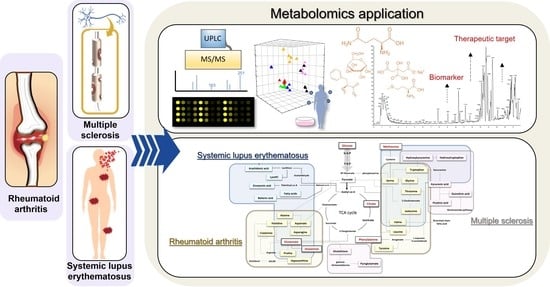
Metabolomics in Autoimmune Diseases: Focus on Rheumatoid Arthritis, Systemic Lupus Erythematous, and Multiple Sclerosis
Abstract
:1. Introduction
2. Application of Metabolomics
2.1. Defining Metabolomics
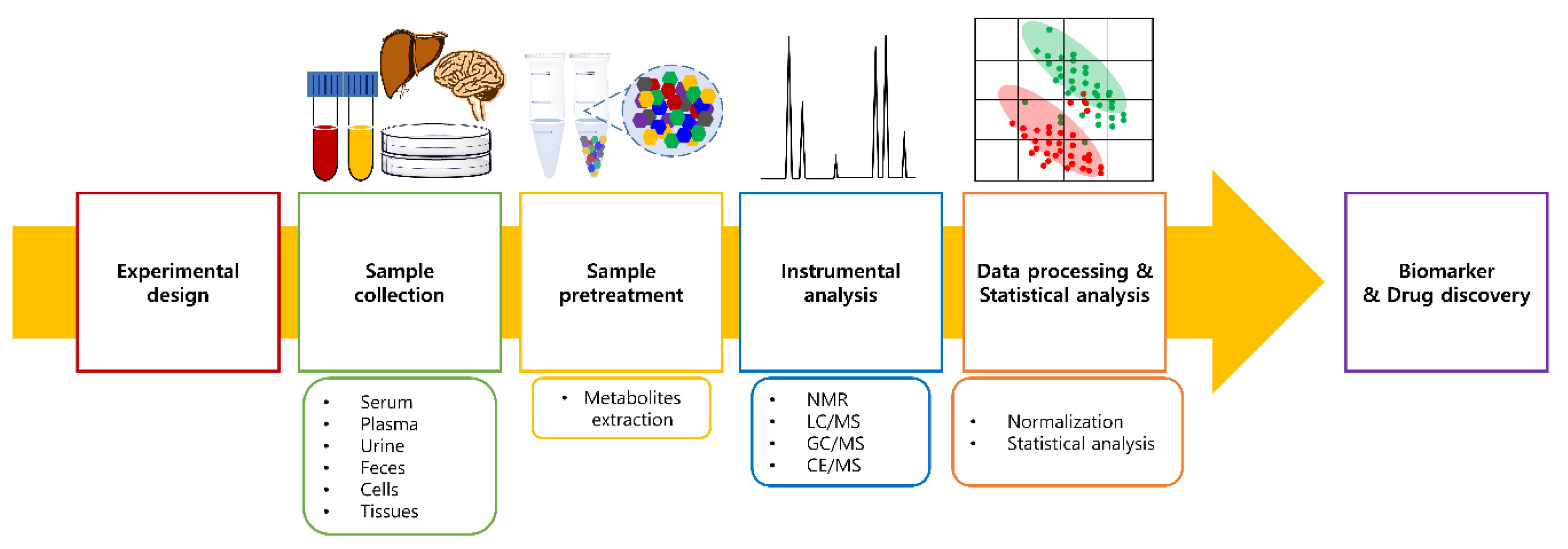
2.2. Metabolomics Workflow
2.2.1. Sample Collection and Pretreatment
2.2.2. Instrumental Analysis
2.2.3. Sample Normalization
2.2.4. Statistical Analysis
3. Metabolomics in Biomarkers of ADs
3.1. Discovery of Biomarkers in ADs
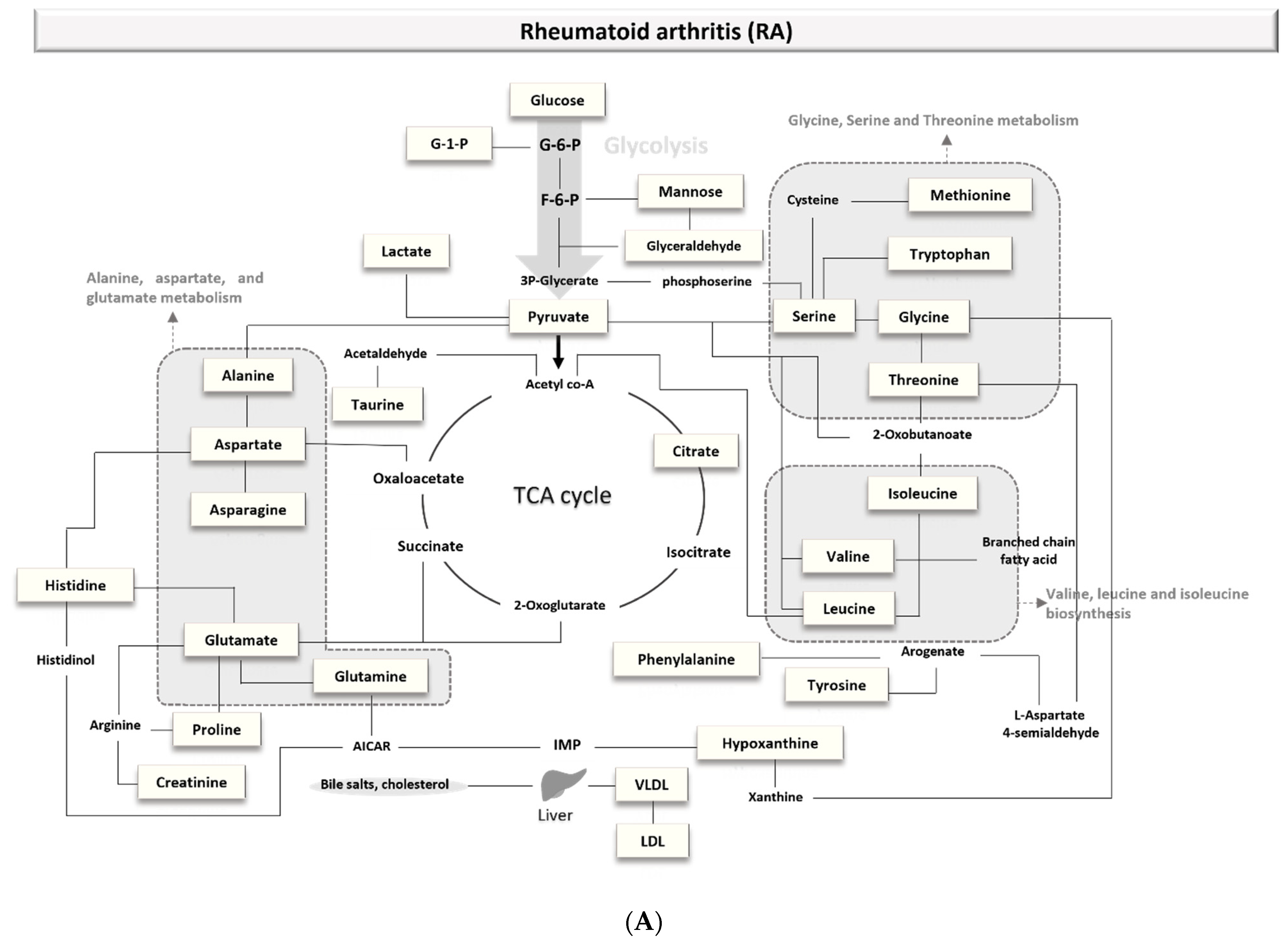
3.1.1. Biomarkers of Rheumatoid Arthritis (RA)
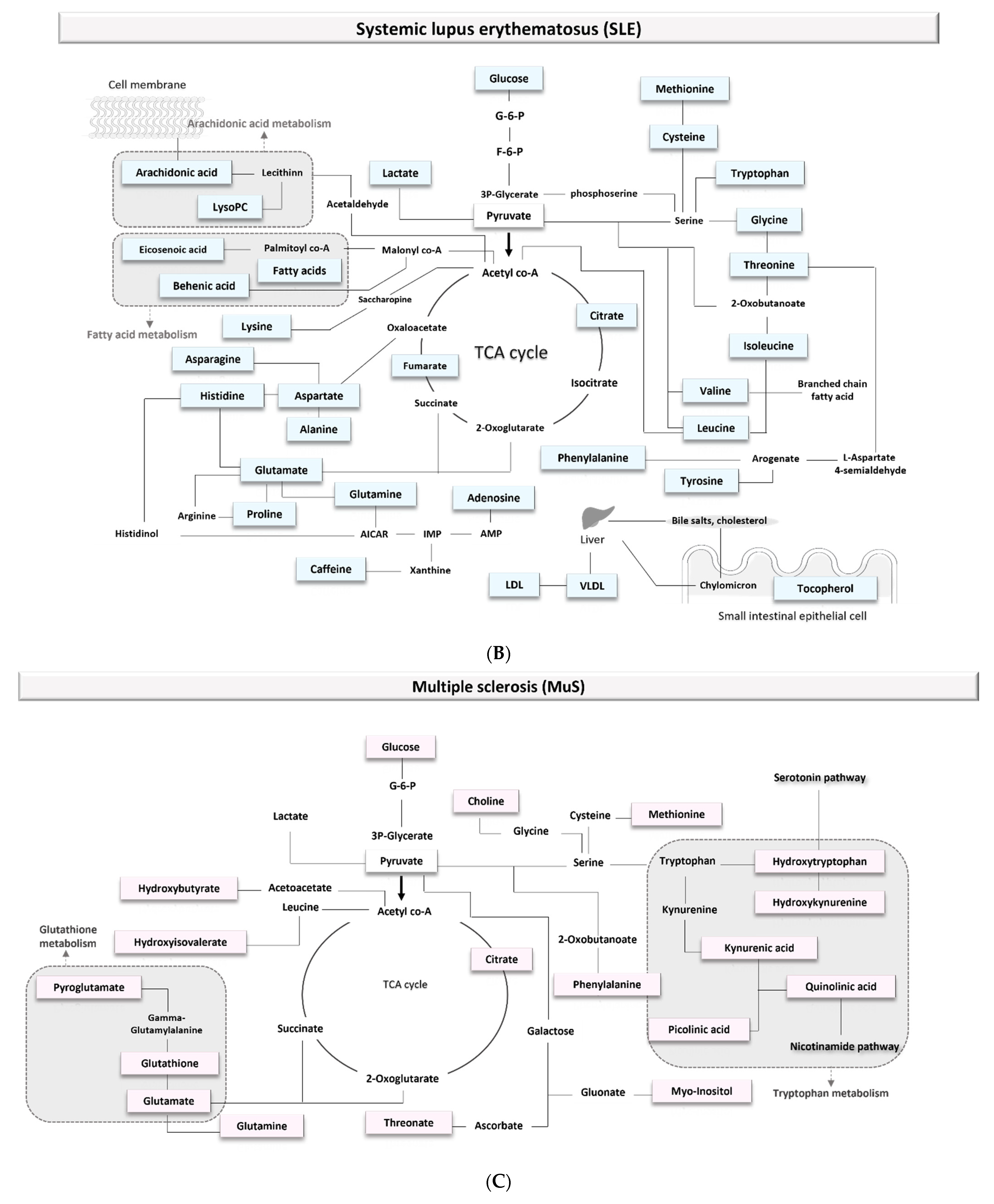
3.1.2. Biomarkers of Multiple Sclerosis (MuS)
3.1.3. Biomarkers of Systemic Lupus Erythematosus (SLE)
3.1.4. Comparing Biomarkers of ADs
3.2. Limitation of Current Biomarkers
4. Metabolomics in Drug Discovery for ADs
4.1. A New Target Discovery
4.1.1. Rheumatoid Arthritis (RA)
4.1.2. Multiple Sclerosis
4.1.3. Systemic Lupus Erythematosus (SLE)
4.2. Metabolomics Applications in Precision Medicine
4.2.1. Rheumatoid Arthritis (RA)
4.2.2. Multiple Sclerosis (MuS)
4.2.3. Systemic Lupus Erythematosus (SLE)
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Johnson, C.; Gonzalez, F.J. Challenges and opportunities of metabolomics. J. Cell. Physiol. 2011, 227, 2975–2981. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [Green Version]
- Johnson, C.H.; Patterson, A.D.; Idle, J.R.; Gonzalez, F.J. Xenobiotic Metabolomics: Major Impact on the Metabolome. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 37–56. [Google Scholar] [CrossRef]
- Pang, H.; Jia, W.; Hu, Z. Emerging Applications of Metabolomics in Clinical Pharmacology. Clin. Pharmacol. Ther. 2019, 106, 544–556. [Google Scholar] [CrossRef]
- Stathopoulou, C.; Nikoleri, D.; Bertsias, G. Immunometabolism: An overview and therapeutic prospects in autoimmune diseases. Immunotherapy 2019, 11, 813–829. [Google Scholar] [CrossRef]
- Kang, J.; Zhu, L.; Lu, J.; Zhang, X. Application of metabolomics in autoimmune diseases: Insight into biomarkers and pathology. J. Neuroimmunol. 2015, 279, 25–32. [Google Scholar] [CrossRef]
- Wang, L.; Wang, F.-S.; Gershwin, M.E. Human autoimmune diseases: A comprehensive update. J. Intern. Med. 2015, 278, 369–395. [Google Scholar] [CrossRef]
- Anaya, J.-M.; Restrepo, P.; Ramírez-Santana, C. The autoimmune ecology: An update. Curr. Opin. Rheumatol. 2018, 30, 350–360. [Google Scholar] [CrossRef]
- Li, P.; Zheng, Y.; Chen, X. Drugs for Autoimmune Inflammatory Diseases: From Small Molecule Compounds to Anti-TNF Biologics. Front. Pharmacol. 2017, 8, 460. [Google Scholar] [CrossRef]
- Tuller, T.; Atar, S.; Ruppin, E.; Gurevich, M.; Achiron, A. Common and specific signatures of gene expression and protein–protein interactions in autoimmune diseases. Genes Immun. 2012, 14, 67–82. [Google Scholar] [CrossRef]
- Liu, E.; Perl, A. Pathogenesis and treatment of autoimmune rheumatic diseases. Curr. Opin. Rheumatol. 2019, 31, 307–315. [Google Scholar] [CrossRef]
- Yang, Z.; Matteson, E.L.; Goronzy, J.J.; Weyand, C.M. T-cell metabolism in autoimmune disease. Arthritis Res. 2015, 17, 29. [Google Scholar] [CrossRef] [Green Version]
- Azad, R.K.; Shulaev, V. Metabolomics technology and bioinformatics for precision medicine. Briefings Bioinform. 2018, 20, 1957–1971. [Google Scholar] [CrossRef]
- Colamatteo, A.; Micillo, T.; Bruzzaniti, S.; Fusco, C.; Garavelli, S.; De Rosa, V.; Galgani, M.; Spagnuolo, M.I.; Di Rella, F.; Puca, A.A.; et al. Metabolism and Autoimmune Responses: The microRNA Connection. Front. Immunol. 2019, 10, 1969. [Google Scholar] [CrossRef]
- Cassotta, M.; Forbes-Hernandez, T.; Cianciosi, D.; Zabaleta, M.E.; Cano, S.S.; Dominguez, I.; Bullon, B.; Regolo, L.; Alvarez-Suarez, J.; Giampieri, F.; et al. Nutrition and Rheumatoid Arthritis in the ‘Omics’ Era. Nutrients 2021, 13, 763. [Google Scholar] [CrossRef]
- Katsila, T.; Konstantinou, E.; Lavda, I.; Malakis, H.; Papantoni, I.; Skondra, L.; Patrinos, G.P. Pharmacometabolomics-aided Pharmacogenomics in Autoimmune Disease. EBioMedicine 2016, 5, 40–45. [Google Scholar] [CrossRef] [Green Version]
- Tsoukalas, D.; Fragoulakis, V.; Papakonstantinou, E.; Antonaki, M.; Vozikis, A.; Tsatsakis, A.; Buga, A.M.; Mitroi, M.; Calina, D. Prediction of Autoimmune Diseases by Targeted Metabolomic Assay of Urinary Organic Acids. Metabolites 2020, 10, 502. [Google Scholar] [CrossRef]
- Zahoor, I.; Rui, B.; Khan, J.; Datta, I.; Giri, S. An emerging potential of metabolomics in multiple sclerosis: A comprehensive overview. Cell. Mol. Life Sci. 2021, 78, 3181–3203. [Google Scholar] [CrossRef]
- López-López, Á.; López-Gonzálvez, Á.; Tejeda, T.C.B.; Barbas, C. A review of validated biomarkers obtained through metabolomics. Expert Rev. Mol. Diagn. 2018, 18, 557–575. [Google Scholar] [CrossRef]
- Chen, L.; Zhong, F.; Zhu, J. Bridging Targeted and Untargeted Mass Spectrometry-Based Metabolomics via Hybrid Approaches. Metabolites 2020, 10, 348. [Google Scholar] [CrossRef]
- Trezzi, J.-P.; Vlassis, N.; Hiller, K. The Role of Metabolomics in the Study of Cancer Biomarkers and in the Development of Diagnostic Tools. Adv. Exp. Med. Biol. 2015, 867, 41–57. [Google Scholar] [CrossRef]
- Gooding, J.R.; Jensen, M.V.; Newgard, C.B. Metabolomics applied to the pancreatic islet. Arch. Biochem. Biophys. 2016, 589, 120–130. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Li, Z.; Lazar, L.; Fang, Z.; Tang, C.; Zhao, J. Metabolomics workflow for lung cancer: Discovery of biomarkers. Clin. Chim. Acta 2019, 495, 436–445. [Google Scholar] [CrossRef]
- Wishart, D.S. Metabolomics for Investigating Physiological and Pathophysiological Processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef]
- Zhou, B.; Xiao, J.F.; Tuli, L.; Ressom, H.W. LC-MS-based metabolomics. Mol. BioSyst. 2012, 8, 470–481. [Google Scholar] [CrossRef] [Green Version]
- Bhargava, P.; A Calabresi, P. Metabolomics in multiple sclerosis. Mult. Scler. J. 2016, 22, 451–460. [Google Scholar] [CrossRef]
- Raterink, R.-J.; Lindenburg, P.W.; Vreeken, R.; Ramautar, R.; Hankemeier, T. Recent developments in sample-pretreatment techniques for mass spectrometry-based metabolomics. Trends Anal. Chem. 2014, 61, 157–167. [Google Scholar] [CrossRef]
- Álvarez-Sánchez, B.; Priego-Capote, F.; de Castro, M.L. Metabolomics analysis II. Preparation of biological samples prior to detection. Trends Anal. Chem. 2010, 29, 120–127. [Google Scholar] [CrossRef]
- Wishart, D.S. Quantitative metabolomics using NMR. Trends Anal. Chem. 2008, 27, 228–237. [Google Scholar] [CrossRef]
- Smolinska, A.; Posma, J.M.; Blanchet, L.; Ampt, K.A.M.; Attali, A.; Tuinstra, T.; Luider, T.; Doskocz, M.; Michiels, P.J.; Girard, F.C.; et al. Simultaneous analysis of plasma and CSF by NMR and hierarchical models fusion. Anal. Bioanal. Chem. 2012, 403, 947–959. [Google Scholar] [CrossRef] [Green Version]
- Broadhurst, D.; Goodacre, R.; Reinke, S.N.; Kuligowski, J.; Wilson, I.D.; Lewis, M.R.; Dunn, W.B. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics 2018, 14, 72. [Google Scholar] [CrossRef] [Green Version]
