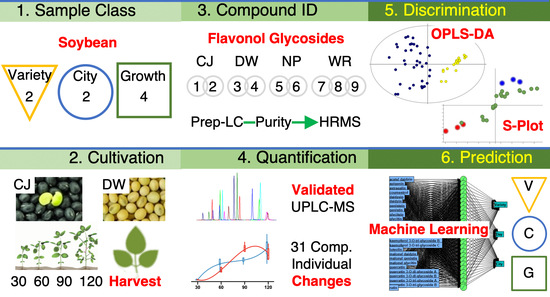
Supervised Statistical Learning Prediction of Soybean Varieties and Cultivation Sites Using Rapid UPLC-MS Separation, Method Validation, and Targeted Metabolomic Analysis of 31 Phenolic Compounds in the Leaves
Abstract
:1. Introduction
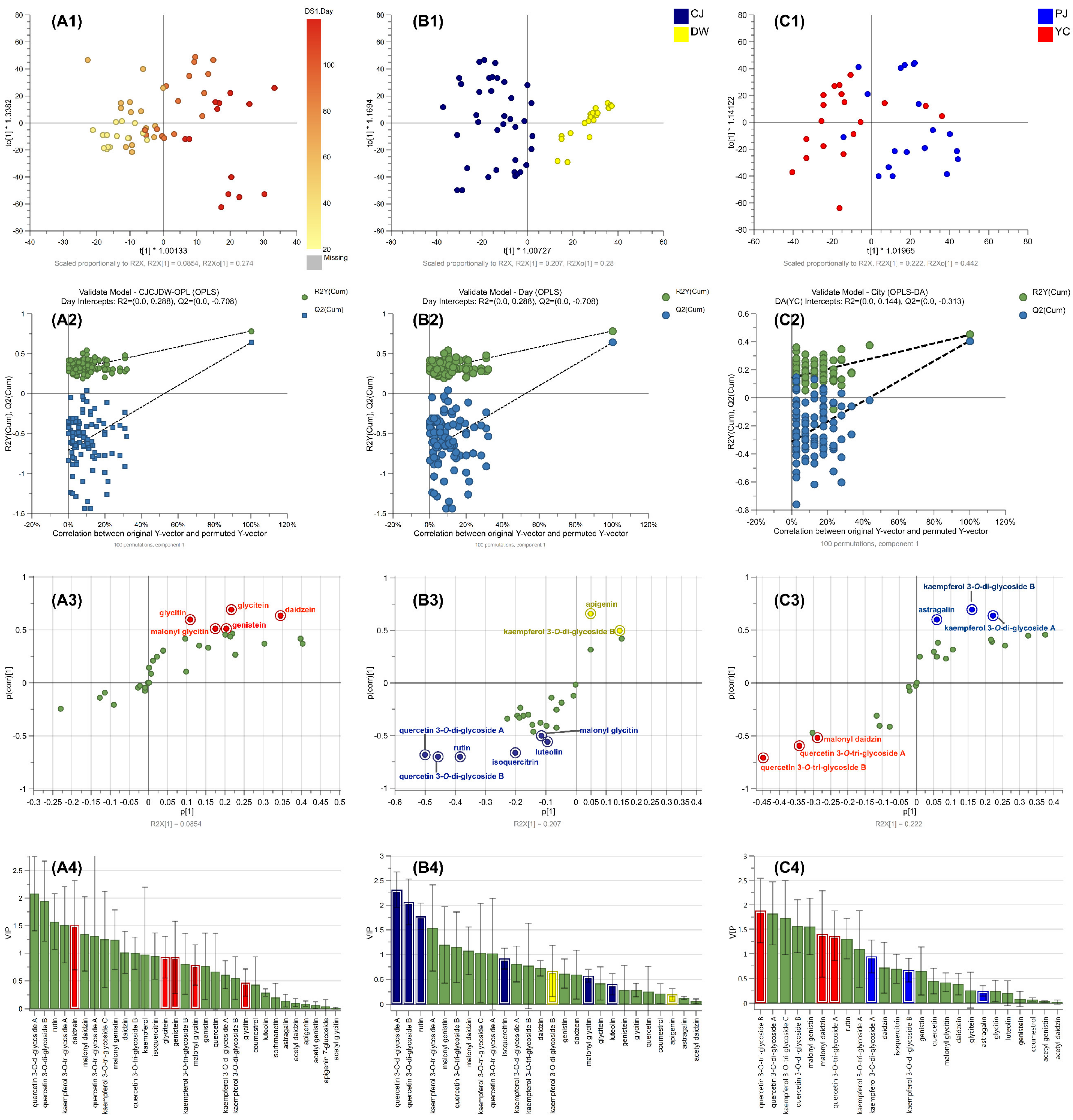
2. Results and Discussion
2.1. Identification of Purified Flavonol Glycosides
2.2. Separation Method Validation
2.3. Changes in 31 Compounds in SLs
2.4. Supervised ML Model Predictions via Targeted Metabolomics
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. SB Seeding, Cultivation, and Leaf Sample Preparation
3.3. Preparation and Identification of Flavonol Glycosides from SLs
3.4. Analytical Conditions for the Quantification of the 31 Compounds
3.5. Stock Solutions and SL Sample Preparations
3.6. Separation Method Validation
3.7. Metabolomic Discrimination Using ML Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carneiro, A.M.; Moreira, E.A.; Bragagnolo, F.S.; Borges, M.S.; Pilon, A.C.; Rinaldo, D.; Funari, C.S. Soya Agricultural Waste as a Rich Source of Isoflavones. Food Res. Int. 2020, 130, 108949. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.L.; Berhow, M.A.; Kim, J.T.; Chi, H.Y.; Lee, S.J.; Chung, I.M. Evaluation of Soyasaponin, Isoflavone, Protein, Lipid, and Free Sugar Accumulation in Developing Soybean Seeds. J. Agric. Food Chem. 2006, 54, 10003–10010. [Google Scholar] [CrossRef] [PubMed]
- Dhaubhadel, S.; McGarvey, B.D.; Williams, R.; Gijzen, M. Isoflavonoid Biosynthesis and Accumulation in Developing Soybean Seeds. Plant Mol. Biol. 2003, 53, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ji, H.S.; Kang, J.H.; Shin, D.H.; Park, H.Y.; Choi, M.S.; Lee, C.H.; Lee, I.K.; Yun, B.S.; Jeong, T.S. Soy Leaf Extract Containing Kaempferol Glycosides and Pheophorbides Improves Glucose Homeostasis by Enhancing Pancreatic Beta-Cell Function and Suppressing Hepatic Lipid Accumulation in db/db Mice. J. Agric. Food Chem. 2015, 63, 7198–7210. [Google Scholar] [CrossRef]
- Ho, H.M.; Chen, R.Y.; Leung, L.K.; Chan, F.L.; Huang, Y.; Chen, Z.-Y. Difference in Flavonoid and Isoflavone Profile between Soybean and Soy Leaf. Biomed. Pharmacother. 2002, 56, 289–295. [Google Scholar] [CrossRef]
- Sugiyama, A.; Yamazaki, Y.; Hamamoto, S.; Takase, H.; Yazaki, K. Synthesis and Secretion of Isoflavones by Field-Grown Soybean. Plant Cell Physiol. 2017, 58, 1594–1600. [Google Scholar] [CrossRef]
- Yun, D.-Y.; Kang, Y.-G.; Kim, M.; Kim, D.; Kim, E.-H.; Hong, Y.-S. Metabotyping of Different Soybean Genotypes and Distinct Metabolism in Their Seeds and Leaves. Food Chem. 2020, 330, 127198. [Google Scholar] [CrossRef]
