V.A.; Kryzer, T.J.; Pittock, S.J.; Verkman, A.S.; Hinson, S.R. IgG marker of optic-spinal multiple sclerosis binds to the aquaporin-4 water channel. J. Exp. Med. 2005, 202, 473–477. [Google Scholar] [CrossRef]
- Popescu, B.F.; Guo, Y.; Jentoft, M.E.; Parisi, J.E.; Lennon, V.A.; Pittock, S.J.; Weinshenker, B.G.; Wingerchuk, D.M.; Giannini, C.; Metz, I.; et al. Diagnostic utility of aquaporin-4 in the analysis of active demyelinating lesions. Neurology 2015, 84, 148–158. [Google Scholar] [CrossRef]
- Lennon, V.A.; Wingerchuk, D.M.; Kryzer, T.J.; Pittock, S.J.; Lucchinetti, C.F.; Fujihara, K.; Nakashima, I.; Weinshenker, B.G. A serum autoantibody marker of neuromyelitis optica: Distinction from multiple sclerosis. Lancet 2004, 364, 2106–2112. [Google Scholar] [CrossRef]
- Jacob, A.; Matiello, M.; Wingerchuk, D.M.; Lucchinetti, C.F.; Pittock, S.J.; Weinshenker, B.G. Neuromyelitis optica: Changing concepts. J. Neuroimmunol. 2007, 187, 126–138. [Google Scholar] [CrossRef]
- Prain, K.; Woodhall, M.; Vincent, A.; Ramanathan, S.; Barnett, M.H.; Bundell, C.S.; Parratt, J.D.E.; Silvestrini, R.A.; Bukhari, W.; Brilot, F.; et al. AQP4 Antibody Assay Sensitivity Comparison in the Era of the 2015 Diagnostic Criteria for NMOSD. Front. Neurol. 2019, 10, 1028. [Google Scholar] [CrossRef]
- Pisani, F.; Sparaneo, A.; Tortorella, C.; Ruggieri, M.; Trojano, M.; Mola, M.G.; Nicchia, G.P.; Frigeri, A.; Svelto, M. Aquaporin-4 autoantibodies in Neuromyelitis Optica: AQP4 isoform-dependent sensitivity and specificity. PLoS ONE 2013, 8, e79185. [Google Scholar] [CrossRef] [PubMed]
- Jarius, S.; Wildemann, B. AQP4 antibodies in neuromyelitis optica: Diagnostic and pathogenetic relevance. Nat. Rev. Neurol. 2010, 6, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Phuan, P.W.; Ratelade, J.; Rossi, A.; Tradtrantip, L.; Verkman, A.S. Complement-dependent cytotoxicity in neuromyelitis optica requires aquaporin-4 protein assembly in orthogonal arrays. J. Biol. Chem. 2012, 287, 13829–13839. [Google Scholar] [CrossRef] [PubMed]
- Crane, J.M.; Lam, C.; Rossi, A.; Gupta, T.; Bennett, J.L.; Verkman, A.S. Binding affinity and specificity of neuromyelitis optica autoantibodies to aquaporin-4 M1/M23 isoforms and orthogonal arrays. J. Biol. Chem. 2011, 286, 16516–16524. [Google Scholar] [CrossRef] [PubMed]
- Sagan, S.A.; Winger, R.C.; Cruz-Herranz, A.; Nelson, P.A.; Hagberg, S.; Miller, C.N.; Spencer, C.M.; Ho, P.P.; Bennett, J.L.; Levy, M.; et al. Tolerance checkpoint bypass permits emergence of pathogenic T cells to neuromyelitis optica autoantigen aquaporin-4. Proc. Natl. Acad. Sci. USA 2016, 113, 14781–14786. [Google Scholar] [CrossRef]
- Weber, M.S.; Derfuss, T.; Metz, I.; Bruck, W. Defining distinct features of anti-MOG antibody associated central nervous system demyelination. Ther Adv. Neurol. Disord 2018, 11, 1756286418762083. [Google Scholar] [CrossRef]
- Kitley, J.; Waters, P.; Woodhall, M.; Leite, M.I.; Murchison, A.; George, J.; Kuker, W.; Chandratre, S.; Vincent, A.; Palace, J. Neuromyelitis optica spectrum disorders with aquaporin-4 and myelin-oligodendrocyte glycoprotein antibodies: A comparative study. JAMA Neurol. 2014, 71, 276–283. [Google Scholar] [CrossRef]
- Sato, D.K.; Callegaro, D.; Lana-Peixoto, M.A.; Waters, P.J.; de Haidar Jorge, F.M.; Takahashi, T.; Nakashima, I.; Apostolos-Pereira, S.L.; Talim, N.; Simm, R.F.; et al. Distinction between MOG antibody-positive and AQP4 antibody-positive NMO spectrum disorders. Neurology 2014, 82, 474–481. [Google Scholar] [CrossRef]
- Dale, R.C.; Tantsis, E.M.; Merheb, V.; Kumaran, R.Y.; Sinmaz, N.; Pathmanandavel, K.; Ramanathan, S.; Booth, D.R.; Wienholt, L.A.; Prelog, K.; et al. Antibodies to MOG have a demyelination phenotype and affect oligodendrocyte cytoskeleton. Neurol. Neuroimmunol. Neuroinflamm. 2014, 1, e12. [Google Scholar] [CrossRef]
- Rostasy, K.; Mader, S.; Hennes, E.M.; Schanda, K.; Gredler, V.; Guenther, A.; Blaschek, A.; Korenke, C.; Pritsch, M.; Pohl, D.; et al. Persisting myelin oligodendrocyte glycoprotein antibodies in aquaporin-4 antibody negative pediatric neuromyelitis optica. Mult. Scler. J. 2013, 19, 1052–1059. [Google Scholar] [CrossRef]
- Narayan, R.; Simpson, A.; Fritsche, K.; Salama, S.; Pardo, S.; Mealy, M.; Paul, F.; Levy, M. MOG antibody disease: A review of MOG antibody seropositive neuromyelitis optica spectrum disorder. Mult. Scler. Relat. Disord 2018, 25, 66–72. [Google Scholar] [CrossRef]
- Ratelade, J.; Asavapanumas, N.; Ritchie, A.M.; Wemlinger, S.; Bennett, J.L.; Verkman, A.S. Involvement of antibody-dependent cell-mediated cytotoxicity in inflammatory demyelination in a mouse model of neuromyelitis optica. Acta Neuropathol. 2013, 126, 699–709. [Google Scholar] [CrossRef]
- Lucchinetti, C.F.; Guo, Y.; Popescu, B.F.; Fujihara, K.; Itoyama, Y.; Misu, T. The pathology of an autoimmune astrocytopathy: Lessons learned from neuromyelitis optica. Brain Pathol. 2014, 24, 83–97. [Google Scholar] [CrossRef]
- Varrin-Doyer, M.; Spencer, C.M.; Schulze-Topphoff, U.; Nelson, P.A.; Stroud, R.M.; Cree, B.A.; Zamvil, S.S. Aquaporin 4-specific T cells in neuromyelitis optica exhibit a Th17 bias and recognize Clostridium ABC transporter. Ann. Neurol. 2012, 72, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Asgari, N.; Flanagan, E.P.; Fujihara, K.; Kim, H.J.; Skejoe, H.P.; Wuerfel, J.; Kuroda, H.; Kim, S.H.; Maillart, E.; Marignier, R.; et al. Disruption of the leptomeningeal blood barrier in neuromyelitis optica spectrum disorder. Neurol. Neuroimmunol. Neuroinflamm. 2017, 4, e343. [Google Scholar] [CrossRef] [PubMed]
- Wingerchuk, D.M.; Pittock, S.J.; Lucchinetti, C.F.; Lennon, V.A.; Weinshenker, B.G. A secondary progressive clinical course is uncommon in neuromyelitis optica. Neurology 2007, 68, 603–605. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, M.C.; Bennett, J.L.; Verkman, A.S. Treatment of neuromyelitis optica: State-of-the-art and emerging therapies. Nat. Rev. Neurol. 2014, 10, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Paul, F.; Murphy, O.; Pardo, S.; Levy, M. Investigational drugs in development to prevent neuromyelitis optica relapses. Expert Opin. Investig. Drugs 2018, 27, 265–271. [Google Scholar] [CrossRef]
- Huang, T.L.; Lin, K.H.; Wang, J.K.; Tsai, R.K. Treatment strategies for neuromyelitis optica. Ci Ji Yi Xue Za Zhi 2018, 30, 204–208. [Google Scholar] [CrossRef]
- Bonnan, M.; Valentino, R.; Olindo, S.; Mehdaoui, H.; Smadja, D.; Cabre, P. Plasma exchange in severe spinal attacks associated with neuromyelitis optica spectrum disorder. Mult. Scler. J. 2009, 15, 487–492. [Google Scholar] [CrossRef]
- Merle, H.; Olindo, S.; Jeannin, S.; Valentino, R.; Mehdaoui, H.; Cabot, F.; Donnio, A.; Hage, R.; Richer, R.; Smadja, D.; et al. Treatment of optic neuritis by plasma exchange (add-on) in neuromyelitis optica. Arch. Ophthalmol. 2012, 130, 858–862. [Google Scholar] [CrossRef] [PubMed]
- Mandler, R.N.; Ahmed, W.; Dencoff, J.E. Devic’s neuromyelitis optica: A prospective study of seven patients treated with prednisone and azathioprine. Neurology 1998, 51, 1219–1220. [Google Scholar] [CrossRef] [PubMed]
- Costanzi, C.; Matiello, M.; Lucchinetti, C.F.; Weinshenker, B.G.; Pittock, S.J.; Mandrekar, J.; Thapa, P.; McKeon, A. Azathioprine: Tolerability, efficacy, and predictors of benefit in neuromyelitis optica. Neurology 2011, 77, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Mealy, M.A.; Wingerchuk, D.M.; Palace, J.; Greenberg, B.M.; Levy, M. Comparison of relapse and treatment failure rates among patients with neuromyelitis optica: Multicenter study of treatment efficacy. JAMA Neurol. 2014, 71, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Torres, J.; Pruitt, A.; Balcer, L.; Galetta, S.; Markowitz, C.; Dahodwala, N. Analysis of the treatment of neuromyelitis optica. J. Neurol. Sci. 2015, 351, 31–35. [Google Scholar] [CrossRef]
