Comparison of Regular, Pure Shift, and Fast 2D NMR Experiments for Determination of the Geographical Origin of Walnuts
Abstract
:1. Introduction
2. Results
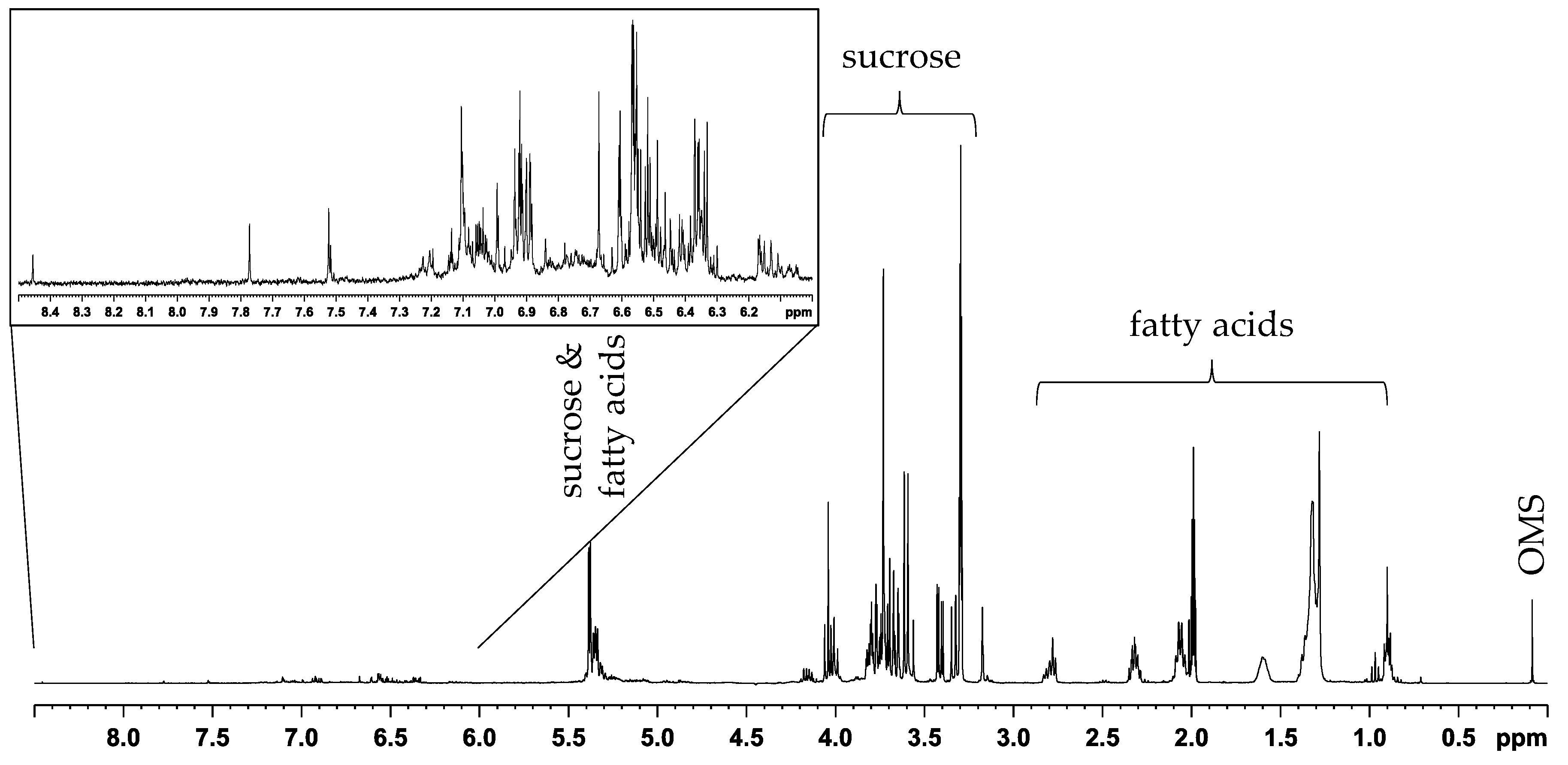
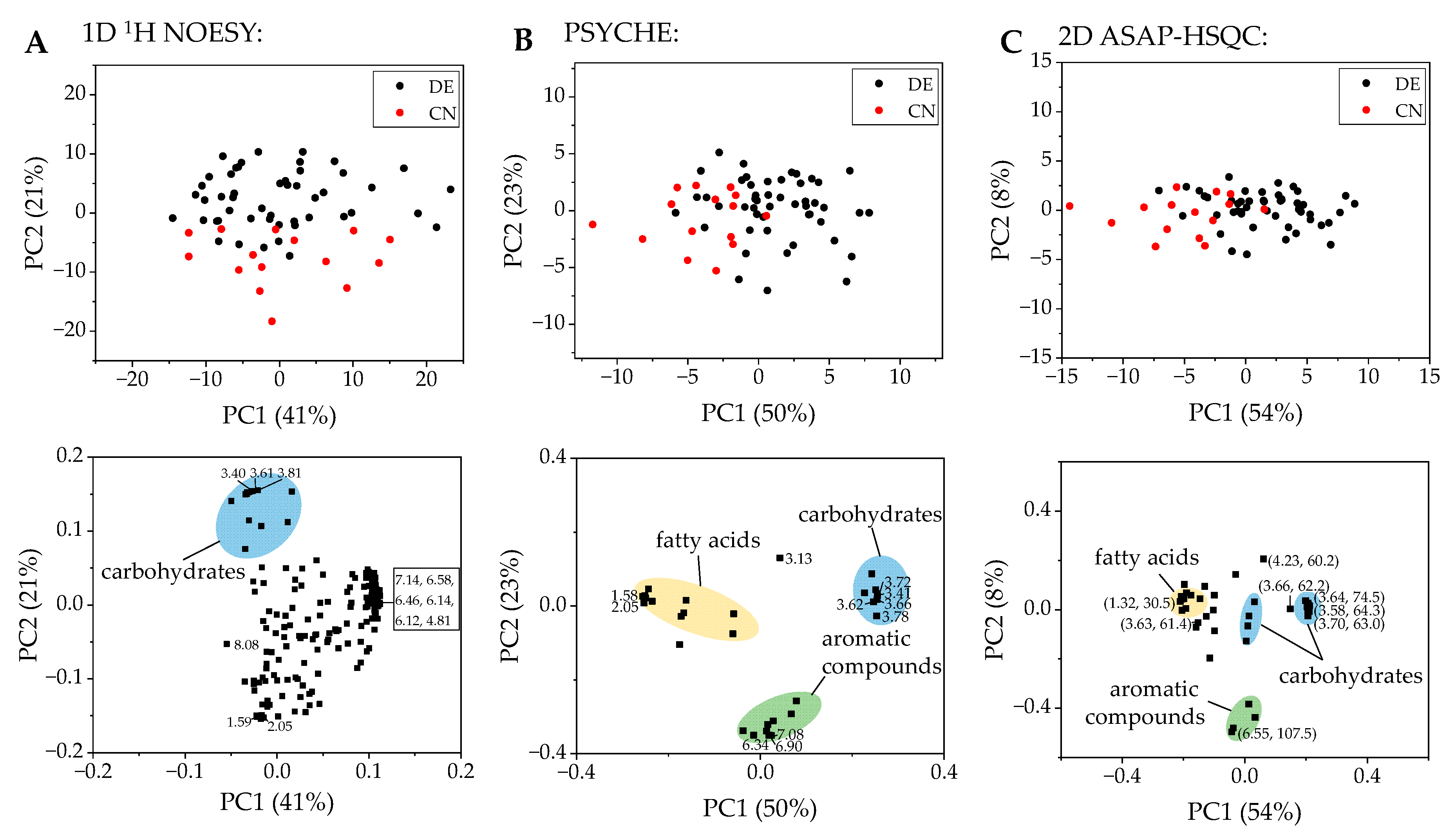
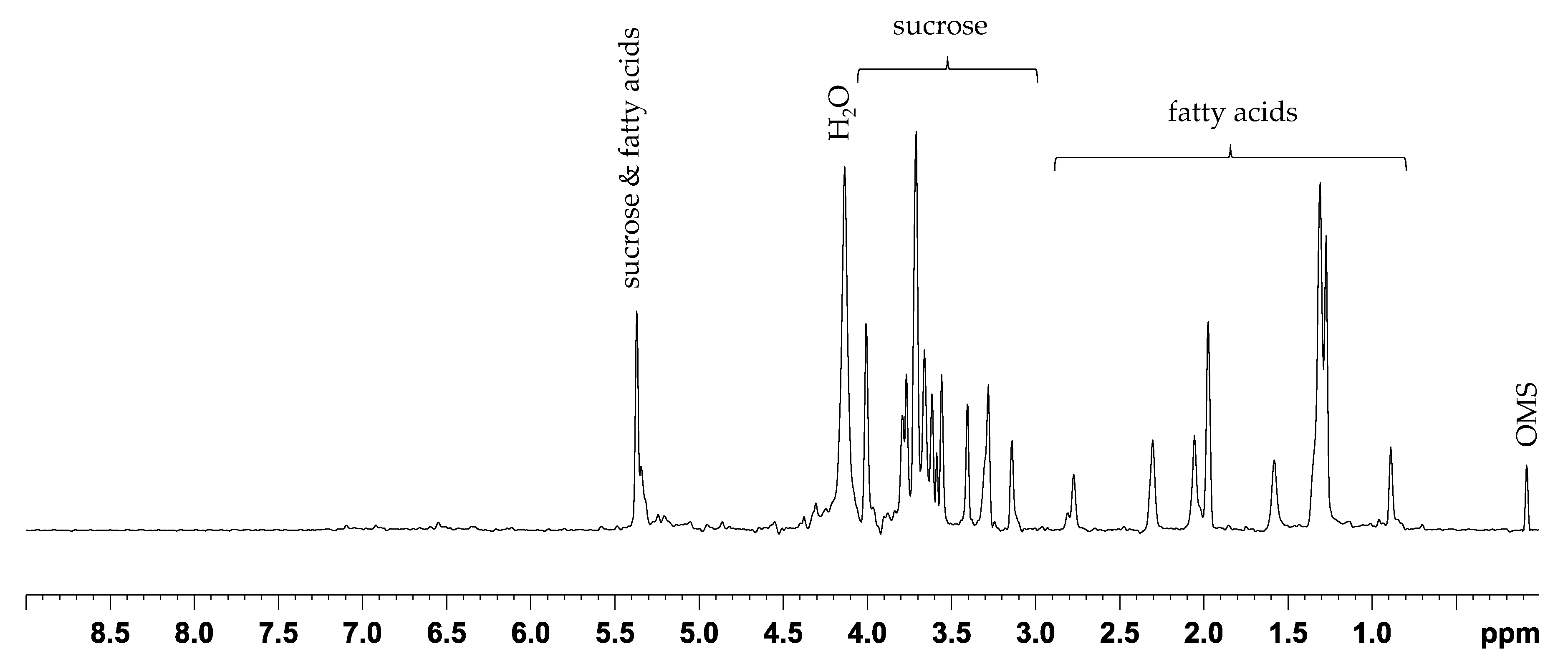
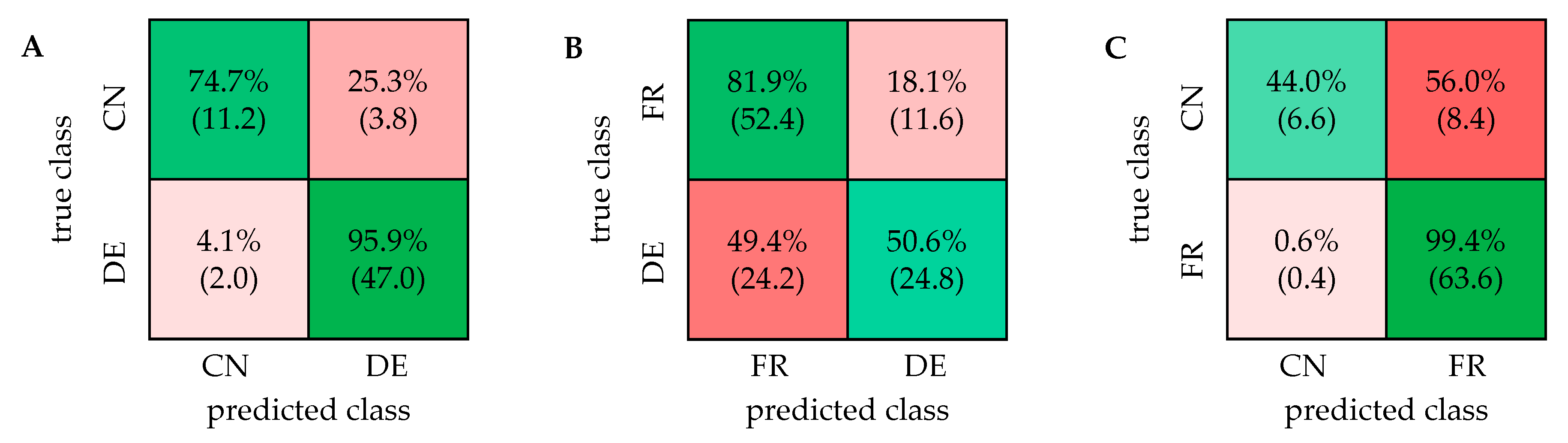
2.1. PCA and Classification Using 1D 1H NOESY Spectra
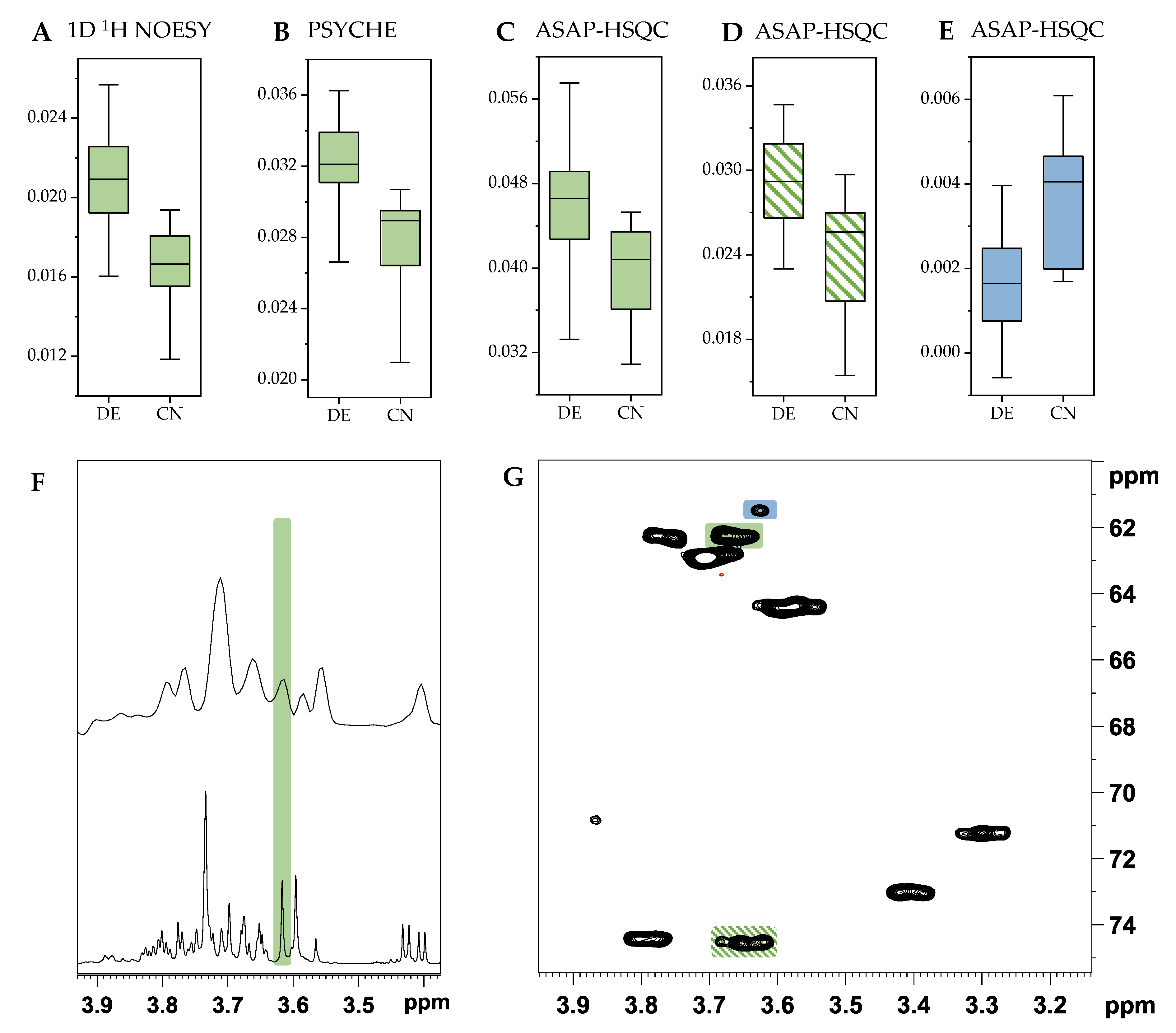
2.2. Comparison with ASAP-HSQC and PSYCHE Spectra
2.3. The Potential of 2D NMR Experiments for Determination of the Geographical Origin of Foods
3. Materials and Methods
3.1. Reagents and Chemicals
3.2. Walnut Samples
3.3. Sample Preparation
3.4. Extraction
3.5. NMR Acquisition
3.6. NMR Data Processing and Analysis
3.7. Multivariate Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Esslinger, S.; Riedl, J.; Fauhl-Hassek, C. Potential and limitations of non-targeted fingerprinting for authentication of food in official control. Food Res. Int. 2014, 60, 189–204. [Google Scholar] [CrossRef]
- Laghi, L.; Picone, G.; Capozzi, F. Nuclear magnetic resonance for foodomics beyond food analysis. TrAC-Trends Anal. Chem. 2014, 59, 93–102. [Google Scholar] [CrossRef]