- Dunn, W.B.; Wilson, I.D.; Nicholls, A.W.; Broadhurst, D. The importance of experimental design and QC samples in large-scale and MS-driven untargeted metabolomic studies of humans. Bioanalysis 2012, 4, 2249–2264. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Li, L. Sample normalization methods in quantitative metabolomics. J. Chromatogr. A 2016, 1430, 80–95. [Google Scholar] [CrossRef]
- Ryan, D.; Robards, K.; Prenzler, P.; Kendall, M. Recent and potential developments in the analysis of urine: A review. Anal. Chim. Acta 2011, 684, 17–29. [Google Scholar] [CrossRef]
- Antonelli, J.; Claggett, B.L.; Henglin, M.; Kim, N.; Ovsak, G.; Deng, K.; Rao, K.; Tyagi, O.; Watrous, J.D.; Lagerborg, K.A.; et al. Statistical Workflow for Feature Selection in Human Metabolomics Data. Metabolites 2019, 9, 143. [Google Scholar] [CrossRef] [Green Version]
- Priori, R.; Scrivo, R.; Brandt, J.; Valerio, M.; Casadei, L.; Valesini, G.; Manetti, C. Metabolomics in rheumatic diseases: The potential of an emerging methodology for improved patient diagnosis, prognosis, and treatment efficacy. Autoimmun. Rev. 2013, 12, 1022–1030. [Google Scholar] [CrossRef]
- Shoenfeld, Y.; Isenberg, D. The mosaic of autoimmunity. Immunol. Today 1989, 10, 123–126. [Google Scholar] [CrossRef]
- Madsen, R.K.; Lundstedt, T.; Gabrielsson, J.; Sennbro, C.-J.; Alenius, G.-M.; Moritz, T.; Rantapää-Dahlqvist, S.; Trygg, J. Diagnostic properties of metabolic perturbations in rheumatoid arthritis. Arthritis Res. 2011, 13, R19. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, X.; Dai, Y.; Wen, J.L.; Wang, L.X. 1H NMR-based metabolomic study of metabolic profiling for systemic lupus erythematosus. Lupus 2011, 20, 1411–1420. [Google Scholar] [CrossRef]
- Young, S.P.; Nessim, M.; Falciani, F.; Trevino, V.; Banerjee, S.P.; Scott, R.; Murray, P.; Wallace, G.R. Metabolomic analysis of human vitreous humor differentiates ocular inflammatory disease. Mol. Vis. 2009, 15, 1210–1217. [Google Scholar]
- Jiang, M.; Chen, T.; Feng, H.; Zhang, Y.; Li, L.; Zhao, A.; Niu, X.; Liang, F.; Wang, M.; Zhan, J.; et al. Serum Metabolic Signatures of Four Types of Human Arthritis. J. Proteome Res. 2013, 12, 3769–3779. [Google Scholar] [CrossRef]
- Yang, X.Y.; Di Zheng, K.; Lin, K.; Zheng, G.; Zou, H.; Wang, J.M.; Lin, Y.Y.; Chuka, C.M.; Ge, R.S.; Zhai, W.; et al. Energy Metabolism Disorder as a Contributing Factor of Rheumatoid Arthritis: A Comparative Proteomic and Metabolomic Study. PLoS ONE 2015, 10, e0132695. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, J.; Hu, C.; Xie, Z.; Li, H.; Wei, S.; Wang, D.; Wen, C.; Xu, G. Exploration of the serum metabolite signature in patients with rheumatoid arthritis using gas chromatography–mass spectrometry. J. Pharm. Biomed. Anal. 2016, 127, 60–67. [Google Scholar] [CrossRef]
- Alonso, R.B.; The IMID Consortium; Julià, A.; Vinaixa, M.; Domènech, E.; Fernandez-Nebro, A.; Cañete, J.D.; Ferrándiz, C.; Tornero, J.; Gisbert, J.P.; et al. Urine metabolome profiling of immune-mediated inflammatory diseases. BMC Med. 2016, 14, 1–12. [Google Scholar] [CrossRef]
- Zabek, A.; Swierkot, J.; Malak, A.; Zawadzka, I.; Deja, S.; Bogunia-Kubik, K.; Mlynarz, P. Application of 1 H NMR-based serum metabolomic studies for monitoring female patients with rheumatoid arthritis. J. Pharm. Biomed. Anal. 2016, 117, 544–550. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Che, N.; Xu, L.; Zhang, Q.; Wang, Q.; Tan, W.; Zhang, M. LC-MS-based serum metabolomics reveals a distinctive signature in patients with rheumatoid arthritis. Clin. Rheumatol. 2018, 37, 1493–1502. [Google Scholar] [CrossRef]
- Urbaniak, B.; Plewa, S.; Klupczynska, A.; Sikorska, D.; Samborski, W.; Kokot, Z.J. Serum free amino acid levels in rheumatoid arthritis according to therapy and physical disability. Cytokine 2019, 113, 332–339. [Google Scholar] [CrossRef]
- He, Z.; Liu, Z.; Gong, L. Biomarker identification and pathway analysis of rheumatoid arthritis based on metabolomics in combination with ingenuity pathway analysis. Proteomics 2021, 21, 2100037. [Google Scholar] [CrossRef]
- Weljie, A.M.; Dowlatabadi, R.; Miller, B.J.; Vogel, A.H.J.; Jirik, F.R. An Inflammatory Arthritis-Associated Metabolite Biomarker Pattern Revealed by 1H NMR Spectroscopy. J. Proteome Res. 2007, 6, 3456–3464. [Google Scholar] [CrossRef]
- Moussallieh, F.-M.; Elbayed, K.; Chanson, J.; Rudolf, G.; Piotto, M.; de Seze, J.; Namer, I.J. Serum analysis by 1H Nuclear Magnetic Resonance spectroscopy: A new tool for distinguishing neuromyelitis optica from multiple sclerosis. Mult. Scler. J. 2014, 20, 558–565. [Google Scholar] [CrossRef]
- Reinke, S.; Broadhurst, D.; Sykes, B.; Baker, G.; Catz, I.; Warren, K.; Power, C.; Reinke, S. Metabolomic profiling in multiple sclerosis: Insights into biomarkers and pathogenesis. Mult. Scler. J. 2014, 20, 1396–1400. [Google Scholar] [CrossRef] [PubMed]
- Pieragostino, D.; D’Alessandro, M.; di Ioia, M.; Rossi, C.; Zucchelli, M.; Urbani, A.; Di Ilio, C.; Lugaresi, A.; Sacchetta, P.; Del Boccio, P. An integrated metabolomics approach for the research of new cerebrospinal fluid biomarkers of multiple sclerosis. Mol. BioSyst. 2015, 11, 1563–1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cocco, E.; Murgia, F.; Lorefice, L.; Barberini, L.; Poddighe, S.; Frau, J.; Fenu, G.; Coghe, G.; Murru, M.R.; Murru, R.; et al. 1H-NMR analysis provides a metabolomic profile of patients with multiple sclerosis. Neurol.-Neuroimmunol. Neuroinflamm. 2016, 3, e185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.J.; Jeong, I.H.; Kong, B.S.; Lee, J.-E.; Kim, K.H.; Lee, D.Y.; Kim, H.J. Disease Type- and Status-Specific Alteration of CSF Metabolome Coordinated with Clinical Parameters in Inflammatory Demyelinating Diseases of CNS. PLoS ONE 2016, 11, e0166277. [Google Scholar] [CrossRef]
- Gebregiworgis, T.; Nielsen, H.H.; Massilamany, C.; Gangaplara, A.; Reddy, J.; Illes, Z.; Powers, R. A Urinary Metabolic Signature for Multiple Sclerosis and Neuromyelitis Optica. J. Proteome Res. 2016, 15, 659–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poddighe, S.; Murgia, F.; Lorefice, L.; Liggi, S.; Cocco, E.; Marrosu, M.G.; Atzori, L. Metabolomic analysis identifies altered metabolic pathways in Multiple Sclerosis. Int. J. Biochem. Cell Biol. 2017, 93, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Jeong, I.H.; Hyun, J.-S.; Kong, B.S.; Kim, H.J.; Park, S.J. Metabolomic profiling of CSF in multiple sclerosis and neuromyelitis optica spectrum disorder by nuclear magnetic resonance. PLoS ONE 2017, 12, e0181758. [Google Scholar] [CrossRef]
- Lim, C.K.; Bilgin, A.; Lovejoy, D.B.; Tan, V.; Bustamante, S.; Taylor, B.V.; Bessede, A.; Brew, B.; Guillemin, G. Kynurenine pathway metabolomics predicts and provides mechanistic insight into multiple sclerosis progression. Sci. Rep. 2017, 7, 41473. [Google Scholar] [CrossRef]