- Trebst, C.; Jarius, S.; Berthele, A.; Paul, F.; Schippling, S.; Wildemann, B.; Borisow, N.; Kleiter, I.; Aktas, O.; Kumpfel, T.; et al. Update on the diagnosis and treatment of neuromyelitis optica: Recommendations of the Neuromyelitis Optica Study Group (NEMOS). J. Neurol. 2014, 261, 1–16. [Google Scholar] [CrossRef]
- Valentino, P.; Marnetto, F.; Granieri, L.; Capobianco, M.; Bertolotto, A. Aquaporin-4 antibody titration in NMO patients treated with rituximab: A retrospective study. Neurol. Neuroimmunol. Neuroinflamm. 2017, 4, e317. [Google Scholar] [CrossRef]
- Cree, B.A.C.; Bennett, J.L.; Kim, H.J.; Weinshenker, B.G.; Pittock, S.J.; Wingerchuk, D.M.; Fujihara, K.; Paul, F.; Cutter, G.R.; Marignier, R.; et al. Inebilizumab for the treatment of neuromyelitis optica spectrum disorder (N-MOmentum): A double-blind, randomised placebo-controlled phase 2/3 trial. Lancet 2019, 394, 1352–1363. [Google Scholar] [CrossRef]
- Frampton, J.E. Inebilizumab: First Approval. Drugs 2020, 80, 1259–1264. [Google Scholar] [CrossRef]
- Pittock, S.J.; Lennon, V.A.; McKeon, A.; Mandrekar, J.; Weinshenker, B.G.; Lucchinetti, C.F.; O’Toole, O.; Wingerchuk, D.M. Eculizumab in AQP4-IgG-positive relapsing neuromyelitis optica spectrum disorders: An open-label pilot study. Lancet Neurol. 2013, 12, 554–562. [Google Scholar] [CrossRef]
- Ringelstein, M.; Ayzenberg, I.; Harmel, J.; Lauenstein, A.S.; Lensch, E.; Stogbauer, F.; Hellwig, K.; Ellrichmann, G.; Stettner, M.; Chan, A.; et al. Long-term Therapy With Interleukin 6 Receptor Blockade in Highly Active Neuromyelitis Optica Spectrum Disorder. JAMA Neurol. 2015, 72, 756–763. [Google Scholar] [CrossRef]
- Schiopu, E.; Chatterjee, S.; Hsu, V.; Flor, A.; Cimbora, D.; Patra, K.; Yao, W.; Li, J.; Streicher, K.; McKeever, K.; et al. Safety and tolerability of an anti-CD19 monoclonal antibody, MEDI-551, in subjects with systemic sclerosis: A phase I, randomized, placebo-controlled, escalating single-dose study. Arthritis Res. Ther. 2016, 18, 131. [Google Scholar] [CrossRef] [PubMed]
- Jarius, S.; Paul, F.; Franciotta, D.; Ruprecht, K.; Ringelstein, M.; Bergamaschi, R.; Rommer, P.; Kleiter, I.; Stich, O.; Reuss, R.; et al. Cerebrospinal fluid findings in aquaporin-4 antibody positive neuromyelitis optica: Results from 211 lumbar punctures. J. Neurol. Sci. 2011, 306, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Storoni, M.; Petzold, A.; Plant, G.T. The use of serum glial fibrillary acidic protein measurements in the diagnosis of neuromyelitis optica spectrum optic neuritis. PLoS ONE 2011, 6, e23489. [Google Scholar] [CrossRef] [PubMed]
- Bergamaschi, R.; Tonietti, S.; Franciotta, D.; Candeloro, E.; Tavazzi, E.; Piccolo, G.; Romani, A.; Cosi, V. Oligoclonal bands in Devic’s neuromyelitis optica and multiple sclerosis: Differences in repeated cerebrospinal fluid examinations. Mult. Scler. J. 2004, 10, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Palace, J.; Leite, M.I.; Nairne, A.; Vincent, A. Interferon Beta treatment in neuromyelitis optica: Increase in relapses and aquaporin 4 antibody titers. Arch. Neurol. 2010, 67, 1016–1017. [Google Scholar] [CrossRef] [PubMed]
- Uzawa, A.; Mori, M.; Hayakawa, S.; Masuda, S.; Kuwabara, S. Different responses to interferon beta-1b treatment in patients with neuromyelitis optica and multiple sclerosis. Eur. J. Neurol. 2010, 17, 672–676. [Google Scholar] [CrossRef]
- Min, J.H.; Kim, B.J.; Lee, K.H. Development of extensive brain lesions following fingolimod (FTY720) treatment in a patient with neuromyelitis optica spectrum disorder. Mult. Scler. J. 2012, 18, 113–115. [Google Scholar] [CrossRef]
- Kitley, J.; Evangelou, N.; Kuker, W.; Jacob, A.; Leite, M.I.; Palace, J. Catastrophic brain relapse in seronegative NMO after a single dose of natalizumab. J. Neurol. Sci. 2014, 339, 223–225. [Google Scholar] [CrossRef]
- Li, J.; Che, N.; Xu, L.; Zhang, Q.; Wang, Q.; Tan, W.; Zhang, M. LC-MS-based serum metabolomics reveals a distinctive signature in patients with rheumatoid arthritis. Clin. Rheumatol. 2018, 37, 1493–1502. [Google Scholar] [CrossRef]
- Pflueger, M.; Seppanen-Laakso, T.; Suortti, T.; Hyotylainen, T.; Achenbach, P.; Bonifacio, E.; Oresic, M.; Ziegler, A.G. Age- and islet autoimmunity-associated differences in amino acid and lipid metabolites in children at risk for type 1 diabetes. Diabetes 2011, 60, 2740–2747. [Google Scholar] [CrossRef]
- Fernandez, D.; Bonilla, E.; Mirza, N.; Niland, B.; Perl, A. Rapamycin reduces disease activity and normalizes T cell activation-induced calcium fluxing in patients with systemic lupus erythematosus. Arthritis Rheum. 2006, 54, 2983–2988. [Google Scholar] [CrossRef]
- Fernandez-Ramos, A.A.; Poindessous, V.; Marchetti-Laurent, C.; Pallet, N.; Loriot, M.A. The effect of immunosuppressive molecules on T-cell metabolic reprogramming. Biochimie 2016, 127, 23–36. [Google Scholar] [CrossRef]
- Yin, Y.; Choi, S.C.; Xu, Z.; Perry, D.J.; Seay, H.; Croker, B.P.; Sobel, E.S.; Brusko, T.M.; Morel, L. Normalization of CD4+ T cell metabolism reverses lupus. Sci. Transl. Med. 2015, 7, 274ra218. [Google Scholar] [CrossRef]
- Leonard, M.M.; Camhi, S.; Huedo-Medina, T.B.; Fasano, A. Celiac Disease Genomic, Environmental, Microbiome, and Metabolomic (CDGEMM) Study Design: Approach to the Future of Personalized Prevention of Celiac Disease. Nutrients 2015, 7, 9325–9336. [Google Scholar] [CrossRef]
- Calabro, A.; Gralka, E.; Luchinat, C.; Saccenti, E.; Tenori, L. A metabolomic perspective on coeliac disease. Autoimmune Dis. 2014, 2014, 756138. [Google Scholar] [CrossRef]
- Lewis, J.S.; Lee, J.A.; Underwood, J.C.; Harris, A.L.; Lewis, C.E. Macrophage responses to hypoxia: Relevance to disease mechanisms. J. Leukoc. Biol. 1999, 66, 889–900. [Google Scholar] [CrossRef]
- Fox, C.J.; Hammerman, P.S.; Thompson, C.B. Fuel feeds function: Energy metabolism and the T-cell response. Nat. Rev. Immunol. 2005, 5, 844–852. [Google Scholar] [CrossRef]
- Mehrpour, M.; Kyani, A.; Tafazzoli, M.; Fathi, F.; Joghataie, M.T. A metabonomics investigation of multiple sclerosis by nuclear magnetic resonance. Magn. Reson. Chem. 2013, 51, 102–109. [Google Scholar] [CrossRef]
- Singh, J.; Cerghet, M.; Poisson, L.M.; Datta, I.; Labuzek, K.; Suhail, H.; Rattan, R.; Giri, S. Urinary and Plasma Metabolomics Identify the Distinct Metabolic Profile of Disease State in Chronic Mouse Model of Multiple Sclerosis. J. Neuroimmune Pharmacol. 2018. [Google Scholar] [CrossRef]
- Lim, C.K.; Bilgin, A.; Lovejoy, D.B.; Tan, V.; Bustamante, S.; Taylor, B.V.; Bessede, A.; Brew, B.J.; Guillemin, G.J. Kynurenine pathway metabolomics predicts and provides mechanistic insight into multiple sclerosis progression. Sci. Rep. 2017, 7, 41473. [Google Scholar] [CrossRef] [PubMed]
- Luque-Cordoba, D.; Luque de Castro, M.D. Metabolomics: A potential way to know the role of vitamin D on multiple sclerosis. J. Pharm. Biomed. Anal. 2017, 136, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Del Boccio, P.; Rossi, C.; di Ioia, M.; Cicalini, I.; Sacchetta, P.; Pieragostino, D. Integration of metabolomics and proteomics in multiple sclerosis: From biomarkers discovery to personalized medicine. Proteomics Clin. Appl. 2016, 10, 470–484. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, P.; Calabresi, P.A. Metabolomics in multiple sclerosis. Mult. Scler. J. 2016, 22, 451–460. [Google Scholar] [CrossRef]