- Bachmann, R.; Klockmann, S.; Haerdter, J.; Fischer, M.; Hackl, T. 1H NMR Spectroscopy for Determination of the Geographical Origin of Hazelnuts. J. Agric. Food Chem. 2018, 66, 11873–11879. [Google Scholar] [CrossRef]
- Sharma, R.; Gogna, N.; Singh, H.; Dorai, K. Fast profiling of metabolite mixtures using chemometric analysis of a speeded-up 2D heteronuclear correlation NMR experiment. RSC Adv. 2017, 7, 29860–29870. [Google Scholar] [CrossRef] [Green Version]
- Mozumder, N.H.M.R.; Lee, Y.R.; Hwang, K.H.; Lee, M.S.; Kim, E.H.; Hong, Y.S. Characterization of tea leaf metabolites dependent on tea (Camellia sinensis) plant age through 1H NMR-based metabolomics. Appl. Biol. Chem. 2020, 63, 1–8. [Google Scholar] [CrossRef]
- Wei, D.; Chen, D.; Lou, Y.; Ye, Y.; Yang, R. Metabolomic profile characteristics of pyropia haitanensis as affected by harvest time. Food Sci. Technol. Res. 2016, 22, 529–536. [Google Scholar] [CrossRef] [Green Version]
- Schmitt, C.; Bastek, T.; Stelzer, A.; Schneider, T.; Fischer, M.; Hackl, T. Detection of Peanut Adulteration in Food Samples by Nuclear Magnetic Resonance Spectroscopy. J. Agric. Food Chem. 2020, 68, 14364–14373. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR spectroscopy for metabolomics research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S. Metabolomics: Applications to food science and nutrition research. Trends Food Sci. Technol. 2008, 19, 482–493. [Google Scholar] [CrossRef]