- Yun, D.Y.; Kang, Y.G.; Kim, E.H.; Kim, M.; Park, N.H.; Choi, H.T.; Go, G.H.; Lee, J.H.; Park, J.S.; Hong, Y.S. Metabolomics Approach for Understanding Geographical Dependence of Soybean Leaf Metabolome. Food Res. Int. 2018, 106, 842–852. [Google Scholar] [CrossRef]
- Jung, J.W.; Park, S.Y.; Oh, S.D.; Jang, Y.; Suh, S.J.; Park, S.K.; Ha, S.H.; Park, S.U.; Kim, J.K. Metabolomic Variability of Different Soybean Genotypes: Beta-Carotene-Enhanced (Glycine max), Wild (Glycine soja), and Hybrid (Glycine max × Glycine soja) Soybeans. Foods 2021, 10, 2421. [Google Scholar] [CrossRef]
- Yilmaz, A.; Rudolph, H.L.; Hurst, J.J.; Wood, T.D. High-Throughput Metabolic Profiling of Soybean Leaves by Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. Anal. Chem. 2016, 88, 1188–1194. [Google Scholar] [CrossRef] [Green Version]
- Lima, M.R.; Diaz, S.O.; Lamego, I.; Grusak, M.A.; Vasconcelos, M.W.; Gil, A.M. Nuclear Magnetic Resonance Metabolomics of Iron Deficiency in Soybean Leaves. J. Proteome Res. 2014, 13, 3075–3087. [Google Scholar] [CrossRef]
- Villate, A.; San Nicolas, M.; Gallastegi, M.; Aulas, P.A.; Olivares, M.; Usobiaga, A.; Etxebarria, N.; Aizpurua-Olaizola, O. Review: Metabolomics as a Prediction Tool for Plants Performance under Environmental Stress. Plant Sci. 2021, 303, 110789. [Google Scholar] [CrossRef]
- Mullard, G.; Allwood, J.W.; Weber, R.; Brown, M.; Begley, P.; Hollywood, K.A.; Jones, M.; Unwin, R.D.; Bishop, P.N.; Cooper, G.J.S.; et al. A New Strategy for MS/MS Data Acquisition Applying Multiple Data Dependent Experiments on Orbitrap Mass Spectrometers in Non-Targeted Metabolomic Applications. Metabolomics 2014, 11, 1068–1080. [Google Scholar] [CrossRef]
- Zhou, P.; Hu, O.; Fu, H.; Ouyang, L.; Gong, X.; Meng, P.; Wang, Z.; Dai, M.; Guo, X.; Wang, Y. UPLC-Q-TOF/MS-Based Untargeted Metabolomics Coupled with Chemometrics Approach for Tieguanyin Tea with Seasonal and Year Variations. Food Chem. 2019, 283, 73–82. [Google Scholar] [CrossRef]
- Lee, E.M.; Park, S.J.; Lee, J.-E.; Lee, B.M.; Shin, B.K.; Kang, D.J.; Choi, H.-K.; Kim, Y.-S.; Lee, D.Y. Highly Geographical Specificity of Metabolomic Traits among Korean Domestic Soybeans (Glycine max). Food Res. Int. 2019, 120, 12–18. [Google Scholar] [CrossRef]
- Yuk, H.; Song, Y.; Curtis-Long, M.J.; Kim, D.; Woo, S.; Lee, Y.; Uddin, Z.; Kim, C.; Park, K. Ethylene Induced a High Accumulation of Dietary Isoflavones and Expression of Isoflavonoid Biosynthetic Genes in Soybean (Glycine max) Leaves. J. Agric. Food Chem. 2016, 64, 7315–7324. [Google Scholar] [CrossRef]
- Lee, M.J.; Chung, I.M.; Kim, H.; Jung, M.Y. High Resolution LC-ESI-TOF-Mass Spectrometry Method for Fast Separation, Identification, and Quantification of 12 Isoflavones in Soybeans and Soybean Products. Food Chem. 2015, 176, 254–262. [Google Scholar] [CrossRef]