- Poisson, L.M.; Suhail, H.; Singh, J.; Datta, I.; Denic, A.; Labuzek, K.; Hoda, M.N.; Shankar, A.; Kumar, A.; Cerghet, M.; et al. Untargeted Plasma Metabolomics Identifies Endogenous Metabolite with Drug-like Properties in Chronic Animal Model of Multiple Sclerosis. J. Biol. Chem. 2015, 290, 30697–30712. [Google Scholar] [CrossRef]
- Dickens, A.M.; Larkin, J.R.; Davis, B.G.; Griffin, J.L.; Claridge, T.D.; Sibson, N.R.; Anthony, D.C. NMR-Based Metabolomics Separates the Distinct Stages of Disease in a Chronic Relapsing Model of Multiple Sclerosis. J. Neuroimmune Pharmacol. 2015, 10, 435–444. [Google Scholar] [CrossRef]
- Pieragostino, D.; D’Alessandro, M.; di Ioia, M.; Rossi, C.; Zucchelli, M.; Urbani, A.; Di Ilio, C.; Lugaresi, A.; Sacchetta, P.; Del Boccio, P. An integrated metabolomics approach for the research of new cerebrospinal fluid biomarkers of multiple sclerosis. Mol. Biosyst. 2015, 11, 1563–1572. [Google Scholar] [CrossRef]
- Mangalam, A.; Poisson, L.; Nemutlu, E.; Datta, I.; Denic, A.; Dzeja, P.; Rodriguez, M.; Rattan, R.; Giri, S. Profile of Circulatory Metabolites in a Relapsing-remitting Animal Model of Multiple Sclerosis using Global Metabolomics. J. Clin. Cell Immunol. 2013, 4. [Google Scholar] [CrossRef]
- Noga, M.J.; Dane, A.; Shi, S.; Attali, A.; van Aken, H.; Suidgeest, E.; Tuinstra, T.; Muilwijk, B.; Coulier, L.; Luider, T.; et al. Metabolomics of cerebrospinal fluid reveals changes in the central nervous system metabolism in a rat model of multiple sclerosis. Metabolomics 2012, 8, 253–263. [Google Scholar] [CrossRef]
- Ibrahim, S.M.; Gold, R. Genomics, proteomics, metabolomics: What is in a word for multiple sclerosis? Curr. Opin. Neurol. 2005, 18, 231–235. [Google Scholar] [CrossRef]
- Rossi, C.; Cicalini, I.; Zucchelli, M.; di Ioia, M.; Onofrj, M.; Federici, L.; Del Boccio, P.; Pieragostino, D. Metabolomic Signature in Sera of Multiple Sclerosis Patients during Pregnancy. Int. J. Mol. Sci. 2018, 19, 3589. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Arguelles, A.; Mendez-Huerta, M.A.; Lozano, C.D.; Ruiz-Arguelles, G.J. Metabolomic profile of insulin resistance in patients with multiple sclerosis is associated to the severity of the disease. Mult. Scler. Relat. Disord 2018, 25, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Stoessel, D.; Stellmann, J.P.; Willing, A.; Behrens, B.; Rosenkranz, S.C.; Hodecker, S.C.; Sturner, K.H.; Reinhardt, S.; Fleischer, S.; Deuschle, C.; et al. Metabolomic Profiles for Primary Progressive Multiple Sclerosis Stratification and Disease Course Monitoring. Front. Hum. Neurosci. 2018, 12, 226. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.H.; Jeong, I.H.; Hyun, J.S.; Kong, B.S.; Kim, H.J.; Park, S.J. Metabolomic profiling of CSF in multiple sclerosis and neuromyelitis optica spectrum disorder by nuclear magnetic resonance. PLoS ONE 2017, 12, e0181758. [Google Scholar] [CrossRef]
- Poddighe, S.; Murgia, F.; Lorefice, L.; Liggi, S.; Cocco, E.; Marrosu, M.G.; Atzori, L. Metabolomic analysis identifies altered metabolic pathways in Multiple Sclerosis. Int. J. Biochem. Cell Biol. 2017, 93, 148–155. [Google Scholar] [CrossRef]
- Villoslada, P.; Alonso, C.; Agirrezabal, I.; Kotelnikova, E.; Zubizarreta, I.; Pulido-Valdeolivas, I.; Saiz, A.; Comabella, M.; Montalban, X.; Villar, L.; et al. Metabolomic signatures associated with disease severity in multiple sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 2017, 4, e321. [Google Scholar] [CrossRef]
- Cocco, E.; Murgia, F.; Lorefice, L.; Barberini, L.; Poddighe, S.; Frau, J.; Fenu, G.; Coghe, G.; Murru, M.R.; Murru, R.; et al. (1)H-NMR analysis provides a metabolomic profile of patients with multiple sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 2016, 3, e185. [Google Scholar] [CrossRef]
- Reinke, S.N.; Broadhurst, D.L.; Sykes, B.D.; Baker, G.B.; Catz, I.; Warren, K.G.; Power, C. Metabolomic profiling in multiple sclerosis: Insights into biomarkers and pathogenesis. Mult. Scler. J. 2014, 20, 1396–1400. [Google Scholar] [CrossRef]
- Vingara, L.K.; Yu, H.J.; Wagshul, M.E.; Serafin, D.; Christodoulou, C.; Pelczer, I.; Krupp, L.B.; Maletic-Savatic, M. Metabolomic approach to human brain spectroscopy identifies associations between clinical features and the frontal lobe metabolome in multiple sclerosis. Neuroimage 2013, 82, 586–594. [Google Scholar] [CrossRef]
- Smolinska, A.; Blanchet, L.; Coulier, L.; Ampt, K.A.; Luider, T.; Hintzen, R.Q.; Wijmenga, S.S.; Buydens, L.M. Interpretation and visualization of non-linear data fusion in kernel space: Study on metabolomic characterization of progression of multiple sclerosis. PLoS ONE 2012, 7, e38163. [Google Scholar] [CrossRef]
- Dickens, A.M.; Larkin, J.R.; Griffin, J.L.; Cavey, A.; Matthews, L.; Turner, M.R.; Wilcock, G.K.; Davis, B.G.; Claridge, T.D.; Palace, J.; et al. A type 2 biomarker separates relapsing-remitting from secondary progressive multiple sclerosis. Neurology 2014, 83, 1492–1499. [Google Scholar] [CrossRef] [PubMed]
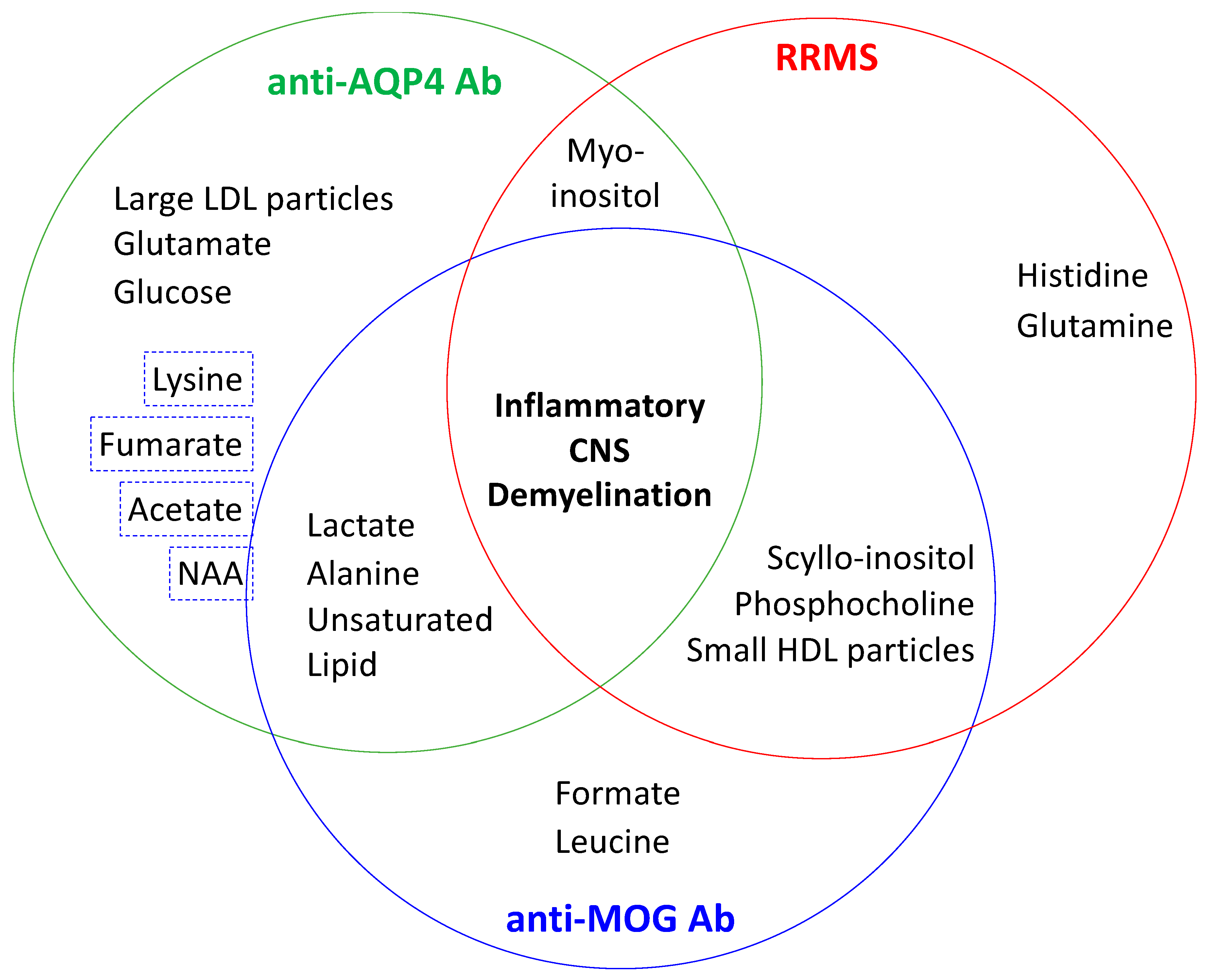
- Jurynczyk, M.; Probert, F.; Yeo, T.; Tackley, G.; Claridge, T.D.W.; Cavey, A.; Woodhall, M.R.; Arora, S.; Winkler, T.; Schiffer, E.; et al. Metabolomics reveals distinct, antibody-independent, molecular signatures of MS, AQP4-antibody and MOG-antibody disease. Acta Neuropathol. Commun. 2017, 5, 95. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo, H.; Brieva, L.; Tatzber, F.; Jove, M.; Cacabelos, D.; Cassanye, A.; Lanau-Angulo, L.; Boada, J.; Serrano, J.C.; Gonzalez, C.; et al. Lipidome analysis in multiple sclerosis reveals protein lipoxidative damage as a potential pathogenic mechanism. J. Neurochem. 2012, 123, 622–634. [Google Scholar] [CrossRef] [PubMed]
- Pruss, H.; Rosche, B.; Sullivan, A.B.; Brommer, B.; Wengert, O.; Gronert, K.; Schwab, J.M. Proresolution lipid mediators in multiple sclerosis-differential, disease severity-dependent synthesis—A clinical pilot trial. PLoS ONE 2013, 8, e55859. [Google Scholar] [CrossRef] [PubMed]
- Moussallieh, F.M.; Elbayed, K.; Chanson, J.B.; Rudolf, G.; Piotto, M.; De Seze, J.; Namer, I.J. Serum analysis by 1H nuclear magnetic resonance spectroscopy: A new tool for distinguishing neuromyelitis optica from multiple sclerosis. Mult. Scler. J. 2014, 20, 558–565. [Google Scholar] [CrossRef]
- Gebregiworgis, T.; Nielsen, H.H.; Massilamany, C.; Gangaplara, A.; Reddy, J.; Illes, Z.; Powers, R. A Urinary Metabolic Signature for Multiple Sclerosis and Neuromyelitis Optica. J. Proteome Res. 2016, 15, 659–666. [Google Scholar] [CrossRef]
- Park, S.J.; Jeong, I.H.; Kong, B.S.; Lee, J.E.; Kim, K.H.; Lee, D.Y.; Kim, H.J. Disease Type- and Status-Specific Alteration of CSF Metabolome Coordinated with Clinical Parameters in Inflammatory Demyelinating Diseases of CNS. PLoS ONE 2016, 11, e0166277. [Google Scholar] [CrossRef]
- Tortorella, C.; Ruggieri, M.; Di Monte, E.; Ceci, E.; Iaffaldano, P.; Direnzo, V.; Mastrapasqua, M.; Frigeri, A.; Amato, M.P.; Hakiki, B.; et al. Serum and CSF N-acetyl aspartate levels differ in multiple sclerosis and neuromyelitis optica. J. Neurol. Neurosurg. Psychiatry 2011, 82, 1355–1359. [Google Scholar] [CrossRef]
- Cha, E.; Lee, K.M.; Park, K.D.; Park, K.S.; Lee, K.W.; Kim, S.M.; Lee, J. Hydroxycholesterol Levels in the Serum and Cerebrospinal Fluid of Patients with Neuromyelitis Optica Revealed by LC-Ag+CIS/MS/MS and LC-ESI/MS/MS with Picolinic Derivatization: Increased Levels and Association with Disability during Acute Attack. PLoS ONE 2016, 11, e0167819. [Google Scholar] [CrossRef]