- Pan, Z.; Raftery, D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal. Bioanal. Chem. 2007, 387, 525–527. [Google Scholar] [CrossRef]
- Cubero-Leon, E.; Peñalver, R.; Maquet, A. Review on metabolomics for food authentication. Food Res. Int. 2014, 60, 95–107. [Google Scholar] [CrossRef]
- Markley, J.L.; Brüschweiler, R.; Edison, A.S.; Eghbalnia, H.R.; Powers, R.; Raftery, D.; Wishart, D.S. The Future of NMR-Based Metabolomics. Curr. Opin. Biotechnol. 2018, 43, 34–40. [Google Scholar] [CrossRef] [Green Version]
- Zangger, K.; Sterk, H. Homonuclear Broadband-Decoupled NMR Spectra. J. Magn. Reson. 1997, 124, 486–489. [Google Scholar] [CrossRef]
- Foroozandeh, M.; Morris, G.A.; Nilsson, M. Psyche Pure Shift NMR Spectroscopy. Chem. Eur. J. 2018, 24, 13988–14000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aue, W.P.; Karhan, J.; Ernst, R.R. Homonuclear broad band decoupling and two-dimensional J-resolved NMR spectroscopy. J. Chem. Phys. 1976, 64, 4226–4227. [Google Scholar] [CrossRef]