- Bystrická, Z.; Laubertová, L.; Durfinova, M.; Paduchová, Z. Methionine metabolism and multiple sclerosis. Biomarkers 2017, 22, 747–754. [Google Scholar] [CrossRef]
- Herman, S.; Åkerfeldt, T.; Spjuth, O.; Burman, J.; Kultima, K. Biochemical Differences in Cerebrospinal Fluid between Secondary Progressive and Relapsing–Remitting Multiple Sclerosis. Cells 2019, 8, 84. [Google Scholar] [CrossRef] [Green Version]
- Rajda, C.; Galla, Z.; Polyák, H.; Maróti, Z.; Babarczy, K.; Pukoli, D.; Vécsei, L. Cerebrospinal Fluid Neurofilament Light Chain Is Associated with Kynurenine Pathway Metabolite Changes in Multiple Sclerosis. Int. J. Mol. Sci. 2020, 21, 2665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tömösi, F.; Kecskeméti, G.; Cseh, E.K.; Szabó, E.; Rajda, C.; Kormány, R.; Szabó, Z.; Vécsei, L.; Janáky, T. A validated UHPLC-MS method for tryptophan metabolites: Application in the diagnosis of multiple sclerosis. J. Pharm. Biomed. Anal. 2020, 185, 113246. [Google Scholar] [CrossRef]
- Wishart, D.S.; Lewis, M.J.; Morrissey, J.A.; Flegel, M.D.; Jeroncic, K.; Xiong, Y.; Cheng, D.; Eisner, R.; Gautam, B.; Tzur, D.; et al. The human cerebrospinal fluid metabolome. J. Chromatogr. B 2008, 871, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Kuzhiumparambil, U.; Bandodkar, S.; Dale, R.C.; Fu, S. Cerebrospinal fluid metabolomics: Detection of neuroinflammation in human central nervous system disease. Clin. Transl. Immunol. 2021, 10, e1318. [Google Scholar] [CrossRef] [PubMed]
- Housley, W.J.; Pitt, D.; Hafler, D.A. Biomarkers in multiple sclerosis. Clin. Immunol. 2015, 161, 51–58. [Google Scholar] [CrossRef]
- Aeinehband, S.; Brenner, P.; Ståhl, S.; Bhat, M.; Fidock, M.D.; Khademi, M.; Olsson, T.; Engberg, G.; Jokinen, J.; Erhardt, S.; et al. Cerebrospinal fluid kynurenines in multiple sclerosis; relation to disease course and neurocognitive symptoms. Brain Behav. Immun. 2016, 51, 47–55. [Google Scholar] [CrossRef] [Green Version]
- Rejdak, K.; Bartosik-Psujek, H.; Dobosz, B.; Kocki, T.; Grieb, P.; Giovannoni, G.; Turski, W.A.; Stelmasiak, Z. Decreased level of kynurenic acid in cerebrospinal fluid of relapsing-onset multiple sclerosis patients. Neurosci. Lett. 2002, 331, 63–65. [Google Scholar] [CrossRef]
- Yan, B.; Huang, J.; Zhang, C.; Hu, X.; Gao, M.; Shi, A.; Zha, W.; Shi, L.; Huang, C.; Yang, L. Serum metabolomic profiling in patients with systemic lupus erythematosus by GC/MS. Mod. Rheumatol. 2016, 26, 914–922. [Google Scholar] [CrossRef]
- Yan, B.; Huang, J.; Dong, F.; Yang, L.; Huang, C.; Gao, M.; Shi, A.; Zha, W.; Shi, L.; Hu, X. Urinary metabolomic study of systemic lupus erythematosus based on gas chromatography/mass spectrometry. Biomed. Chromatogr. 2016, 30, 1877–1881. [Google Scholar] [CrossRef]
- Bengtsson, A.A.; Trygg, J.; Wuttge, D.M.; Sturfelt, G.; Theander, E.; Donten, M.; Moritz, T.; Sennbro, C.-J.; Torell, F.; Lood, C.; et al. Metabolic Profiling of Systemic Lupus Erythematosus and Comparison with Primary Sjögren’s Syndrome and Systemic Sclerosis. PLoS ONE 2016, 11, e0159384. [Google Scholar] [CrossRef]
- Guleria, A.; Pratap, A.; Dubey, D.; Rawat, A.; Chaurasia, S.; Sukesh, E.; Phatak, S.; Ajmani, S.; Kumar, U.; Khetrapal, C.L.; et al. NMR based serum metabolomics reveals a distinctive signature in patients with Lupus Nephritis. Sci. Rep. 2016, 6, 35309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, T.H.; Kim, H.-A.; Jung, J.-Y.; Baek, W.-Y.; Lee, H.-S.; Park, H.J.; Min, J.; Paik, M.-J.; Lee, G.; Suh, C.-H. Analysis of the free fatty acid metabolome in the plasma of patients with systemic lupus erythematosus and fever. Metabolomics 2017, 14, 14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yin, X.; Wang, H.; Wu, X.; Li, X.; Li, Y.; Zhang, X.; Fu, C.; Li, H.; Qiu, Y. Fecal Metabolomics and Potential Biomarkers for Systemic Lupus Erythematosus. Front. Immunol. 2019, 10, 976. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liang, L.; Deng, X.; Zhong, L. Lipidomic and metabolomic profiling reveals novel candidate biomarkers in active systemic lupus erythematosus. Int. J. Clin. Exp. Pathol 2019, 12, 857–866. [Google Scholar]
- Yan, R.; Jiang, H.; Gu, S.; Feng, N.; Zhang, N.; Lv, L.; Liu, F. Fecal Metabolites Were Altered, Identified as Biomarkers and Correlated with Disease Activity in Patients with Systemic Lupus Erythematosus in a GC-MS-Based Metabolomics Study. Front. Immunol. 2020, 11, 2138. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, X.; Yin, X.; Wang, H.; Fu, C.; Wang, H.; Li, K.; Li, Y.; Zhang, X.; Liang, H.; et al. Metabolomic profiling reveals serum L-pyroglutamic acid as a potential diagnostic biomarker for systemic lupus erythematosus. Rheumatology 2021, 60, 598–606. [Google Scholar] [CrossRef]
- Scavuzzi, B.M.; Simão, A.N.C.; Iriyoda, T.M.V.; Lozovoy, M.A.B.; Stadtlober, N.P.; Santos, L.F.D.R.F.; Flauzino, T.; De Medeiros, F.A.; de Sá, M.C.; Consentin, L.; et al. Increased lipid and protein oxidation and lowered anti-oxidant defenses in systemic lupus erythematosus are associated with severity of illness, autoimmunity, increased adhesion molecules, and Th1 and Th17 immune shift. Immunol. Res. 2018, 66, 158–171. [Google Scholar] [CrossRef]
- Lu, L.; Hu, C.; Zhao, Y.; He, L.; Zhou, J.; Li, H.; Du, Y.; Wang, Y.; Wen, C.; Han, X.; et al. Shotgun Lipidomics Revealed Altered Profiles of Serum Lipids in Systemic Lupus Erythematosus Closely Associated with Disease Activity. Biomolecules 2018, 8, 105. [Google Scholar] [CrossRef] [Green Version]
- Samuelsson, B. Arachidonic acid metabolism: Role in inflammation. Z. Rheumatol. 1991, 50 (Suppl. S1), 3–6. [Google Scholar]
- Calder, P.C. Fatty acids and inflammation: The cutting edge between food and pharma. Eur. J. Pharmacol. 2011, 668 (Suppl. S1), S50–S58. [Google Scholar] [CrossRef]
- Ruddick, J.P.; Evans, A.K.; Nutt, D.J.; Lightman, S.; Rook, G.; Lowry, C.A. Tryptophan metabolism in the central nervous system: Medical implications. Expert Rev. Mol. Med. 2006, 8, 1–27. [Google Scholar] [CrossRef]
- Gaetani, L.; Boscaro, F.; Pieraccini, G.; Calabresi, P.; Romani, L.; Di Filippo, M.; Zelante, T. Host and Microbial Tryptophan Metabolic Profiling in Multiple Sclerosis. Front. Immunol. 2020, 11, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nourbakhsh, B.; Nunan-Saah, J.; Maghzi, A.-H.; Julian, L.J.; Spain, R.; Jin, C.; Lazar, A.; Pelletier, D.; Waubant, E. Longitudinal associations between MRI and cognitive changes in very early MS. Mult. Scler. Relat. Disord. 2016, 5, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, D.R.; Patel, R.; Kirsch, D.G.; Lewis, C.A.; Vander Heiden, M.G.; Locasale, J.W. Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J. Clin. 2021, 71, 333. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Yan, G.; Wang, P.; Wang, X. Metabolomics for Biomarker Discovery: Moving to the Clinic. BioMed Res. Int. 2015, 2015, 354671. [Google Scholar] [CrossRef]