- Waniewski, R.A.; Martin, D.L. Preferential utilization of acetate by astrocytes is attributable to transport. J. Neurosci. 1998, 18, 5225–5233. [Google Scholar] [CrossRef]
- Rae, C.; Fekete, A.D.; Kashem, M.A.; Nasrallah, F.A.; Broer, S. Metabolism, compartmentation, transport and production of acetate in the cortical brain tissue slice. Neurochem. Res. 2012, 37, 2541–2553. [Google Scholar] [CrossRef] [PubMed]
- Rowlands, B.D.; Klugmann, M.; Rae, C.D. Acetate metabolism does not reflect astrocytic activity, contributes directly to GABA synthesis, and is increased by silent information regulator 1 activation. J. Neurochem. 2017, 140, 903–918. [Google Scholar] [CrossRef]
- Albrecht, J.; Sidoryk-Wegrzynowicz, M.; Zielinska, M.; Aschner, M. Roles of glutamine in neurotransmission. Neuron. Glia Biol. 2010, 6, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.B.; de Graaf, R.A.; Mason, G.F.; Rothman, D.L.; Shulman, R.G.; Behar, K.L. The contribution of GABA to glutamate/glutamine cycling and energy metabolism in the rat cortex in vivo. Proc. Natl. Acad. Sci. USA 2005, 102, 5588–5593. [Google Scholar] [CrossRef]
- Wyss, M.T.; Magistretti, P.J.; Buck, A.; Weber, B. Labeled acetate as a marker of astrocytic metabolism. Br. J. Pharmacol. 2011, 31, 1668–1674. [Google Scholar] [CrossRef] [PubMed]
- Nave, K.A.; Werner, H.B. Myelination of the nervous system: Mechanisms and functions. Annu Rev. Cell Dev. Biol. 2014, 30, 503–533. [Google Scholar] [CrossRef]
- Namboodiri, A.M.; Peethambaran, A.; Mathew, R.; Sambhu, P.A.; Hershfield, J.; Moffett, J.R.; Madhavarao, C.N. Canavan disease and the role of N-acetylaspartate in myelin synthesis. Mol. Cell Endocrinol. 2006, 252, 216–223. [Google Scholar] [CrossRef]
- Kim, M.H.; Kang, S.G.; Park, J.H.; Yanagisawa, M.; Kim, C.H. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 2013, 145, 396–406. [Google Scholar] [CrossRef]
- Marino, E.; Richards, J.L.; McLeod, K.H.; Stanley, D.; Yap, Y.A.; Knight, J.; McKenzie, C.; Kranich, J.; Oliveira, A.C.; Rossello, F.J.; et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat. Immunol. 2017, 18, 552–562. [Google Scholar] [CrossRef] [PubMed]
- Erny, D.; Hrabe de Angelis, A.L.; Jaitin, D.; Wieghofer, P.; Staszewski, O.; David, E.; Keren-Shaul, H.; Mahlakoiv, T.; Jakobshagen, K.; Buch, T.; et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015, 18, 965–977. [Google Scholar] [CrossRef]
- Kim, C.H. B cell-helping functions of gut microbial metabolites. Microb. Cell 2016, 3, 529–531. [Google Scholar] [CrossRef]
- Park, J.; Goergen, C.J.; HogenEsch, H.; Kim, C.H. Chronically Elevated Levels of Short-Chain Fatty Acids Induce T Cell-Mediated Ureteritis and Hydronephrosis. J. Immunol. 2016, 196, 2388–2400. [Google Scholar] [CrossRef]
- Park, J.; Kim, M.; Kang, S.G.; Jannasch, A.H.; Cooper, B.; Patterson, J.; Kim, C.H. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal. Immunol. 2015, 8, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Asarat, M.; Apostolopoulos, V.; Vasiljevic, T.; Donkor, O. Short-Chain Fatty Acids Regulate Cytokines and Th17/Treg Cells in Human Peripheral Blood Mononuclear Cells in vitro. Immunol. Investig. 2016, 45, 205–222. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Hinson, S.R.; Romero, M.F.; Popescu, B.F.; Lucchinetti, C.F.; Fryer, J.P.; Wolburg, H.; Fallier-Becker, P.; Noell, S.; Lennon, V.A. Molecular outcomes of neuromyelitis optica (NMO)-IgG binding to aquaporin-4 in astrocytes. Proc. Natl. Acad. Sci. USA 2012, 109, 1245–1250. [Google Scholar] [CrossRef]
- Hinson, S.R.; Clift, I.C.; Luo, N.; Kryzer, T.J.; Lennon, V.A. Autoantibody-induced internalization of CNS AQP4 water channel and EAAT2 glutamate transporter requires astrocytic Fc receptor. Proc. Natl. Acad. Sci. USA 2017, 114, 5491–5496. [Google Scholar] [CrossRef]
- Zeng, X.N.; Sun, X.L.; Gao, L.; Fan, Y.; Ding, J.H.; Hu, G. Aquaporin-4 deficiency down-regulates glutamate uptake and GLT-1 expression in astrocytes. Mol. Cell Neurosci. 2007, 34, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Pietzke, M.; Meiser, J.; Vazquez, A. Formate metabolism in health and disease. Mol. Metab. 2020, 33, 23–37. [Google Scholar] [CrossRef]
- Ma, E.H.; Bantug, G.; Griss, T.; Condotta, S.; Johnson, R.M.; Samborska, B.; Mainolfi, N.; Suri, V.; Guak, H.; Balmer, M.L.; et al. Serine Is an Essential Metabolite for Effector T Cell Expansion. Cell Metab. 2017, 25, 345–357. [Google Scholar] [CrossRef]
- Ananieva, E.A.; Powell, J.D.; Hutson, S.M. Leucine Metabolism in T Cell Activation: mTOR Signaling and Beyond. Adv. Nutr. 2016, 7, 798S–805S. [Google Scholar] [CrossRef] [PubMed]
- Cree, B.A.; Spencer, C.M.; Varrin-Doyer, M.; Baranzini, S.E.; Zamvil, S.S. Gut microbiome analysis in neuromyelitis optica reveals overabundance of Clostridium perfringens. Ann. Neurol. 2016, 80, 443–447. [Google Scholar] [CrossRef]
- Macfarlane, S.; Macfarlane, G.T. Regulation of short-chain fatty acid production. Proc. Nutr. Soc. 2003, 62, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Bittner, C.X.; Loaiza, A.; Ruminot, I.; Larenas, V.; Sotelo-Hitschfeld, T.; Gutierrez, R.; Cordova, A.; Valdebenito, R.; Frommer, W.B.; Barros, L.F. High resolution measurement of the glycolytic rate. Front. Neuroenergetics 2010, 2. [Google Scholar] [CrossRef] [PubMed]
- Belanger, M.; Allaman, I.; Magistretti, P.J. Brain energy metabolism: Focus on astrocyte-neuron metabolic cooperation. Cell Metab. 2011, 14, 724–738. [Google Scholar] [CrossRef]
- Lovatt, D.; Sonnewald, U.; Waagepetersen, H.S.; Schousboe, A.; He, W.; Lin, J.H.; Han, X.; Takano, T.; Wang, S.; Sim, F.J.; et al. The transcriptome and metabolic gene signature of protoplasmic astrocytes in the adult murine cortex. J. Neurosci. 2007, 27, 12255–12266. [Google Scholar] [CrossRef]
- Pellerin, L.; Magistretti, P.J. Glutamate uptake into astrocytes stimulates aerobic glycolysis: A mechanism coupling neuronal activity to glucose utilization. Proc. Natl. Acad. Sci. USA 1994, 91, 10625–10629. [Google Scholar] [CrossRef]
- Magistretti, P.J. Role of glutamate in neuron-glia metabolic coupling. Am. J. Clin. Nutr. 2009, 90, 875S–880S. [Google Scholar] [CrossRef]
- Machler, P.; Wyss, M.T.; Elsayed, M.; Stobart, J.; Gutierrez, R.; von Faber-Castell, A.; Kaelin, V.; Zuend, M.; San Martin, A.; Romero-Gomez, I.; et al. In Vivo Evidence for a Lactate Gradient from Astrocytes to Neurons. Cell Metab. 2016, 23, 94–102. [Google Scholar] [CrossRef]
- Genc, S.; Kurnaz, I.A.; Ozilgen, M. Astrocyte-neuron lactate shuttle may boost more ATP supply to the neuron under hypoxic conditions--in silico study supported by in vitro expression data. BMC Syst. Biol. 2011, 5, 162. [Google Scholar] [CrossRef]
- Yabu, M.; Shime, H.; Hara, H.; Saito, T.; Matsumoto, M.; Seya, T.; Akazawa, T.; Inoue, N. IL-23-dependent and -independent enhancement pathways of IL-17A production by lactic acid. Int. Immunol. 2011, 23, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Haas, R.; Smith, J.; Rocher-Ros, V.; Nadkarni, S.; Montero-Melendez, T.; D’Acquisto, F.; Bland, E.J.; Bombardieri, M.; Pitzalis, C.; Perretti, M.; et al. Lactate Regulates Metabolic and Pro-inflammatory Circuits in Control of T Cell Migration and Effector Functions. PLoS Biol. 2015, 13, e1002202. [Google Scholar] [CrossRef] [PubMed]
- Shime, H.; Yabu, M.; Akazawa, T.; Kodama, K.; Matsumoto, M.; Seya, T.; Inoue, N. Tumor-secreted lactic acid promotes IL-23/IL-17 proinflammatory pathway. J. Immunol. 2008, 180, 7175–7183. [Google Scholar] [CrossRef] [PubMed]