- Zangger, K. Pure shift NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2015, 86–87, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bo, Y.; Feng, J.; Xu, J.J.; Huang, Y.; Cai, H.; Cui, X.; Dong, J.; Ding, S.; Chen, Z. High-resolution pure shift NMR spectroscopy offers better metabolite discrimination in food quality analysis. Food Res. Int. 2019, 125, 108574. [Google Scholar] [CrossRef] [PubMed]
- Foroozandeh, M.; Adams, R.W.; Meharry, N.J.; Jeannerat, D.; Nilsson, M.; Morris, G.A. Ultrahigh-resolution NMR spectroscopy. Angew. Chem. Int. Ed. 2014, 53, 6990–6992. [Google Scholar] [CrossRef] [Green Version]
- Castañar, L.; Parella, T. Broadband 1H homodecoupled NMR experiments: Recent developments, methods and applications. Magn. Reson. Chem. 2015, 53, 399–426. [Google Scholar] [CrossRef] [PubMed]
- Santacruz, L.; Hurtado, D.X.; Doohan, R.; Thomas, O.P.; Puyana, M.; Tello, E. Metabolomic study of soft corals from the Colombian Caribbean: PSYCHE and 1H-NMR comparative analysis. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Lopez, J.M.; Cabrera, R.; Maruenda, H. Ultra-Clean Pure Shift 1H-NMR applied to metabolomics profiling. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Schulze-Sünninghausen, D.; Becker, J.; Luy, B. Rapid Heteronuclear Single Quantum Correlation NMR Spectra at Natural Abundance. J. Am. Chem. Soc. 2014, 136, 1242–1245. [Google Scholar] [CrossRef]