- Berman, D.M.; Rodrigues, L.M.; Nicklas, B.J.; Ryan, A.S.; Dennis, K.E.; Goldberg, A.P. Racial Disparities in Metabolism, Central Obesity, and Sex Hormone-Binding Globulin in Postmenopausal Women. J. Clin. Endocrinol. Metab. 2001, 86, 97–103. [Google Scholar] [CrossRef]
- Sharp, T.A.; Bell, M.; Grunwald, G.K.; Schmitz, K.H.; Sidney, S.; Lewis, C.E.; Tolan, K.; Hill, J.O. Differences in Resting Metabolic Rate between White and African-American Young Adults. Obes. Res. 2002, 10, 726–732. [Google Scholar] [CrossRef] [PubMed]
- Preissner, S.C.; Hoffmann, M.F.; Preissner, R.; Dunkel, M.; Gewiess, A.; Preissner, S. Polymorphic Cytochrome P450 Enzymes (CYPs) and Their Role in Personalized Therapy. PLoS ONE 2013, 8, e82562. [Google Scholar] [CrossRef]
- McGraw, J.; Waller, D. Cytochrome P450 variations in different ethnic populations. Expert Opin. Drug Metab. Toxicol. 2012, 8, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Grover, S.; Kukreti, R. Functional genetic polymorphisms from phase-II drug metabolizing enzymes. CNS Neurosci. Ther. 2012, 18, 705–706. [Google Scholar] [CrossRef]
- Zhang, T.; Mohan, C. Caution in studying and interpreting the lupus metabolome. Arthritis Res. 2020, 22, 1–10. [Google Scholar] [CrossRef]
- Porter, L.; Shoushtarizadeh, A.; Jelinek, G.A.; Brown, C.R.; Lim, C.K.; De Livera, A.M.; Jacobs, K.R.; Weiland, T.J. Metabolomic Biomarkers of Multiple Sclerosis: A Systematic Review. Front. Mol. Biosci. 2020, 7, 574133. [Google Scholar] [CrossRef]
- Lee, M.Y.; Hu, T. Computational Methods for the Discovery of Metabolic Markers of Complex Traits. Metabolites 2019, 9, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bental, M.; Deutsch, P.C. Metabolic changes in activated T cells: An NMR study of human peripheral blood lymphocytes. Magn. Reson. Med. 1993, 29, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Caro-Maldonado, A.; Wang, R.; Nichols, A.G.; Kuraoka, M.; Milasta, S.; Sun, L.D.; Gavin, A.L.; Abel, E.D.; Kelsoe, G.; Green, D.R.; et al. Metabolic Reprogramming Is Required for Antibody Production That Is Suppressed in Anergic but Exaggerated in Chronically BAFF-Exposed B Cells. J. Immunol. 2014, 192, 3626–3636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Chess, L. Regulation of Immune Responses by T Cells. N. Engl. J. Med. 2006, 354, 1166–1176. [Google Scholar] [CrossRef] [Green Version]
- Weyand, C.M.; Goronzy, J.J. Immunometabolism in the development of rheumatoid arthritis. Immunol. Rev. 2020, 294, 177–187. [Google Scholar] [CrossRef]
- Garcia-Carbonell, R.; Divakaruni, A.S.; Lodi, A.; Vicente-Suarez, I.; Saha, A.; Cheroutre, H.; Boss, G.R.; Tiziani, S.; Murphy, A.N.; Guma, M. Critical Role of Glucose Metabolism in Rheumatoid Arthritis Fibroblast-like Synoviocytes. Arthritis Rheumatol. 2016, 68, 1614–1626. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Lopez, E.; Cheng, A.; Guma, M. Can Metabolic Pathways Be Therapeutic Targets in Rheumatoid Arthritis? J. Clin. Med. 2019, 8, 753. [Google Scholar] [CrossRef] [Green Version]
- Goronzy, J.J.; Weyand, C.M. Developments in the scientific understanding of rheumatoid arthritis. Arthritis Res. Ther. 2009, 11, 249. [Google Scholar] [CrossRef] [Green Version]
- Weyand, C.M.; Fujii, H.; Shao, L.; Goronzy, J.J. Rejuvenating the immune system in rheumatoid arthritis. Nat. Rev. Rheumatol. 2009, 5, 583–588. [Google Scholar] [CrossRef]
- Gosselt, H.R.; Muller, I.B.; Jansen, G.; Van Weeghel, M.; Vaz, F.M.; Hazes, J.M.W.; Heil, S.G.; De Jonge, R. Identification of Metabolic Biomarkers in Relation to Methotrexate Response in Early Rheumatoid Arthritis. J. Pers. Med. 2020, 10, 271. [Google Scholar] [CrossRef]
- Guma, M.; Tiziani, S.; Firestein, M.G.G.S. Metabolomics in rheumatic diseases: Desperately seeking biomarkers. Nat. Rev. Rheumatol. 2016, 12, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Saegusa, J.; Sendo, S.; Okano, T.; Akashi, K.; Irino, Y.; Morinobu, A. Glutaminase 1 plays a key role in the cell growth of fibroblast-like synoviocytes in rheumatoid arthritis. Arthritis Res. 2017, 19, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narasimhan, R.; Coras, R.; Rosenthal, S.B.; Sweeney, S.R.; Lodi, A.; Tiziani, S.; Boyle, D.; Kavanaugh, A.; Guma, M. Serum metabolomic profiling predicts synovial gene expression in rheumatoid arthritis. Arthritis Res. 2018, 20, 164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volchenkov, R.; Cao, M.D.; Elgstøen, K.; Goll, G.; Eikvar, K.; Bjørneboe, O.; Bathen, T.; Holen, H.; Kvien, T.; Skålhegg, B. Metabolic profiling of synovial tissue shows altered glucose and choline metabolism in rheumatoid arthritis samples. Scand. J. Rheumatol. 2017, 46, 160–161. [Google Scholar] [CrossRef] [PubMed]
- Guma, M.; Sanchez-Lopez, E.; Lodi, A.; Garcia-Carbonell, R.; Tiziani, S.; Karin, M.; Lacal, J.C.; Firestein, G.S. Choline kinase inhibition in rheumatoid arthritis. Ann. Rheum. Dis. 2015, 74, 1399–1407. [Google Scholar] [CrossRef]
- Xu, D.; Liang, J.; Lin, J.; Yu, C. PKM2: A Potential Regulator of Rheumatoid Arthritis via Glycolytic and Non-Glycolytic Pathways. Front. Immunol. 2019, 10, 2919. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Yue, Y.; Cheng, W.; Li, J.; Hu, Y.; Qin, L.; Zhang, P. Hypoxia-inducible factor: A potential therapeutic target for rheumatoid arthritis. Curr. Drug Targets 2013, 14, 700–707. [Google Scholar] [CrossRef]
- Chang, X.; Wei, C. Glycolysis and rheumatoid arthritis. Int. J. Rheum. Dis. 2011, 14, 217–222. [Google Scholar] [CrossRef]
- Hitchon, C.A.; El-Gabalawy, H.S.; Bezabeh, T. Characterization of synovial tissue from arthritis patients: A proton magnetic resonance spectroscopic investigation. Rheumatol. Int. 2009, 29, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, S.E.; O’Neill, L. HIF1α and metabolic reprogramming in inflammation. J. Clin. Investig. 2016, 126, 3699–3707. [Google Scholar] [CrossRef] [PubMed]
- Courtnay, R.; Ngo, D.C.; Malik, N.; Ververis, K.; Tortorella, S.M.; Karagiannis, T.C. Cancer metabolism and the Warburg effect: The role of HIF-1 and PI3K. Mol. Biol. Rep. 2015, 42, 841–851. [Google Scholar] [CrossRef] [PubMed]
- Dehne, N.; Brüne, B. HIF-1 in the inflammatory microenvironment. Exp. Cell Res. 2009, 315, 1791–1797. [Google Scholar] [CrossRef]
- Belisario, D.C.; Kopecka, J.; Pasino, M.; Akman, M.; De Smaele, E.; Donadelli, M.; Riganti, C. Hypoxia Dictates Metabolic Rewiring of Tumors: Implications for Chemoresistance. Cells 2020, 9, 2598. [Google Scholar] [CrossRef]
- Deng, W.; Feng, X.; Li, X.; Wang, D.; Sun, L. Hypoxia-inducible factor 1 in autoimmune diseases. Cell. Immunol. 2016, 303, 7–15. [Google Scholar] [CrossRef]
- Lee, Y.-Z.; Guo, H.-C.; Zhao, G.-H.; Yang, C.-W.; Chang, H.-Y.; Yang, R.-B.; Chen, L.; Lee, S.-J. Tylophorine-based compounds are therapeutic in rheumatoid arthritis by targeting the caprin-1 ribonucleoprotein complex and inhibiting expression of associated c-Myc and HIF-1α. Pharmacol. Res. 2020, 152, 104581. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Wang, C.; Xia, W.-R.; Zheng, J.-Y.; Yang, J.; Liu, B.; Liu, J.-Q.; Liu, L.-F. Succinate induces synovial angiogenesis in rheumatoid arthritis through metabolic remodeling and HIF-1α/VEGF axis. Free Radic. Biol. Med. 2018, 126, 1–14. [Google Scholar] [CrossRef]