- Morishima, T.; Aoyama, M.; Iida, Y.; Yamamoto, N.; Hirate, H.; Arima, H.; Fujita, Y.; Sasano, H.; Tsuda, T.; Katsuya, H.; et al. Lactic acid increases aquaporin 4 expression on the cell membrane of cultured rat astrocytes. Neurosci. Res. 2008, 61, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, H.; Hu, X.; Peng, F.; Yang, Y. Serum lipoprotein levels in patients with neuromyelitis optica elevated but had little correlation with clinical presentations. Clin. Neurol. Neurosurg. 2010, 112, 478–481. [Google Scholar] [CrossRef] [PubMed]
- Pokharel, S.M.; Shil, N.K.; Gc, J.B.; Colburn, Z.T.; Tsai, S.Y.; Segovia, J.A.; Chang, T.H.; Bandyopadhyay, S.; Natesan, S.; Jones, J.C.R.; et al. Integrin activation by the lipid molecule 25-hydroxycholesterol induces a proinflammatory response. Nat. Commun. 2019, 10, 1482. [Google Scholar] [CrossRef]
- Rosen, Y.; Lenkinski, R.E. Recent advances in magnetic resonance neurospectroscopy. Neurotherapeutics 2007, 4, 330–345. [Google Scholar] [CrossRef]
- Pichiecchio, A.; Tavazzi, E.; Poloni, G.; Ponzio, M.; Palesi, F.; Pasin, M.; Piccolo, L.; Tosello, D.; Romani, A.; Bergamaschi, R.; et al. Advanced magnetic resonance imaging of neuromyelitis optica: A multiparametric approach. Mult. Scler. J. 2012, 18, 817–824. [Google Scholar] [CrossRef]
- Duan, Y.; Liu, Z.; Liu, Y.; Huang, J.; Ren, Z.; Sun, Z.; Chen, H.; Dong, H.; Ye, J.; Li, K. Metabolic changes in normal-appearing white matter in patients with neuromyelitis optica and multiple sclerosis: A comparative magnetic resonance spectroscopy study. Acta Radiol. 2017. [Google Scholar] [CrossRef]
- Bichuetti, D.B.; Rivero, R.L.; de Oliveira, E.M.; Oliveira, D.M.; de Souza, N.A.; Nogueira, R.G.; Abdala, N.; Gabbai, A. White matter spectroscopy in neuromyelitis optica: A case control study. J. Neurol. 2008, 255, 1895–1899. [Google Scholar] [CrossRef]
- Aboul-Enein, F.; Krssak, M.; Hoftberger, R.; Prayer, D.; Kristoferitsch, W. Diffuse white matter damage is absent in neuromyelitis optica. AJNR Am. J. Neuroradiol. 2010, 31, 76–79. [Google Scholar] [CrossRef] [PubMed]
- de Seze, J.; Blanc, F.; Kremer, S.; Collongues, N.; Fleury, M.; Marcel, C.; Namer, I.J. Magnetic resonance spectroscopy evaluation in patients with neuromyelitis optica. J. Neurol. Neurosurg. Psychiatry 2010, 81, 409–411. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Spraggins, J.M.; Rose, K.L.; Schey, K.L. High spatial resolution imaging mass spectrometry of human optic nerve lipids and proteins. J. Am. Soc. Mass Spectrom. 2015, 26, 940–947. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, J.; McKinney, K.Q.; Pavlopoulos, A.J.; Han, M.H.; Kim, S.H.; Kim, H.J.; Hwang, S. Exosomal proteome analysis of cerebrospinal fluid detects biosignatures of neuromyelitis optica and multiple sclerosis. Clin. Chim. Acta 2016, 462, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.F.; Lu, Q.Y.; Hu, S.; Chen, Y.; Liu, X.L.; Yang, Y.; Ding, M.P. Proteomics comparison of the sera from multiple sclerosis patients and neuromyelitis optica patients. Genet. Mol. Res. 2014, 13, 9292–9299. [Google Scholar] [CrossRef]
- Jiang, S.; Wu, J.; Yang, Y.; Liu, J.; Ding, Y.; Ding, M. Proteomic analysis of the cerebrospinal fluid in multiple sclerosis and neuromyelitis optica patients. Mol. Med. Rep. 2012, 6, 1081–1086. [Google Scholar] [CrossRef]
- Bai, S.; Liu, S.; Guo, X.; Qin, Z.; Wang, B.; Li, X.; Qin, Y.; Liu, Y.H. Proteome analysis of biomarkers in the cerebrospinal fluid of neuromyelitis optica patients. Mol. Vis. 2009, 15, 1638–1648. [Google Scholar]
- Bai, S.; Liu, S.; Guo, X.; Qin, Z.; Wang, B.; Li, X.; Qin, Y. Proteome analysis of haptoglobin in cerebrospinal fluid of neuromyelitis optica. Mol. Biol. Rep. 2010, 37, 1619–1625. [Google Scholar] [CrossRef]
- Nielsen, H.H.; Beck, H.C.; Kristensen, L.P.; Burton, M.; Csepany, T.; Simo, M.; Dioszeghy, P.; Sejbaek, T.; Grebing, M.; Heegaard, N.H.; et al. The Urine Proteome Profile Is Different in Neuromyelitis Optica Compared to Multiple Sclerosis: A Clinical Proteome Study. PLoS ONE 2015, 10, e0139659. [Google Scholar] [CrossRef]
- Komori, M.; Matsuyama, Y.; Nirasawa, T.; Thiele, H.; Becker, M.; Alexandrov, T.; Saida, T.; Tanaka, M.; Matsuo, H.; Tomimoto, H.; et al. Proteomic pattern analysis discriminates among multiple sclerosis-related disorders. Ann. Neurol. 2012, 71, 614–623. [Google Scholar] [CrossRef]
- Kowarik, M.C.; Dzieciatkowska, M.; Wemlinger, S.; Ritchie, A.M.; Hemmer, B.; Owens, G.P.; Bennett, J.L. The cerebrospinal fluid immunoglobulin transcriptome and proteome in neuromyelitis optica reveals central nervous system-specific B cell populations. J. Neuroinflamm. 2015, 12, 19. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.H.; Tseng, M.Y.; Ro, L.S.; Lyu, R.K.; Tai, Y.H.; Chang, H.S.; Wu, Y.R.; Huang, C.C.; Hsu, W.C.; Kuo, H.C.; et al. Analyses of haptoglobin level in the cerebrospinal fluid and serum of patients with neuromyelitis optica and multiple sclerosis. Clin. Chim. Acta 2013, 417, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Miller, Y.I.; Altamentova, S.M.; Shaklai, N. Oxidation of low-density lipoprotein by hemoglobin stems from a heme-initiated globin radical: Antioxidant role of haptoglobin. Biochemistry 1997, 36, 12189–12198. [Google Scholar] [CrossRef]
- Schaer, D.J.; Buehler, P.W.; Alayash, A.I.; Belcher, J.D.; Vercellotti, G.M. Hemolysis and free hemoglobin revisited: Exploring hemoglobin and hemin scavengers as a novel class of therapeutic proteins. Blood 2013, 121, 1276–1284. [Google Scholar] [CrossRef]
- Bertaggia, E.; Scabia, G.; Dalise, S.; Lo Verso, F.; Santini, F.; Vitti, P.; Chisari, C.; Sandri, M.; Maffei, M. Haptoglobin is required to prevent oxidative stress and muscle atrophy. PLoS ONE 2014, 9, e100745. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.K.; Ferraro, B.; Moore, K.; Halliwell, B. Role of haptoglobin in free hemoglobin metabolism. Redox Rep. 2001, 6, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.F.; Lin, C.C.; Huang, H.Y.; Liu, H.C.; Mao, S.J. Antioxidant role of human haptoglobin. Proteomics 2004, 4, 2221–2228. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.K.; Pavlotsky, N.; Tauber, A.I. Specific binding of haptoglobin to human neutrophils and its functional consequences. J. Leukoc Biol. 1990, 47, 142–148. [Google Scholar] [CrossRef]
- Wagner, L.; Gessl, A.; Parzer, S.B.; Base, W.; Waldhausl, W.; Pasternack, M.S. Haptoglobin phenotyping by newly developed monoclonal antibodies. Demonstration of haptoglobin uptake into peripheral blood neutrophils and monocytes. J. Immunol. 1996, 156, 1989–1996. [Google Scholar]
- Huntoon, K.M.; Russell, L.; Tracy, E.; Barbour, K.W.; Li, Q.; Shrikant, P.A.; Berger, F.G.; Garrett-Sinha, L.A.; Baumann, H. A unique form of haptoglobin produced by murine hematopoietic cells supports B-cell survival, differentiation and immune response. Mol. Immunol. 2013, 55, 345–354. [Google Scholar] [CrossRef]
- Arredouani, M.; Matthijs, P.; Van Hoeyveld, E.; Kasran, A.; Baumann, H.; Ceuppens, J.L.; Stevens, E. Haptoglobin directly affects T cells and suppresses T helper cell type 2 cytokine release. Immunology 2003, 108, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Huntoon, K.M.; Wang, Y.; Eppolito, C.A.; Barbour, K.W.; Berger, F.G.; Shrikant, P.A.; Baumann, H. The acute phase protein haptoglobin regulates host immunity. J. Leukoc Biol. 2008, 84, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Song, S.; Sun, G.; Strong, R.; Zhang, J.; Grotta, J.C.; Aronowski, J. Neuroprotective role of haptoglobin after intracerebral hemorrhage. J. Neurosci. 2009, 29, 15819–15827. [Google Scholar] [CrossRef] [PubMed]