- Jung, Y.S.; Rha, C.S.; Baik, M.Y.; Baek, N.I.; Kim, D.O. A Brief History and Spectroscopic Analysis of Soy Isoflavones. Food Sci. Biotechnol. 2020, 29, 1605–1617. [Google Scholar] [CrossRef]
- Seiber, J.N.; Molyneux, R.J.; Schieberle, P. Targeted Metabolomics: A New Section in the Journal of Agricultural and Food Chemistry. J. Agric. Food Chem. 2014, 62, 22–23. [Google Scholar] [CrossRef]
- Liebal, U.W.; Phan, A.N.T.; Sudhakar, M.; Raman, K.; Blank, L.M. Machine Learning Applications for Mass Spectrometry-Based Metabolomics. Metabolites 2020, 10, 243. [Google Scholar] [CrossRef]
- Song, H.-H.; Ryu, H.W.; Lee, K.J.; Jeong, I.Y.; Kim, D.S.; Oh, S.-R. Metabolomics Investigation of Flavonoid Synthesis in Soybean Leaves Depending on the Growth Stage. Metabolomics 2014, 10, 833–841. [Google Scholar] [CrossRef]
- Li, H.; Kim, U.H.; Yoon, J.H.; Ji, H.S.; Park, H.M.; Park, H.Y.; Jeong, T.S. Suppression of Hyperglycemia and Hepatic Steatosis by Black-Soybean-Leaf Extract Via Enhanced Adiponectin-Receptor Signaling and AMPK Activation. J. Agric. Food Chem. 2019, 67, 90–101. [Google Scholar] [CrossRef]
- Eidhammer, I.; Barsnes, H.; Eide, G.E.; Martens, L. Targeted Quantification-Selected Reaction Monitoring. In Computational and Statistical Methods for Protein Quantification by Mass Spectrometry; Wiley Online Books; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013; pp. 218–234. [Google Scholar]
- Rha, C.-S.; Choi, Y.-M.; Kim, J.-C.; Kim, D.-O. Cost-Effective Simultaneous Separation and Quantification of Phenolics in Green and Processed Tea Using HPLC–UV–ESI Single-Quadrupole MS Detector and Python Script. Separations 2021, 8, 45. [Google Scholar] [CrossRef]
- Baranowska, I.; Magiera, S. Analysis of Isoflavones and Flavonoids in Human Urine by UHPLC. Anal. Bioanal. Chem. 2011, 399, 3211–3219. [Google Scholar] [CrossRef] [Green Version]
- Svoboda, P.; Vlckova, H.; Novakova, L. Development and Validation of UHPLC-MS/MS Method for Determination of Eight Naturally Occurring Catechin Derivatives in Various Tea Samples and the Role of Matrix Effects. J. Pharm. Biomed. Anal. 2015, 114, 62–70. [Google Scholar] [CrossRef]
- Tripathi, P.; Rabara, R.C.; Reese, R.N.; Miller, M.A.; Rohila, J.S.; Subramanian, S.; Shen, Q.J.; Morandi, D.; Bucking, H.; Shulaev, V.; et al. A Toolbox of Genes, Proteins, Metabolites and Promoters for Improving Drought Tolerance in Soybean Includes the Metabolite Coumestrol and Stomatal Development Genes. BMC Genom. 2016, 17, 102. [Google Scholar] [CrossRef] [Green Version]
- Yun, D.Y.; Kang, Y.G.; Yun, B.; Kim, E.H.; Kim, M.; Park, J.S.; Lee, J.H.; Hong, Y.S. Distinctive Metabolism of Flavonoid between Cultivated and Semiwild Soybean Unveiled through Metabolomics Approach. J. Agric. Food Chem. 2016, 64, 5773–5783. [Google Scholar] [CrossRef]