- Chemin, K.; Gerstner, C.; Malmström, V. Effector Functions of CD4+ T Cells at the Site of Local Autoimmune Inflammation—Lessons From Rheumatoid Arthritis. Front. Immunol. 2019, 10, 353. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Fujii, H.; Mohan, S.V.; Goronzy, J.J.; Weyand, C.M. Phosphofructokinase deficiency impairs ATP generation, autophagy, and redox balance in rheumatoid arthritis T cells. J. Exp. Med. 2013, 210, 2119–2134. [Google Scholar] [CrossRef] [Green Version]
- Weyand, C.M.; Wu, B.; Goronzy, J.J. The metabolic signature of T cells in rheumatoid arthritis. Curr. Opin. Rheumatol. 2020, 32, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Wen, Z.; Li, Y.; Matteson, E.L.; Hong, J.; Goronzy, J.J.; Weyand, C.M. Metabolic control of the scaffold protein TKS5 in tissue-invasive, proinflammatory T cells. Nat. Immunol. 2017, 18, 1025–1034. [Google Scholar] [CrossRef] [PubMed]
- Kostoglou-Athanassiou, I.; Athanassiou, L.; Athanassiou, P. The Effect of Omega-3 Fatty Acids on Rheumatoid Arthritis. Mediterr. J. Rheumatol. 2020, 31, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Veselinovic, M.; Vasiljevic, D.; Vucic, V.; Arsic, A.; Petrovic, S.; Tomic-Lucic, A.; Savic, M.; Zivanovic, S.; Stojic, V.; Jakovljevic, V. Clinical Benefits of n-3 PUFA and ɤ-Linolenic Acid in Patients with Rheumatoid Arthritis. Nutrients 2017, 9, 325. [Google Scholar] [CrossRef] [PubMed]
- Lourdudoss, C.; Wolk, A.; Nise, L.; Alfredsson, L.; Van Vollenhoven, R. Are dietary vitamin D, omega-3 fatty acids and folate associated with treatment results in patients with early rheumatoid arthritis? Data from a Swedish population-based prospective study. BMJ Open 2017, 7, e016154. [Google Scholar] [CrossRef]
- Rajaei, E.; Mowla, K.; Ghorbani, A.; Bahadoram, S.; Bahadoram, M.; Dargahi, M. The Effect of Omega-3 Fatty Acids in Patients with Active Rheumatoid Arthritis Receiving DMARDs Therapy: Double-Blind Randomized Controlled Trial. Glob. J. Health Sci. 2015, 8, 18–25. [Google Scholar] [CrossRef] [Green Version]
- Kremer, J.M.; Lawrence, D.A.; Jubiz, W.; Digiacomo, R.; Rynes, R.; Bartholomew, L.E.; Sherman, M. Dietary fish oil and olive oil supplementation in patients with Rheumatoid Arthritis clinical and immunologic effects. Arthritis Rheum. 1990, 33, 810–820. [Google Scholar] [CrossRef]
- Bhargava, P.; Anthony, D. Metabolomics in multiple sclerosis disease course and progression. Mult. Scler. J. 2020, 26, 591–598. [Google Scholar] [CrossRef]
- Hayes, C.E.; Ntambi, J.M. Multiple Sclerosis: Lipids, Lymphocytes, and Vitamin D. Immunometabolism 2020, 10, 19. [Google Scholar] [CrossRef]
- Lazzarino, G.; Amorini, A.M.; Petzold, A.; Gasperini, C.; Ruggieri, S.; Quartuccio, M.E.; Lazzarino, G.; Di Stasio, E.; Tavazzi, B. Serum Compounds of Energy Metabolism Impairment Are Related to Disability, Disease Course and Neuroimaging in Multiple Sclerosis. Mol. Neurobiol. 2017, 54, 7520–7533. [Google Scholar] [CrossRef]
- Pihl-Jensen, G.; Tsakiri, A.; Frederiksen, J.L. Statin Treatment in Multiple Sclerosis: A Systematic Review and Meta-Analysis. CNS Drugs 2015, 29, 277–291. [Google Scholar] [CrossRef]
- Biernacki, T.; Sandi, D.; Bencsik, K.; Vécsei, L. Kynurenines in the Pathogenesis of Multiple Sclerosis: Therapeutic Perspectives. Cells 2020, 9, 1564. [Google Scholar] [CrossRef]
- Badawy, A.A.-B. Kynurenine Pathway of Tryptophan Metabolism: Regulatory and Functional Aspects. Int. J. Tryptophan Res. 2017, 10, 1178646917691938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Negrotto, L.; Correale, J. Amino Acid Catabolism in Multiple Sclerosis Affects Immune Homeostasis. J. Immunol. 2017, 198, 1900–1909. [Google Scholar] [CrossRef] [Green Version]
- Annus, A.; Vecsei, L. Kynurenine System and Multiple Sclerosis, Pathomechanism and Drug Targets with An Emphasis on Laquinimod. Curr. Drug Targets 2018, 19, 805–814. [Google Scholar] [CrossRef]
- Rajda, C.; Majláth, Z.; Pukoli, D.; Vécsei, L. Kynurenines and Multiple Sclerosis: The Dialogue between the Immune System and the Central Nervous System. Int. J. Mol. Sci. 2015, 16, 18270–18282. [Google Scholar] [CrossRef]
- Hubler, Z.; Allimuthu, D.; Bederman, I.; Elitt, M.; Madhavan, M.; Allan, K.C.; Shick, H.E.; Garrison, E.; Karl, M.T.; Factor, D.C.; et al. Accumulation of 8,9-unsaturated sterols drives oligodendrocyte formation and remyelination. Nat. Cell Biol. 2018, 560, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, P.; Smith, M.D.; Mische, L.; Harrington, E.P.; Fitzgerald, K.C.; Martin, K.A.; Kim, S.; Reyes, A.A.A.; Gonzalez-Cardona, J.; Volsko, C.; et al. Bile acid metabolism is altered in multiple sclerosis and supplementation ameliorates neuroinflammation. J. Clin. Investig. 2020, 130, 3467–3482. [Google Scholar] [CrossRef] [Green Version]
- Klotz, L.; Eschborn, M.; Lindner, M.; Liebmann, M.; Herold, M.; Janoschka, C.; Garrido, B.T.; Schulte-Mecklenbeck, A.; Gross, C.C.; Breuer, J.; et al. Teriflunomide treatment for multiple sclerosis modulates T cell mitochondrial respiration with affinity-dependent effects. Sci. Transl. Med. 2019, 11, eaao5563. [Google Scholar] [CrossRef] [Green Version]
- Ding, H.; Mohan, C. Promises and challenges of metabolomics in SLE. Nat. Rev. Rheumatol. 2016, 12, 627–628. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Xie, C.; Han, J.; Ye, Y.; Weiel, J.; Li, Q.; Blanco, I.; Ahn, C.; Olsen, N.; Putterman, C.; et al. Metabolic Disturbances Associated with Systemic Lupus Erythematosus. PLoS ONE 2012, 7, e37210. [Google Scholar] [CrossRef] [Green Version]
- Shah, D.; Sah, S.; Nath, S.K. Interaction between glutathione and apoptosis in systemic lupus erythematosus. Autoimmun. Rev. 2013, 12, 741–751. [Google Scholar] [CrossRef] [Green Version]
- Shah, D.; Aggarwal, A.; Bhatnagar, A.; Kiran, R.; Wanchu, A. Association between T lymphocyte sub-sets apoptosis and peripheral blood mononuclear cells oxidative stress in systemic lupus erythematosus. Free Radic. Res. 2011, 45, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Gergely, P.; Grossman, C.; Niland, B.; Puskas, F.; Neupane, H.; Allam, F.; Banki, K.; Phillips, P.E.; Perl, A. Mitochondrial hyperpolarization and ATP depletion in patients with systemic lupus erythematosus. Arthritis Rheum. 2002, 46, 175–190. [Google Scholar] [CrossRef]
- Lai, Z.-W.; Hanczko, R.; Bonilla, E.; Caza, T.N.; Clair, B.; Bartos, A.; Miklossy, G.; Jimah, J.; Doherty, E.; Tily, H.; et al. N-acetylcysteine reduces disease activity by blocking mammalian target of rapamycin in T cells from systemic lupus erythematosus patients: A randomized, double-blind, placebo-controlled trial. Arthritis Rheum. 2012, 64, 2937–2946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Battaglia, M.; Stabilini, A.; Migliavacca, B.; Horejs-Hoeck, J.; Kaupper, T.; Roncarolo, M.-G. Rapamycin Promotes Expansion of Functional CD4+CD25+FOXP3+ Regulatory T Cells of Both Healthy Subjects and Type 1 Diabetic Patients. J. Immunol. 2006, 177, 8338–8347. [Google Scholar] [CrossRef] [Green Version]