- Delanghe, J.R.; Langlois, M.R.; De Buyzere, M.L. Haptoglobin polymorphism: A key factor in the proatherogenic role of B cells? Atherosclerosis 2011, 217, 80–82. [Google Scholar] [CrossRef]
- Gruys, E.; Toussaint, M.J.; Niewold, T.A.; Koopmans, S.J. Acute phase reaction and acute phase proteins. J. Zhejiang Univ. Sci. B 2005, 6, 1045–1056. [Google Scholar] [CrossRef]
- Arguelles, S.; Venero, J.L.; Garcia-Rodriguez, S.; Tomas-Camardiel, M.; Ayala, A.; Cano, J.; Machado, A. Use of haptoglobin and transthyretin as potential biomarkers for the preclinical diagnosis of Parkinson’s disease. Neurochem. Int. 2010, 57, 227–234. [Google Scholar] [CrossRef]
- Marchi, N.; Fazio, V.; Cucullo, L.; Kight, K.; Masaryk, T.; Barnett, G.; Vogelbaum, M.; Kinter, M.; Rasmussen, P.; Mayberg, M.R.; et al. Serum transthyretin monomer as a possible marker of blood-to-CSF barrier disruption. J. Neurosci. 2003, 23, 1949–1955. [Google Scholar] [CrossRef]
- Roemer, S.F.; Parisi, J.E.; Lennon, V.A.; Benarroch, E.E.; Lassmann, H.; Bruck, W.; Mandler, R.N.; Weinshenker, B.G.; Pittock, S.J.; Wingerchuk, D.M.; et al. Pattern-specific loss of aquaporin-4 immunoreactivity distinguishes neuromyelitis optica from multiple sclerosis. Brain 2007, 130, 1194–1205. [Google Scholar] [CrossRef]
- Tzartos, J.S.; Stergiou, C.; Kilidireas, K.; Zisimopoulou, P.; Thomaidis, T.; Tzartos, S.J. Anti-aquaporin-1 autoantibodies in patients with neuromyelitis optica spectrum disorders. PLoS ONE 2013, 8, e74773. [Google Scholar] [CrossRef]
- Long, Y.; Zheng, Y.; Shan, F.; Chen, M.; Fan, Y.; Zhang, B.; Gao, C.; Gao, Q.; Yang, N. Development of a cell-based assay for the detection of anti-aquaporin 1 antibodies in neuromyelitis optica spectrum disorders. J. Neuroimmunol. 2014, 273, 103–110. [Google Scholar] [CrossRef]
- Brill, L.; Goldberg, L.; Karni, A.; Petrou, P.; Abramsky, O.; Ovadia, H.; Ben-Hur, T.; Karussis, D.; Vaknin-Dembinsky, A. Increased anti-KIR4.1 antibodies in multiple sclerosis: Could it be a marker of disease relapse? Mult. Scler. J. 2015, 21, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, F.; Schaller, K.L.; Owens, G.P.; Cotleur, A.C.; Kellner, D.; Takeshita, Y.; Obermeier, B.; Kryzer, T.J.; Sano, Y.; Kanda, T.; et al. Glucose-regulated protein 78 autoantibody associates with blood-brain barrier disruption in neuromyelitis optica. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Wingerchuk, D.M.; Banwell, B.; Bennett, J.L.; Cabre, P.; Carroll, W.; Chitnis, T.; de Seze, J.; Fujihara, K.; Greenberg, B.; Jacob, A.; et al. International consensus diagnostic criteria for neuromyelitis optica spectrum disorders. Neurology 2015, 85, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Correale, J.; Fiol, M. Chitinase effects on immune cell response in neuromyelitis optica and multiple sclerosis. Mult. Scler. J. 2011, 17, 521–531. [Google Scholar] [CrossRef]
- Saadoun, S.; Waters, P.; Owens, G.P.; Bennett, J.L.; Vincent, A.; Papadopoulos, M.C. Neuromyelitis optica MOG-IgG causes reversible lesions in mouse brain. Acta Neuropathol. Commun. 2014, 2, 35. [Google Scholar] [CrossRef]
- Hakobyan, S.; Luppe, S.; Evans, D.R.; Harding, K.; Loveless, S.; Robertson, N.P.; Morgan, B.P. Plasma complement biomarkers distinguish multiple sclerosis and neuromyelitis optica spectrum disorder. Mult. Scler. J. 2017, 23, 946–955. [Google Scholar] [CrossRef]
- Yoshikura, N.; Kimura, A.; Hayashi, Y.; Inuzuka, T. Anti-C1q autoantibodies in patients with neuromyelitis optica spectrum disorders. J. Neuroimmunol. 2017, 310, 150–157. [Google Scholar] [CrossRef]
- Nytrova, P.; Potlukova, E.; Kemlink, D.; Woodhall, M.; Horakova, D.; Waters, P.; Havrdova, E.; Zivorova, D.; Vincent, A.; Trendelenburg, M. Complement activation in patients with neuromyelitis optica. J. Neuroimmunol. 2014, 274, 185–191. [Google Scholar] [CrossRef]
- Tuzun, E.; Kurtuncu, M.; Turkoglu, R.; Icoz, S.; Pehlivan, M.; Birisik, O.; Eraksoy, M.; Akman-Demir, G. Enhanced complement consumption in neuromyelitis optica and Behcet’s disease patients. J. Neuroimmunol. 2011, 233, 211–215. [Google Scholar] [CrossRef]
- Kuroda, H.; Fujihara, K.; Takano, R.; Takai, Y.; Takahashi, T.; Misu, T.; Nakashima, I.; Sato, S.; Itoyama, Y.; Aoki, M. Increase of complement fragment C5a in cerebrospinal fluid during exacerbation of neuromyelitis optica. J. Neuroimmunol. 2013, 254, 178–182. [Google Scholar] [CrossRef]
- Doi, H.; Matsushita, T.; Isobe, N.; Matsuoka, T.; Minohara, M.; Ochi, H.; Kira, J.I. Hypercomplementemia at relapse in patients with anti-aquaporin-4 antibody. Mult. Scler. J. 2009, 15, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, K.; Wang, C.; Qiu, W.; Lu, Z.; Hu, X. Increased soluble C5b-9 in CSF of neuromyelitis optica. Scand. J. Immunol. 2014, 79, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Melamed, E.; Levy, M.; Waters, P.J.; Sato, D.K.; Bennett, J.L.; John, G.R.; Hooper, D.C.; Saiz, A.; Bar-Or, A.; Kim, H.J.; et al. Update on biomarkers in neuromyelitis optica. Neurol. Neuroimmunol. Neuroinflamm. 2015, 2, e134. [Google Scholar] [CrossRef] [PubMed]
- Uzawa, A.; Mori, M.; Kuwabara, S. Cytokines and chemokines in neuromyelitis optica: Pathogenetic and therapeutic implications. Brain Pathol. 2014, 24, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.H.; Ro, L.S.; Lyu, R.K.; Chen, C.M. Biomarkers for neuromyelitis optica. Clin. Chim. Acta 2015, 440, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Uzawa, A.; Mori, M.; Ito, M.; Uchida, T.; Hayakawa, S.; Masuda, S.; Kuwabara, S. Markedly increased CSF interleukin-6 levels in neuromyelitis optica, but not in multiple sclerosis. J. Neurol. 2009, 256, 2082–2084. [Google Scholar] [CrossRef]
- Correale, J.; Fiol, M. Activation of humoral immunity and eosinophils in neuromyelitis optica. Neurology 2004, 63, 2363–2370. [Google Scholar] [CrossRef]
- Wang, H.; Wang, K.; Zhong, X.; Qiu, W.; Dai, Y.; Wu, A.; Hu, X. Cerebrospinal fluid BAFF and APRIL levels in neuromyelitis optica and multiple sclerosis patients during relapse. J. Clin. Immunol. 2012, 32, 1007–1011. [Google Scholar] [CrossRef]
- Uzawa, A.; Mori, M.; Arai, K.; Sato, Y.; Hayakawa, S.; Masuda, S.; Taniguchi, J.; Kuwabara, S. Cytokine and chemokine profiles in neuromyelitis optica: Significance of interleukin-6. Mult. Scler. J. 2010, 16, 1443–1452. [Google Scholar] [CrossRef]
- Wang, H.; Wang, K.; Zhong, X.; Dai, Y.; Qiu, W.; Wu, A.; Hu, X. Notable increased cerebrospinal fluid levels of soluble interleukin-6 receptors in neuromyelitis optica. Neuroimmunomodulation 2012, 19, 304–308. [Google Scholar] [CrossRef]
- Matsushita, T.; Tateishi, T.; Isobe, N.; Yonekawa, T.; Yamasaki, R.; Matsuse, D.; Murai, H.; Kira, J. Characteristic cerebrospinal fluid cytokine/chemokine profiles in neuromyelitis optica, relapsing remitting or primary progressive multiple sclerosis. PLoS ONE 2013, 8, e61835. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Wang, H.; Dai, Y.; Wu, A.; Bao, J.; Xu, W.; Cheng, C.; Lu, Z.; Qiu, W.; Hu, X. Cerebrospinal fluid levels of CXCL13 are elevated in neuromyelitis optica. J. Neuroimmunol. 2011, 240-241, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, E.; Piccio, L.; Mikesell, R.J.; Klawiter, E.C.; Parks, B.J.; Naismith, R.T.; Cross, A.H. CXCL13 is a biomarker of inflammation in multiple sclerosis, neuromyelitis optica, and other neurological conditions. Mult. Scler. J. 2013, 19, 1204–1208. [Google Scholar] [CrossRef] [PubMed]
- Ishizu, T.; Osoegawa, M.; Mei, F.J.; Kikuchi, H.; Tanaka, M.; Takakura, Y.; Minohara, M.; Murai, H.; Mihara, F.; Taniwaki, T.; et al. Intrathecal activation of the IL-17/IL-8 axis in opticospinal multiple sclerosis. Brain 2005, 128, 988–1002. [Google Scholar] [CrossRef]