- Kupce, E.; Freeman, R. Fast multidimensional NMR by polarization sharing. Magn. Reson. Chem. 2007, 45, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Schulze-Sünninghausen, D.; Becker, J.; Koos, M.R.M.; Luy, B. Improvements, extensions, and practical aspects of rapid ASAP-HSQC and ALSOFAST-HSQC pulse sequences for studying small molecules at natural abundance. J. Magn. Reson. 2017, 281, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Le Guennec, A.; Giraudeau, P.; Caldarelli, S. Evaluation of fast 2D NMR for metabolomics. Anal. Chem. 2014, 86, 5946–5954. [Google Scholar] [CrossRef]
- Puig-Castellví, F.; Pérez, Y.; Piña, B.; Tauler, R.; Alfonso, I. Comparative analysis of 1H NMR and 1H-13C HSQC NMR metabolomics to understand the effects of medium composition in yeast growth. Anal. Chem. 2018, 90, 12422–12430. [Google Scholar] [CrossRef] [PubMed]
- Popescu, R.; Ionete, R.E.; Botoran, O.R.; Costinel, D.; Bucura, F.; Geana, E.I.; Alabedallat, Y.F.J.; Botu, M. 1H NMR profiling and carbon isotope discrimination as tools for the comparative assessment of walnut (Juglans regia L.) cultivars with various geographical and genetic origins—A preliminary study. Molecules 2019, 24, 1378. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Wang, H.; Zhao, Q.; Ouyang, J.; Wu, Y. Rapid detection of authenticity and adulteration of walnut oil by FTIR and fluorescence spectroscopy: A comparative study. Food Chem. 2015, 181, 25–30. [Google Scholar] [CrossRef]
- Esteki, M.; Farajmand, B.; Amanifar, S.; Barkhordari, R.; Ahadiyan, Z.; Dashtaki, E.; Mohammadlou, M.; Heyden, Y.V. Classification and authentication of Iranian walnuts according to their geographical origin based on gas chromatographic fatty acid fingerprint analysis using pattern recognition methods. Chemom. Intell. Lab. Syst. 2017, 171, 251–258. [Google Scholar] [CrossRef]
- Beckonert, O.; Keun, H.C.; Ebbels, T.M.D.; Bundy, J.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007, 2, 2692–2703. [Google Scholar] [CrossRef]
- McKay, R.T. How the 1D-NOESY Suppresses Solvent Signal in Metabonomics NMR Spectroscopy: An Examination of the Pulse Sequence Components and Evolution. Concepts Magn. Reson. Part A 2011, 38, 197–220. [Google Scholar] [CrossRef]
- Schmitt, C.; Schneider, T.; Rumask, L.; Fischer, M.; Hackl, T. Food Profiling: Determination of the Geographical Origin of Walnuts by 1H NMR Spectroscopy Using the Polar Extract. J. Agric. Food Chem. 2020. [Google Scholar] [CrossRef]
- Varma, S.; Simon, R. Bias in error estimation when using cross-validation for model selection. BMC Bioinform. 2006, 7, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Westerhuis, J.A.; Hoefsloot, H.C.J.; Smit, S.; Vis, D.J.; Smilde, A.K.; Velzen, E.J.J.; Duijnhoven, J.P.M.; Dorsten, F.A. Assessment of PLSDA cross validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef] [Green Version]
- Krstajic, D.; Buturovic, L.J.; Leahy, D.E.; Thomas, S. Cross-validation pitfalls when selecting and assessing regression and classification models. J. Cheminform. 2014, 6, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Goodpaster, A.M.; Romick-Rosendale, L.E.; Kennedy, M.A. Statistical significance analysis of nuclear magnetic resonance-based metabonomics data. Anal. Biochem. 2010, 401, 134–143. [Google Scholar] [CrossRef]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nuc. Acids Res. 2020, 48, D440–D444. [Google Scholar] [CrossRef] [Green Version]
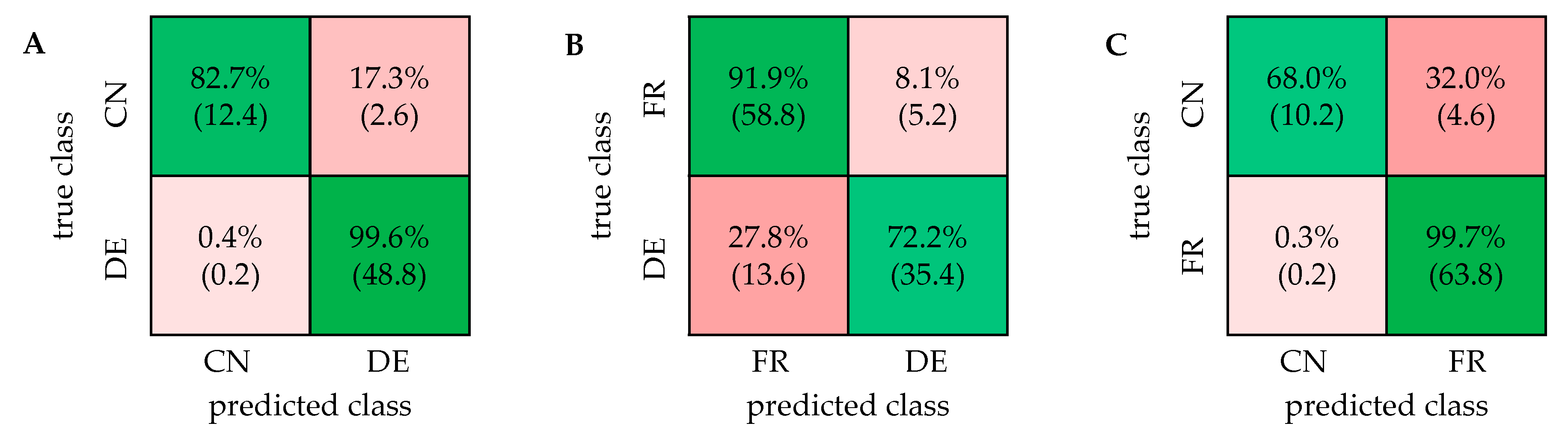


| Model | 1D 1H NOESY | PSYCHE | ASAP-HSQC |
|---|---|---|---|