- Valencia, X.; Yarboro, C.; Illei, G.; Lipsky, P.E. Deficient CD4+CD25high T Regulatory Cell Function in Patients with Active Systemic Lupus Erythematosus. J. Immunol. 2007, 178, 2579–2588. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.; Yin, T.; Shinozaki, K.; Lampe, J.W.; Stevens, J.F.; Becker, L.B.; Kim, J. Comprehensive analysis of phospholipids in the brain, heart, kidney, and liver: Brain phospholipids are least enriched with polyunsaturated fatty acids. Mol. Cell. Biochem. 2018, 442, 187–201. [Google Scholar] [CrossRef]
- Katsuyama, T.; Tsokos, G.C.; Moulton, V.R. Aberrant T Cell Signaling and Subsets in Systemic Lupus Erythematosus. Front. Immunol. 2018, 9, 1088. [Google Scholar] [CrossRef]
- Kono, M.; Yoshida, N.; Tsokos, G.C. Amino Acid Metabolism in Lupus. Front. Immunol. 2021, 12, 623844. [Google Scholar] [CrossRef]
- Kono, M.; Yoshida, N.; Maeda, K.; Tsokos, G.C. Transcriptional factor ICER promotes glutaminolysis and the generation of Th17 cells. Proc. Natl. Acad. Sci. USA 2018, 115, 2478–2483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, Y.; Choi, S.-C.; Xu, Z.; Perry, D.J.; Seay, H.; Croker, B.P.; Sobel, E.S.; Brusko, T.M.; Morel, L. Normalization of CD4 + T cell metabolism reverses lupus. Sci. Transl. Med. 2015, 7, 274ra18. [Google Scholar] [CrossRef] [Green Version]
- Teng, X.; Brown, J.; Choi, S.; Li, W.; Morel, L. Metabolic determinants of lupus pathogenesis. Immunol. Rev. 2020, 295, 167–186. [Google Scholar] [CrossRef] [PubMed]
- Kono, M.; Yoshida, N.; Maeda, K.; Suárez-Fueyo, A.; Kyttaris, V.C.; Tsokos, G.C. Glutaminase 1 Inhibition Reduces Glycolysis and Ameliorates Lupus-like Disease in MRL / lpr Mice and Experimental Autoimmune Encephalomyelitis. Arthritis Rheumatol. 2019, 71, 1869–1878. [Google Scholar] [CrossRef] [PubMed]
- Puchades-Carrasco, L. Metabolomics Applications in Precision Medicine: An Oncological Perspective. Curr. Top. Med. Chem. 2017, 17, 2740–2751. [Google Scholar] [CrossRef] [Green Version]
- Lequerré, T.; Rottenberg, P.; Derambure, C.; Cosette, P.; Vittecoq, O. Predictors of treatment response in rheumatoid arthritis. Jt. Bone Spine 2019, 86, 151–158. [Google Scholar] [CrossRef]
- Kaddurah-Daouk, R.; Weinshilboum, R.; Pharmacometabolomics Research Network. Metabolomic Signatures for Drug Response Phenotypes: Pharmacometabolomics Enables Precision Medicine. Clin. Pharmacol. Ther. 2015, 98, 71–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piquette-Miller, M.; Grant, D.M. The Art and Science of Personalized Medicine. Clin. Pharmacol. Ther. 2007, 81, 311–315. [Google Scholar] [CrossRef]
- Balashova, E.E.; Maslov, D.L.; Lokhov, P.G. A Metabolomics Approach to Pharmacotherapy Personalization. J. Pers. Med. 2018, 8, 28. [Google Scholar] [CrossRef] [Green Version]
- Kaddurah-Daouk, R.; Kristal, B.S.; Weinshilboum, R.M. Metabolomics: A Global Biochemical Approach to Drug Response and Disease. Annu. Rev. Pharmacol. Toxicol. 2008, 48, 653–683. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Chen, Z.; Yang, S.; Wang, Y.; Yu, L.; Zhang, B.; Rao, Z.; Gao, J.; Tu, S. 1H NMR-based metabolomic analysis for identifying serum biomarkers to evaluate methotrexate treatment in patients with early rheumatoid arthritis. Exp. Ther. Med. 2012, 4, 165–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Tu, S.; Hu, Y.; Wang, Y.; Xia, Y.; Jiang, Y. Prediction of response of collagen-induced arthritis rats to methotrexate: An 1H-NMR-based urine metabolomic analysis. Acta Acad. Med. Wuhan 2012, 32, 438–443. [Google Scholar] [CrossRef]
- Kapoor, S.R.; Filer, A.; Fitzpatrick, M.; Fisher, B.A.; Taylor, P.C.; Buckley, C.D.; McInnes, I.; Raza, K.; Young, S.P. Metabolic Profiling Predicts Response to Anti-Tumor Necrosis Factor α Therapy in Patients with Rheumatoid Arthritis. Arthritis Rheum. 2013, 65, 1448–1456. [Google Scholar] [CrossRef] [PubMed]
- Priori, R.; Casadei, L.; Valerio, M.; Scrivo, R.; Valesini, G.; Manetti, C. 1H-NMR-Based Metabolomic Study for Identifying Serum Profiles Associated with the Response to Etanercept in Patients with Rheumatoid Arthritis. PLoS ONE 2015, 10, e0138537. [Google Scholar] [CrossRef]
- Cuppen, B.V.J.; Fu, J.; van Wietmarschen, H.A.; Harms, A.C.; Koval, S.; Marijnissen, A.C.A.; Peeters, J.J.W.; Bijlsma, J.W.J.; Tekstra, J.; van Laar, J.M.; et al. Exploring the Inflammatory Metabolomic Profile to Predict Response to TNF-α Inhibitors in Rheumatoid Arthritis. PLoS ONE 2016, 11, e0163087. [Google Scholar] [CrossRef] [Green Version]
- Tatar, Z.; Migné, C.; Pétéra, M.; Gaudin, P.; Lequerré, T.; Marotte, H.; Tebib, J.; Guillot, E.P.; Soubrier, M. Variations in the metabolome in response to disease activity of rheumatoid arthritis. BMC Musculoskelet. Disord. 2016, 17, 353. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.; Cuppen, B.V.J.; Welsing, P.M.J.; Van Wietmarschen, H.; Harms, A.C.; Berger, R.; Koval, S.; Fritsch-Stork, R.D.E.; Bijlsma, J.W.J.; Hankemeier, T.; et al. Differences between serum polar lipid profiles of male and female rheumatoid arthritis patients in response to glucocorticoid treatment. Inflammopharmacology 2016, 24, 397–402. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, S.; Saegusa, J.; Onishi, A.; Morinobu, A. Biomarkers identified by serum metabolomic analysis to predict biologic treatment response in rheumatoid arthritis patients. Rheumatology 2019, 58, 2153–2161. [Google Scholar] [CrossRef]
- Artacho, A.; Isaac, S.; Nayak, R.; Flor-Duro, A.; Alexander, M.; Koo, I.; Manasson, J.; Smith, P.B.; Rosenthal, P.; Homsi, Y.; et al. The Pretreatment Gut Microbiome Is Associated with Lack of Response to Methotrexate in New-Onset Rheumatoid Arthritis. Arthritis Rheumatol. 2021, 73, 931–942. [Google Scholar] [CrossRef]
- Maciejewski, M.; Sands, C.; Nair, N.; Ling, S.; Verstappen, S.; Hyrich, K.; Barton, A.; Ziemek, D.; Lewis, M.R.; Plant, D. Prediction of response of methotrexate in patients with rheumatoid arthritis using serum lipidomics. Sci. Rep. 2021, 11, 1–6. [Google Scholar] [CrossRef]
- Dudka, I.; Chachaj, A.; Sebastian, A.; Tański, W.; Stenlund, H.; Gröbner, G.; Szuba, A. Metabolomic profiling reveals plasma GlycA and GlycB as a potential biomarkers for treatment efficiency in rheumatoid arthritis. J. Pharm. Biomed. Anal. 2021, 197, 113971. [Google Scholar] [CrossRef] [PubMed]
- Lorefice, L.; Murgia, F.; Fenu, G.; Frau, J.; Coghe, G.; Murru, M.R.; Tranquilli, S.; Visconti, A.; Marrosu, M.G.; Atzori, L.; et al. Assessing the Metabolomic Profile of Multiple Sclerosis Patients Treated with Interferon Beta 1a by 1H-NMR Spectroscopy. Neurotherapeutics 2019, 16, 797–807. [Google Scholar] [CrossRef] [Green Version]
- Stürner, K.H.; Werz, O.; Koeberle, A.; Otto, M.; Pless, O.; Leypoldt, F.; Paul, F.; Heesen, C. Lipid Mediator Profiles Predict Response to Therapy with an Oral Frankincense Extract in Relapsing-Remitting Multiple Sclerosis. Sci. Rep. 2020, 10, 8776. [Google Scholar] [CrossRef] [PubMed]