- Okada, K.; Matsushita, T.; Kira, J.; Tsuji, S. B-cell activating factor of the TNF family is upregulated in neuromyelitis optica. Neurology 2010, 74, 177–178. [Google Scholar] [CrossRef]
- Takano, R.; Misu, T.; Takahashi, T.; Sato, S.; Fujihara, K.; Itoyama, Y. Astrocytic damage is far more severe than demyelination in NMO: A clinical CSF biomarker study. Neurology 2010, 75, 208–216. [Google Scholar] [CrossRef]
- Uzawa, A.; Mori, M.; Masuda, S.; Kuwabara, S. Markedly elevated soluble intercellular adhesion molecule 1, soluble vascular cell adhesion molecule 1 levels, and blood-brain barrier breakdown in neuromyelitis optica. Arch. Neurol. 2011, 68, 913–917. [Google Scholar] [CrossRef]
- Uzawa, A.; Mori, M.; Sawai, S.; Masuda, S.; Muto, M.; Uchida, T.; Ito, S.; Nomura, F.; Kuwabara, S. Cerebrospinal fluid interleukin-6 and glial fibrillary acidic protein levels are increased during initial neuromyelitis optica attacks. Clin. Chim. Acta 2013, 421, 181–183. [Google Scholar] [CrossRef]
- Uzawa, A.; Mori, M.; Masuda, H.; Ohtani, R.; Uchida, T.; Sawai, S.; Kuwabara, S. Interleukin-6 analysis of 572 consecutive CSF samples from neurological disorders: A special focus on neuromyelitis optica. Clin. Chim. Acta 2017, 469, 144–149. [Google Scholar] [CrossRef]
- Uchida, T.; Mori, M.; Uzawa, A.; Masuda, H.; Muto, M.; Ohtani, R.; Kuwabara, S. Increased cerebrospinal fluid metalloproteinase-2 and interleukin-6 are associated with albumin quotient in neuromyelitis optica: Their possible role on blood-brain barrier disruption. Mult. Scler. J. 2017, 23, 1072–1084. [Google Scholar] [CrossRef]
- Hosokawa, T.; Nakajima, H.; Doi, Y.; Sugino, M.; Kimura, F.; Hanafusa, T.; Takahashi, T. Increased serum matrix metalloproteinase-9 in neuromyelitis optica: Implication of disruption of blood-brain barrier. J. Neuroimmunol. 2011, 236, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Masuda, H.; Mori, M.; Uchida, T.; Uzawa, A.; Ohtani, R.; Kuwabara, S. Soluble CD40 ligand contributes to blood-brain barrier breakdown and central nervous system inflammation in multiple sclerosis and neuromyelitis optica spectrum disorder. J. Neuroimmunol. 2017, 305, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Zhong, X.; Wang, H.; Xu, W.; Cheng, C.; Dai, Y.; Bao, J.; Qiu, W.; Lu, Z.; Hu, X. Cerebrospinal fluid IL-21 levels in Neuromyelitis Optica and multiple sclerosis. Can. J. Neurol. Sci. 2012, 39, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Narikawa, K.; Misu, T.; Fujihara, K.; Nakashima, I.; Sato, S.; Itoyama, Y. CSF chemokine levels in relapsing neuromyelitis optica and multiple sclerosis. J. Neuroimmunol. 2004, 149, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Yanagawa, K.; Kawachi, I.; Toyoshima, Y.; Yokoseki, A.; Arakawa, M.; Hasegawa, A.; Ito, T.; Kojima, N.; Koike, R.; Tanaka, K.; et al. Pathologic and immunologic profiles of a limited form of neuromyelitis optica with myelitis. Neurology 2009, 73, 1628–1637. [Google Scholar] [CrossRef]
- Kang, H.; Cao, S.; Chen, T.; Jiang, Z.; Liu, Z.; Li, Z.; Wei, Y.; Ai, N.; Xu, Q.; Lin, Q.; et al. The poor recovery of neuromyelitis optica spectrum disorder is associated with a lower level of CXCL12 in the human brain. J. Neuroimmunol. 2015, 289, 56–61. [Google Scholar] [CrossRef]
- Tingjun, C.; Zhaohui, L.; Zhaocai, J.; Zihao, L.; Quangang, X.; Dehui, H.; Qing, L.; Shihui, W. Changes of CXCL12, CXCL14 and PDGF levels in the brain of patients with idiopathic demyelinating optic neuritis and neuromyelitis optica. J. Neuroimmunol. 2015, 279, 1–6. [Google Scholar] [CrossRef]
- Wang, H.; Wang, C.; Qiu, W.; Lu, Z.; Hu, X.; Wang, K. Cerebrospinal fluid light and heavy neurofilaments in neuromyelitis optica. Neurochem. Int. 2013, 63, 805–808. [Google Scholar] [CrossRef]
- Misu, T.; Takano, R.; Fujihara, K.; Takahashi, T.; Sato, S.; Itoyama, Y. Marked increase in cerebrospinal fluid glial fibrillar acidic protein in neuromyelitis optica: An astrocytic damage marker. J. Neurol. Neurosurg. Psychiatry 2009, 80, 575–577. [Google Scholar] [CrossRef]
- Uzawa, A.; Mori, M.; Masuda, S.; Muto, M.; Kuwabara, S. CSF high-mobility group box 1 is associated with intrathecal inflammation and astrocytic damage in neuromyelitis optica. J. Neurol. Neurosurg. Psychiatry 2013, 84, 517–522. [Google Scholar] [CrossRef]
- Wang, H.; Wang, K.; Wang, C.; Xu, F.; Zhong, X.; Qiu, W.; Hu, X. Cerebrospinal fluid high-mobility group box protein 1 in neuromyelitis optica and multiple sclerosis. Neuroimmunomodulation 2013, 20, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Quan, C.; Yu, H.; Qiao, J.; Xiao, B.; Zhao, G.; Wu, Z.; Li, Z.; Lu, C. Impaired regulatory function and enhanced intrathecal activation of B cells in neuromyelitis optica: Distinct from multiple sclerosis. Mult. Scler. J. 2013, 19, 289–298. [Google Scholar] [CrossRef]
- Zhang, D.Q.; Jia, K.; Wang, R.; Li, T.; Zhao, N.; Yang, L.N.; Yang, L. Decreased serum IL-27 and IL-35 levels are associated with disease severity in neuromyelitis optica spectrum disorders. J. Neuroimmunol. 2016, 293, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Uzawa, A.; Mori, M.; Uchida, T.; Masuda, H.; Ohtani, R.; Kuwabara, S. Increased levels of CSF CD59 in neuromyelitis optica and multiple sclerosis. Clin. Chim. Acta 2016, 453, 131–133. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Jia, K.; Zhang, D.Q.; Li, T.; Li, L.M.; Zhang, L.J.; Wang, J.; Gao, C.L.; Sun, L.S.; Shi, F.D.; et al. Increased resistin levels in the serum and cerebrospinal fluid of patients with neuromyelitis optica. Clin. Chim. Acta 2016, 456, 176–179. [Google Scholar] [CrossRef]
- Herges, K.; de Jong, B.A.; Kolkowitz, I.; Dunn, C.; Mandelbaum, G.; Ko, R.M.; Maini, A.; Han, M.H.; Killestein, J.; Polman, C.; et al. Protective effect of an elastase inhibitor in a neuromyelitis optica-like disease driven by a peptide of myelin oligodendroglial glycoprotein. Mult. Scler. J. 2012, 18, 398–408. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.H.; Dai, Y.Q.; Qiu, W.; Lu, Z.Q.; Peng, F.H.; Wang, Y.G.; Bao, J.; Li, Y.; Hu, X.Q. Interleukin-17-secreting T cells in neuromyelitis optica and multiple sclerosis during relapse. J. Clin. Neurosci. 2011, 18, 1313–1317. [Google Scholar] [CrossRef]
- Michael, B.D.; Elsone, L.; Griffiths, M.J.; Faragher, B.; Borrow, R.; Solomon, T.; Jacob, A. Post-acute serum eosinophil and neutrophil-associated cytokine/chemokine profile can distinguish between patients with neuromyelitis optica and multiple sclerosis; and identifies potential pathophysiological mechanisms—A pilot study. Cytokine 2013, 64, 90–96. [Google Scholar] [CrossRef]
- Yang, T.; Wang, S.; Zheng, Q.; Wang, L.; Li, Q.; Wei, M.; Du, Z.; Fan, Y. Increased plasma levels of epithelial neutrophil-activating peptide 78/CXCL5 during the remission of Neuromyelitis optica. BMC Neurol. 2016, 16, 96. [Google Scholar] [CrossRef]
- Alves-Leon, S.V.; Pimentel, M.L.; Sant’Anna, G.; Malfetano, F.R.; Estrada, C.D.; Quirico-Santos, T. Immune system markers of neuroinflammation in patients with clinical diagnose of neuromyelitis optica. Arq. Neuropsiquiatr. 2008, 66, 678–684. [Google Scholar] [CrossRef]
- Penton-Rol, G.; Cervantes-Llanos, M.; Martinez-Sanchez, G.; Cabrera-Gomez, J.A.; Valenzuela-Silva, C.M.; Ramirez-Nunez, O.; Casanova-Orta, M.; Robinson-Agramonte, M.A.; Lopategui-Cabezas, I.; Lopez-Saura, P.A. TNF-alpha and IL-10 downregulation and marked oxidative stress in Neuromyelitis Optica. J. Inflamm. (Lond.) 2009, 6, 18. [Google Scholar] [CrossRef]