| DE/CN | 95.9% (±0.8%) | 90.9% (±1.5%) | 86.3% (±0.6%) |
| FR/DE | 83.4% (±2.0%) | 68.3% (±2.8%) | 58.9% (±1.3%) |
| FR/CN | 93.7% (±1.1%) | 88.9% (±1.2%) | 88.9% (±1.2%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watermann, S.; Schmitt, C.; Schneider, T.; Hackl, T. Comparison of Regular, Pure Shift, and Fast 2D NMR Experiments for Determination of the Geographical Origin of Walnuts. Metabolites 2021, 11, 39. https://doi.org/10.3390/metabo11010039
Watermann S, Schmitt C, Schneider T, Hackl T. Comparison of Regular, Pure Shift, and Fast 2D NMR Experiments for Determination of the Geographical Origin of Walnuts. Metabolites. 2021; 11(1):39. https://doi.org/10.3390/metabo11010039
Chicago/Turabian StyleWatermann, Stephanie, Caroline Schmitt, Tobias Schneider, and Thomas Hackl. 2021. "Comparison of Regular, Pure Shift, and Fast 2D NMR Experiments for Determination of the Geographical Origin of Walnuts" Metabolites 11, no. 1: 39. https://doi.org/10.3390/metabo11010039
APA StyleWatermann, S., Schmitt, C., Schneider, T., & Hackl, T. (2021). Comparison of Regular, Pure Shift, and Fast 2D NMR Experiments for Determination of the Geographical Origin of Walnuts. Metabolites, 11(1), 39. https://doi.org/10.3390/metabo11010039