- Waddington, K.E.; Papadaki, A.; Coelewij, L.; Adriani, M.; Nytrova, P.; Havrdova, E.K.; Fogdell-Hahn, A.; Farrell, R.; Dönnes, P.; Pineda-Torra, I.; et al. Using Serum Metabolomics to Predict Development of Anti-drug Antibodies in Multiple Sclerosis Patients Treated with IFNβ. Front. Immunol. 2020, 11, 1527. [Google Scholar] [CrossRef] [PubMed]
- Signoriello, E.; Iardino, P.; Casertano, S.; De Lucia, D.; Pucciarelli, A.; Puoti, G.; Chiosi, E.; Lus, G. 12-months prospective Pentraxin-3 and metabolomic evaluation in multiple sclerosis patients treated with glatiramer acetate. J. Neuroimmunol. 2020, 348, 577385. [Google Scholar] [CrossRef]
- Guleria, A.; Phatak, S.; Dubey, D.; Kumar, S.; Zanwar, A.; Chaurasia, S.; Kumar, U.; Gupta, R.; Aggarwal, A.; Kumar, D.; et al. NMR-Based Serum Metabolomics Reveals Reprogramming of Lipid Dysregulation Following Cyclophosphamide-Based Induction Therapy in Lupus Nephritis. J. Proteome Res. 2018, 17, 2440–2448. [Google Scholar] [CrossRef]
- Ganguly, S.; Kumar, U.; Gupta, N.; Guleria, A.; Majumdar, S.; Phatak, S.; Chaurasia, S.; Kumar, S.; Aggarwal, A.; Kumar, D.; et al. Nuclear magnetic resonance–based targeted profiling of urinary acetate and citrate following cyclophosphamide therapy in patients with lupus nephritis. Lupus 2020, 29, 782–786. [Google Scholar] [CrossRef]
- Ma, X.; Xu, S. TNF inhibitor therapy for rheumatoid arthritis. Biomed. Rep. 2012, 1, 177–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, B.; Cronstein, B. Methotrexate mechanism in treatment of rheumatoid arthritis. Jt. Bone Spine 2019, 86, 301–307. [Google Scholar] [CrossRef]
- Filipi, M.; Jack, S. Interferons in the Treatment of Multiple Sclerosis. Int. J. MS Care 2020, 22, 165–172. [Google Scholar] [CrossRef] [Green Version]
- Patti, F. Standardised Frankincense extract: New possible therapeutic option for patients with relapsing-remitting multiple sclerosis. J. Neurol. Neurosurg. Psychiatry 2017, 89, 327. [Google Scholar] [CrossRef]
- Stürner, K.H.; Stellmann, J.-P.; Dörr, J.; Paul, F.; Friede, T.; Schammler, S.; Reinhardt, S.; Gellissen, S.; Weissflog, G.; Faizy, T.D.; et al. A standardised frankincense extract reduces disease activity in relapsing-remitting multiple sclerosis (the SABA phase IIa trial). J. Neurol. Neurosurg. Psychiatry 2017, 89, 330–338. [Google Scholar] [CrossRef] [Green Version]
- Petri, M. Cyclophosphamide: New approaches for systemic lupus erythematosus. Lupus 2004, 13, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Suhre, K.; Shin, S.Y.; Petersen, A.K.; Mohney, R.P.; Meredith, D.; Wagele, B.; Altmaier, E.; Gram, C.; Deloukas, P.; Erdmann, J.; et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature 2011, 477, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Newgard, C.B.; Attie, A.D. Getting biological about the genetics of diabetes. Nat. Med. 2010, 16, 388–391. [Google Scholar] [CrossRef] [PubMed]
- Menni, C.; Zierer, J.; Valdes, A.M.; Spector, T.D. Mixing omics: Combining genetics and metabolomics to study rheumatic diseases. Nat. Rev. Rheumatol. 2017, 13, 174–181. [Google Scholar] [CrossRef] [Green Version]
- Chu, X.; Jaeger, M.; Beumer, J.; Bakker, O.B.; Aguirre-Gamboa, R.; Oosting, M.; Smeekens, S.P.; Moorlag, S.; Mourits, V.P.; Koeken, V.A.C.M.; et al. Integration of metabolomics, genomics, and immune phenotypes reveals the causal roles of metabolites in disease. Genome Biol. 2021, 22, 1–22. [Google Scholar] [CrossRef]
- Blanchet, L.; Smolinska, A.; Attali, A.; Stoop, M.P.; Ampt, K.A.M.; Van Aken, H.; Suidgeest, E.; Tuinstra, T.; Wijmenga, S.S.; Luider, T.; et al. Fusion of metabolomics and proteomics data for biomarkers discovery: Case study on the experimental autoimmune encephalomyelitis. BMC Bioinform. 2011, 12, 254. [Google Scholar] [CrossRef] [Green Version]
| Date | Sample | Instruments | Upregulated | Downregulated | Ref. |
|---|---|---|---|---|---|
| 2011 | Plasma | GC-MS LC-MS | Glyceric acid, D-ribofuranose, Hypoxanthine | Histidine, threonic acid, methionine, cholesterol, asparagine, threonine | [38] |
| 2011 | Serum | 1H NMR | Glucose, glycoprotein, lactate, VLDL, LDL | Valine, tyrosine, pyruvate, lysine, phenylalanine, HDL, cholesterol, isoleucine, histidine, alanine, phosphocholine, glycerol, glutamine, glutamate, creatinine, citrate | [39] |
| 2009 | Serum | 1H NMR | 3-hydroxybutyrate, lactate, acetylglycine, taurine, glucose | LDL, alanine, methylguanidine | [40] |
| 2013 | Serum | GC/QTOF-MS LC/QTOF-MS | Lactic acid, dihydroxyfumaric acid, glyceraldehyde, aspartic acid, homoserine | 4,8-dimethylnonanoyl carnitine | [41] |
| 2015 | Synovial fluid | GC/TOF-MS | Lactic acid, carnitine, diglycerol, pipecolinic acid beta-mannosylglycerate, | Valine, citric acid, gluconic lactone, glucose, glucose-1-phosphate, mannose, 5-methoxytryptamine, D-glucose, ribitol | [42] |
| 2016 | Serum | GC-MS | Docosahexaenoate, palmitelaidate, oleate, trans-9-octadecenoate, D-mannose, glycerol, ribose | 2-Ketoisocaproate, isoleucine, leucine, serine, phenylalanine, pyroglutamate, methionine, proline, threonine, valine, urate | [43] |
| 2016 | Urine | 1H NMR | Tyrosine | N-acetyl amino acids, citrate, alanine | [44] |
| 2016 | Serum | 1H NMR | 3-hydroxyisobutyrate, acetate, NAC, acetoacetate, acetone | Isoleucine, lactate, alanine, creatinine, valine, histidine | [45] |
| 2018 | Serum | LC-MS | 4-methoxyphenylacetic acid, glutamic acid, L-leucine, L-phenylalanine, L-tryptophan, L-proline, glyceraldehyde, fumaric acid, cholesterol | Capric acid, argininosuccinic acid, bilirubin | [46] |
| 2019 | Serum | LC-MS | Glutamine | Taurine, asparagine, serine, glycine, ethanolamine, aspartic acid, proline, threonine, sarcosine, alanine, valine, histidine, arginine, leucine, ornithine, methionine, tryptophan, phenylalanine | [47] |
| 2021 | Plasma | GC-MS | L-cysteine, citric acid, L-glutamine | [48] |
| Date | Sample | Instruments | Group | Upregulated | Downregulated | Ref. |
|---|---|---|---|---|---|---|
| 2014 | Serum | 1H NMR | MuS | Lysine | L-Glutamine, valine | [50] |
| 2014 | CSF | 1H NMR | MuS | Threonate, choline, myo-inositol | Phenylalanine, mannose, citrate, 3-hydroxybutyrate, 2-hydroxyisovalerate | [51] |
| 2015 | CSF | MALDI-TOF-MS, LC-MS/MS | MuS | L-glutamate | [52] | |
| 2016 | Serum | 1H NMR | MuS | Alanine, acetoacetate, acetone, choline, 3-hydroxybutyrate | Tryptophan, 5-hydroxytryptophan, glycerol, glucose | [53] |
| 2016 | CSF | GC/MS | MuS | 1-Monopalmitin, 1-monostearin, pentadecanoic acid, oleic acid, methionine, valine, phenylalanine, tyrosine, leucine, proline, threose, isoleucine, putrescine, oxoproline, | [54] | |
| 2016 | Urine | 1H NMR | MuS | Trimethylamine N-oxide, 3-hydroxyisovalerate, hippurate, malonate | Creatinine, 3-hydroxybutyrate, methylmalonate | [55] |
| 2017 | Plasma | GC-MS | MuS | L-asparagine, L-ornithine, L-glutamate, L-glutamine | Pyroglutamate, fructose, myo-inositol, threonate, phosphate | [56] |