- Li, Y.; Wang, H.; Long, Y.; Lu, Z.; Hu, X. Increased memory Th17 cells in patients with neuromyelitis optica and multiple sclerosis. J. Neuroimmunol. 2011, 234, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Li, R.; Dai, Y.; Wu, A.; Wang, H.; Cheng, C.; Qiu, W.; Lu, Z.; Zhong, X.; Shu, Y.; et al. IL-22 secreting CD4+ T cells in the patients with neuromyelitis optica and multiple sclerosis. J. Neuroimmunol. 2013, 261, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Lee, C.L.; Chen, S.Y.; Chen, J.C.; Yang, C.W.; Chen, S.J.; Tsai, C.P. Distinct serum cytokine profiles in neuromyelitis optica and multiple sclerosis. J. Interferon Cytokine Res. 2013, 33, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Storoni, M.; Verbeek, M.M.; Illes, Z.; Marignier, R.; Teunissen, C.E.; Grabowska, M.; Confavreux, C.; Plant, G.T.; Petzold, A. Serum GFAP levels in optic neuropathies. J. Neurol. Sci. 2012, 317, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yang, T.; Wan, J.; Zhang, Y.; Fan, Y. Elevated C-X-C motif ligand 13 and B-cell-activating factor levels in neuromyelitis optica during remission. Brain Behav. 2017, 7, e00648. [Google Scholar] [CrossRef]
- Zhong, X.; Wang, H.; Ye, Z.; Qiu, W.; Lu, Z.; Li, R.; Shu, Y.; Chang, Y.; Hu, X. Serum concentration of CD40L is elevated in inflammatory demyelinating diseases. J. Neuroimmunol. 2016, 299, 66–69. [Google Scholar] [CrossRef]
- Barros, P.O.; Cassano, T.; Hygino, J.; Ferreira, T.B.; Centuriao, N.; Kasahara, T.M.; Andrade, R.M.; Linhares, U.C.; Andrade, A.F.; Vasconcelos, C.C.; et al. Prediction of disease severity in neuromyelitis optica by the levels of interleukin (IL)-6 produced during remission phase. Clin. Exp. Immunol. 2016, 183, 480–489. [Google Scholar] [CrossRef]
- Wang, H.; Wang, K.; Wang, C.; Xu, F.; Qiu, W.; Hu, X. Increased plasma interleukin-32 expression in patients with neuromyelitis optica. J. Clin. Immunol. 2013, 33, 666–670. [Google Scholar] [CrossRef]
- Wang, K.C.; Tsai, C.P.; Lee, C.L.; Chen, S.Y.; Chin, L.T.; Chen, S.J. Elevated plasma high-mobility group box 1 protein is a potential marker for neuromyelitis optica. Neuroscience 2012, 226, 510–516. [Google Scholar] [CrossRef]

| Endpoint | Change | Biopsy | NMO Patients | Disease State a | AQP4-Ab | Control b | Method c | Ref. | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Female | Male | Relapse | Remission | + | − | ||||||
| 1-Monostearin | ↑ | CSF | 43 | 6 | 32 | 17 | NR | NR | 12 HC | GC-TOF-MS | [87] |
| 1-Monopalmitin | ↑ | CSF | 43 | 6 | 32 | 17 | NR | NR | 12 HC | GC-TOF-MS | [87] |
| 2-Hydroxybutyrate | ↑ | CSF | 51 | 6 | 36 | 21 | NR | NR | 17 HC | 1D & 2D 1H-NMR | [74] |
| 3-Hydroxybutyrate | ↓ | Urine | 6 | 3 | ⎯ | 9 | 9 | ⎯ | 8 RRMS, 7 HC | 1D 1H-NMR | [86] |
| 3-Hydroxyisovalerate | ↓ | Urine | 6 | 3 | ⎯ | 9 | 9 | ⎯ | 8 RRMS | 1D 1H-NMR | [86] |
| 3-Hydroxypropionic acid | ↓ | CSF | 43 | 6 | 32 | 17 | NR | NR | 12 HC | GC-TOF-MS | [87] |
| 25-Hydroxycholesterol | ↑ | CSF | 19 | 7 | 26 | ⎯ | 26 | ⎯ | 23 ONND | LC-ESI/MS/MS + PE | [89] |
| 27-Hydroxycholesterol | ↑ | CSF | 19 | 7 | 26 | ⎯ | 26 | ⎯ | 23 ONND | LC-ESI/MS/MS + PE | [89] |
| Acetate | ↑ | Serum | 32 | 12 | NR | NR | 22 | 22 | 47 RRMS, 42 HC | 1H HRMAS NMR | [85] |
| ↓ | CSF | 51 | 6 | 36 | 21 | NR | NR | 17 HC | 1D & 2D 1H-NMR | [74] | |
| Acetone | ↑ | CSF | 51 | 6 | 36 | 21 | NR | NR | 17 HC | 1D & 2D1H-NMR | [74] |
| Alanine | ↑ | Plasma | 46 | 8 | NR | NR | 56 | ⎯ | 34 RRMS + 20 MOG+ | 1D 1H-NOESY | [82] |
| Butane-2,3-Diol | ↓ | CSF | 43 | 6 | 32 | 17 | NR | NR | 12 HC | GC-TOF-MS | [87] |
| Citrate | ↓ | CSF | 51 | 6 | 36 | 21 | NR | NR | 50 MS | 1D & 2D 1H-NMR | [74] |
| Creatinine | ↓ | Urine | 6 | 3 | ⎯ | 9 | 9 | ⎯ | 8 RRMS, 7 HC | 1D 1H-NMR | [86] |
| Formate | ↑ | CSF | 51 | 6 | 36 | 21 | NR | NR | 17 HC | 1D & 2D 1H-NMR | [74] |
| (MOG-Ab+) * | ↑ | Plasma | 46 | 8 | NR | NR | 56 | ⎯ | 34 RRMS + AQPA-Ab+ | 1D 1H-NOESY | [82] |
| Fumaric Acid | ↑ | CSF | 43 | 6 | 32 | 17 | NR | NR | 12 HC, 54 RRMS | GC-TOF-MS | [87] |
| Glucose | ↓ | CSF | 51 | 6 | 36 | 21 | NR | NR | 17 HC | 1D & 2D 1H-NMR | [74] |
| ↑ | Plasma | 46 | 8 | NR | NR | 56 | ⎯ | 34 RRMS + 20 MOG+ | 1D 1H-NOESY | [82] | |
| Glutamate | ↑ | Serum | 32 | 12 | NR | NR | 22 | 22 | 47 RRMS, 42 HC | 1H HRMAS NMR | [85] |
| Glutamine | ↓ | Serum | 32 | 12 | NR | NR | 22 | 22 | 47 RRMS, 42 HC | 1H HRMAS NMR | [85] |
| Histidine | ↓ | Plasma | 46 | 8 | NR | NR | 56 | ⎯ | 34 RRMS + 20 MOG+ | 1D 1H-NOESY | [82] |
| Inosine | ↓ | CSF | 43 | 6 | 32 | 17 | NR | NR | 12 HC | GC-TOF-MS | [87] |
| Lactate/Lactic Acid | ↑ | CSF | 51 | 6 | 36 | 21 | NR | NR | 50 MS, 17 HC | 1D & 2D 1H NMR | [74] |
| ↑ | Plasma | 46 | 8 | NR | NR | 56 | ⎯ | 34 RRMS | 1D 1H-NOESY | [82] | |
| ↑ | Serum | 32 | 12 | NR | NR | 22 | 22 | 47 RRMS, 42 HC | 1H HRMAS NMR | [85] | |
| ↑ | CSF | 43 | 6 | 32 | 17 | NR | NR | 12 HC, 54 RRMS | GC-TOF-MS | [87] | |
| ↓ | Urine | 6 | 3 | ⎯ | 9 | 9 | ⎯ | 8 RRMS, 7 HC | 1D 1H-NMR | [86] | |
| Large LDL particles, Concentration | ↑ | Plasma | 46 | 8 | NR | NR | 56 | ⎯ | 34 RRMS + 20 MOG+ | 1D 1H-NOESY | [82] |
| Large LDL particles, Size | ↑ | Plasma | 46 | 8 | NR | NR | 56 | ⎯ | 34 RRMS + 20 MOG+ | 1D 1H-NOESY | [82] |
| Leucine (MOG-Ab+) * | ↑ | Plasma | 46 | 8 | NR | NR | 56 | ⎯ | 34 RRMS + AQPA-Ab+ | 1D 1H-NOESY | [82] |
| Lysine | ↓ | Plasma | 46 | 8 | NR | NR | 56 | ⎯ | 34 RRMS | 1D 1H-NOESY | [82] |
| ↑ | Serum | 32 | 12 | NR | NR | 22 | 22 | 47 RRMS | 1H HRMAS NMR | [85] | |
| ↓ | Serum | 32 | 12 | NR | NR | 22 | 22 | 42 HC | 1H HRMAS NMR | [85] | |
| Methylmalonate | ↓ | Urine | 6 | 3 | ⎯ | 9 | 9 | ⎯ | 8 RRMS, 7 HC | 1D 1H-NMR | [86] |
| Myo-Inositol (MOG-Ab+) * | ↓ | Plasma | 46 | 8 | NR | NR | 56 | ⎯ | 34 RRMS | 1D 1H-NOESY | [82] |
| NAA | ↓ | CSF | 24 | 8 | 10 | 22 | 13 | 19 | 48 RRMS | HPLC-MS/MS | [88] |
| ↓ | Serum | 24 | 8 | 10 | 22 | 13 | 19 | 48 RRMS | HPLC-MS/MS | [88] | |
| Oxaloacetate | ↑ | Urine | 6 | 3 | ⎯ | 9 | 9 | ⎯ | 7 HC | 1D 1H-NMR | [86] |
| Phosphocholine/lipoprotein Lipoprotein | ↓ | Plasma | 46 | 8 | NR | NR | 56 | ⎯ | 34 RRMS + 20 MOG+ | 1D 1H-NOESY | [82] |
| Pyroglutamate | ↑ | CSF | 51 | 6 | 36 | 21 | NR | NR | 17 HC | 1D & 2D 1H-NMR | [74] |
| Salicylaldehyde | ↑ | CSF | 43 | 6 | 32 | 17 | NR | NR | 12 HC | GC-TOF-MS | [87] |
| Scyllo-Inositol | ↓ | Serum | 32 | 12 | NR | NR | 22 | 22 | 47 RRMS | 1H HRMAS NMR | [85] |
| ↓ | Plasma | 46 | 8 | NR | NR | 56 | ⎯ | 34 RRMS + 20 MOG+ | 1D 1H-NOESY | [82] | |
| Small HDL Particles | ↓ | Plasma | 46 | 8 | NR | NR | 56 | ⎯ | 34 RRMS + 20 MOG+ | 1D 1H-NOESY | [82] |
| Threose | ↓ | CSF | 43 | 6 | 32 | 17 | NR | NR | 12 HC | GC-TOF-MS | [87] |
| Unsaturated Lipid | ↑ | Plasma | 46 | 8 | NR | NR | 56 | ⎯ | 34 RRMS | 1D 1H-NOESY | [82] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thoman, M.E.; McKarns, S.C. Metabolomic Profiling in Neuromyelitis Optica Spectrum Disorder Biomarker Discovery. Metabolites 2020, 10, 374. https://doi.org/10.3390/metabo10090374
Thoman ME, McKarns SC. Metabolomic Profiling in Neuromyelitis Optica Spectrum Disorder Biomarker Discovery. Metabolites. 2020; 10(9):374. https://doi.org/10.3390/metabo10090374
Chicago/Turabian StyleThoman, Maxton E., and Susan C. McKarns. 2020. "Metabolomic Profiling in Neuromyelitis Optica Spectrum Disorder Biomarker Discovery" Metabolites 10, no. 9: 374. https://doi.org/10.3390/metabo10090374
APA StyleThoman, M. E., & McKarns, S. C. (2020). Metabolomic Profiling in Neuromyelitis Optica Spectrum Disorder Biomarker Discovery. Metabolites, 10(9), 374. https://doi.org/10.3390/metabo10090374