| 2017 | CSF | NMR | MuS | Pyroglutamate, 2-hydroxybutyrate, formate | Glucose, acetate, citrate | [57] |
| 2017 | CSF | UHPLC-FLD, GC/MS | MuS | L-glutamine, lactate | [58] | |
| Serum | RRMS | Kynurenic acid, picolinic acid | ||||
| PPMS | 3-hydroxykynurenine, quinolinic acid | Kynurenic acid, picolinic acid | ||||
| SPMS | 3-hydroxykynurenine, quinolinic acid | Kynurenic acid, picolinic acid | ||||
| 2017 | Serum | HPLC-ECD | SPMS, RRMS | Methionine, glutathione | [59] | |
| 2019 | CSF | UPLC-HRMS | SPMS | Trigonelline, citrulline, O-Succinyl-homoserine, N6-(delta2-isopentenyl)-adenine, pipecolate, 1-methyladenosine, 4-acetamidobutanoate, 5-hydroxytryptophan, kynurenate N-acetylserotonin | 3-methoxytyramine, caffeine | [60] |
| 2020 | CSF | LC-MS/MS | MuS | Kynurenine, quinolinic acid, neopterin, kynurenic acid | tryptophan, 5-hydroxy-indolacetic acid, piconilic acid | [61] |
| 2020 | CSF | LC-MS | MuS | 3-hydroxykynurenine, quinolinic acid | L-kynurenine, picolinic acid | [62] |
| Serum | MuS | quinolinic acid | 5-hydroxyindoleacetic acid |
| Date | Sample | Instruments | Upregulated | Downregulated | Ref. |
|---|---|---|---|---|---|
| 2011 | Serum | 1H NMR | N-acetyl glycoprotein, VLDL, LDL | Valine, tyrosine, phenylalanine, lysine, isoleucine, histidine, glutamine, alanine, citrate, creatinine, creatine, pyruvate, HDL, cholesterol, glycerol, formate | [39] |
| 2016 | Serum | GC-MS | Methionine, glutamate, cystine, 1-monopalmitin, 1-monolinolein, 1-monoolein, 2-hydroxyisobutyrate | Tryptophan, alanine, proline, glycine, serine, threonine, aspartate, glutamine, asparagine, lysine, histidine, tyrosine, valine, leucine, isoleucine, fumarate, threonate, 2-hydroxyisovalerate, carbohydrates, 2-keto-3-methylvalerate, 2-ketoisocaproate, fatty acids, aminomalonate, alpha-tocopherol | [68] |
| 2016 | Urine | GC-MS | Valine, leucine, fumarate, malate, cystine, pyroglutamate, cysteine, tryptophan, threonate, uracil, urate, pseudouridine, xanthine, glyceric acid, myo-inositol, p-cresol, glutarate, hydroxyisobutyrate, dihydroxybutyrate, 3,4,5-trihydroxypentanoic acid | [69] | |
| 2016 | Serum | GC-MS | Urea, cystine, threonine, naproxen, glucose | Lysine, fumaric acid, malic acid, methionine, tyrosine, alanine, cysteine, tryptophan asparagine, threonic acid, histidine, citric acid, lactic acid, caffeine, theobromine | [70] |
| 2016 | Serum | 1H NMR | Acetate, NAG, glucose | Leucine, valine, alanine, glutamate, citrate, choline, proline, glycine, lactate, LDL, VLDL | [71] |
| 2017 | Plasma | GC-MS | Myristic acids, palmitoleic acids, oleic acids, eicosenoic acids | Caproic acid, caprylic acid, linoleic acid, stearic acid, arachidonic acid, eicosanoic acid, behenic acid, lignoceric acid, hexacosanoic acid | [72] |
| 2019 | Feces | LC-MS | Proline, L-tyrosine, L-methionine, L-asparagine, Dl-pipecolinic acid, glycyl-L proline, L-carnosine, xanthurenic acid, kynurenic acid, 1,2-dioleoyl-rac-glycerol, lysoPE 16:0, lysoPC 22:5, PG 27:2, MG 22:6, MG 16:5 | D-Ala-D-ala, lauryl diethanolamide, SQDG 26:5, adenosine, mucic acid, adenosine 5′-diphosphate, trigonelline thiamine pyrophosphate | [73] |
| 2019 | Serum | LC-MS | Ceramide, trimethylamine n-oxide, xanthine | Acylcarnitine, caffeine, hydrocortisone, itaconic acid, serotonin | [74] |
| 2020 | Feces | GC-MS | Triethylene glycol, erucamide, leucic acid, 1-phenyl-1,2-ethanediol, pyrimidine, 4-aminobutanoic acid, vaccenic acid, L-valine, L-ornithine, L-phenylalanine, L-leucine, lactic acid, arachidic acid, behenic acid, putrescine, benzoic acid, erucic acid, n-(4-aminobutyl) acetamide | 2,4-di-tert-butylphenol, phosphoric acid, Glyceric acid, (Z)-13-octadecenoic acid, γ-tocopherol | [75] |
| 2021 | Serum | LC-MS | MG 20:2, L-pyroglutamic acid | Arachidonic acid, adenosine, SM 24:1, MG 17:0, lysoPE 18:0, lysoPE 16:0, lysoPC 20:0, lysoPC 18:0 | [76] |
| Disease | Year | Treatment | Sample | Instruments | Biomarker | Ref. |
|---|---|---|---|---|---|---|
| RA | 2012 | MTX | Serum | 1H-NMR | α-oxoglutarate, glycine, citrate, aspartate, acetate, alanine, cholesterol, cysteine, histidine, hypoxanthine, lactate, glutamine, methionine, serine, taurine, tryptophan, trimethylamine-N-oxide, uracil, uric acid | [161] |
| 2012 | Anti-TNF | Urine | 1H-NMR | Uric acid, taurine, histidine, methionine, glycine, uracil, acetate, α-oxoglutarate, aspartate, tryptophan, hypoxanthine, TMAO, methionine, acetate | [162] | |
| 2013 | Infliximab or ETA | Urine | NMR | Histamine, glutamine, xanthurenic acid, ethanolamine | [163] | |
| 2015 | ETA | Serum | 1H-NMR | Isoleucine, leucine, valine, alanine, glutamine, tyrosine, glucose | [164] | |
| 2016 | 5 TNFis | Serum | LC-MS | Sn1-LPC(18:3-ω3/ω6), sn1-LPC(15:0), ethanolamine, lysine | [165] | |
| 2016 | Anti-TNF | Plasma | TOF-MS | D-glucose, D-fructose, sucrose, maltos | [166] | |
| 2016 | Glucocorticoids | Serum | LC-MS | Lysophospholipids | [167] | |
| 2020 | TNFis or ABT | Serum | CE-TOF-MS | Glycerol 3-phosphate, betonicine, N-Acetylalanine, hexanoic acid, taurine (TNFis) 3-Aminobutyric acid, citric acid, quinic acid (ABT) | [168] | |
| 2020 | MTX | Fecal | NMR, LC-MS | Bacteria-produced metabolites | [169] | |
| 2021 | MTX | Serum | UPLC–MS | no effect (lipidomics) | [170] | |
| 2021 | DMARDs | Plasma | NMR/MS | N-acetylgalactosamine, N-acetylneuraminic acid | [171] | |
| MuS | 2019 | IFN ß | Plasma | NMR | Lactate, acetone, 3-OH-butyrate, tryptophan, citrate, lysine, glucose | [172] |
| 2020 | SFE | Plasma | MRI | 12- and 15-lipoxygenase products | [173] | |
| 2020 | IFNβ formulations | Serum | NMR | 29 metabolites (e.g., TG, XL-VLDL-PL, etc.) | [174] | |
| 2020 | Glatiramer acetate | Serum | 1H-NMR | Lactate, tyrosine, hypoxanthine, hydroxyproline, ADP, citrulline, ornithine, tryptophan | [175] | |
| SLE | 2018 | Cyclophosphamide + prednisolone | Serum | NMR | Lipid metabolites and acetate | [176] |
| 2020 | Cyclophosphamide | Urine | NMR | Citrate | [177] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoon, N.; Jang, A.-K.; Seo, Y.; Jung, B.H. Metabolomics in Autoimmune Diseases: Focus on Rheumatoid Arthritis, Systemic Lupus Erythematous, and Multiple Sclerosis. Metabolites 2021, 11, 812. https://doi.org/10.3390/metabo11120812
Yoon N, Jang A-K, Seo Y, Jung BH. Metabolomics in Autoimmune Diseases: Focus on Rheumatoid Arthritis, Systemic Lupus Erythematous, and Multiple Sclerosis. Metabolites. 2021; 11(12):812. https://doi.org/10.3390/metabo11120812
Chicago/Turabian StyleYoon, Naeun, Ah-Kyung Jang, Yerim Seo, and Byung Hwa Jung. 2021. "Metabolomics in Autoimmune Diseases: Focus on Rheumatoid Arthritis, Systemic Lupus Erythematous, and Multiple Sclerosis" Metabolites 11, no. 12: 812. https://doi.org/10.3390/metabo11120812
APA StyleYoon, N., Jang, A.-K., Seo, Y., & Jung, B. H. (2021). Metabolomics in Autoimmune Diseases: Focus on Rheumatoid Arthritis, Systemic Lupus Erythematous, and Multiple Sclerosis. Metabolites, 11(12), 812. https://doi.org/10.3390/metabo11120812