- Mierziak, J.; Kostyn, K.; Kulma, A. Flavonoids as Important Molecules of Plant Interactions with the Environment. Molecules 2014, 19, 16240–16265. [Google Scholar] [CrossRef]
- Peiretti, P.; Karamać, M.; Janiak, M.; Longato, E.; Meineri, G.; Amarowicz, R.; Gai, F. Phenolic Composition and Antioxidant Activities of Soybean (Glycine max (L.) Merr.) Plant during Growth Cycle. Agronomy 2019, 9, 153. [Google Scholar] [CrossRef] [Green Version]
- Clarke, J.D.; Alexander, D.C.; Ward, D.P.; Ryals, J.A.; Mitchell, M.W.; Wulff, J.E.; Guo, L. Assessment of Genetically Modified Soybean in Relation to Natural Variation in the Soybean Seed Metabolome. Sci. Rep. 2013, 3, 3082. [Google Scholar] [CrossRef]
- Ballin, N.Z.; Laursen, K.H. To Target or Not to Target? Definitions and Nomenclature for Targeted Versus Non-Targeted Analytical Food Authentication. Trends Food Sci. Technol. 2019, 86, 537–543. [Google Scholar] [CrossRef]
- Wang, H.; Cao, X.; Yuan, Z.; Guo, G. Untargeted Metabolomics Coupled with Chemometrics Approach for Xinyang Maojian Green Tea with Cultivar, Elevation and Processing Variations. Food Chem. 2021, 352, 129359. [Google Scholar] [CrossRef] [PubMed]
- Triba, M.N.; Le Moyec, L.; Amathieu, R.; Goossens, C.; Bouchemal, N.; Nahon, P.; Rutledge, D.N.; Savarin, P. PLS/OPLS Models in Metabolomics: The Impact of Permutation of Dataset Rows on the K-fold Cross-Validation Quality Parameters. Mol. Biosyst. 2015, 11, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Akhatou, I.; Sayago, A.; Gonzalez-Dominguez, R.; Fernandez-Recamales, A. Application of Targeted Metabolomics to Investigate Optimum Growing Conditions to Enhance Bioactive Content of Strawberry. J. Agric. Food Chem. 2017, 65, 9559–9567. [Google Scholar] [CrossRef]
- SAS Institute. JMP 13 Predictive and Specialized Modeling; SAS Institute: Cary, NC, USA, 2017. [Google Scholar]
- Cho, J.-W.; Lee, J.-J.; Oh, Y.-J.; Lee, J.-D.; Lee, S.-B. Effects of Planting Densities and Maturing Types on Growth and Yield of Soybean in Paddy Field. Korean J. Crop Sci. 2004, 49, 105–109. [Google Scholar]
- Singh, S.; Singh, R.; Banerjee, S.; Negi, A.S.; Shanker, K. Determination of Anti-Tubercular Agent in Mango Ginger (Curcuma amada Roxb.) by Reverse Phase HPLC-PDA-MS. Food Chem. 2012, 131, 375–379. [Google Scholar] [CrossRef]
- Fabre, H.; Le Bris, A.; Blanchin, M.D. Evaluation of Different Techniques for Peak Purity Assessment on a Diode-Array Detector in Liquid Chromatography. J. Chromatogr. A 1995, 697, 81–88. [Google Scholar] [CrossRef]
- Magnusson, B.; Örnemark, U. Eurachem Guide: The Fitness for Purpose of Analytical Methods: A Laboratory Guide to Method Validation and Related Topics, 2nd ed.; LGC: Teddington, Middlesex, UK, 2014; Available online: http://www.eurachem.org (accessed on 23 November 2020).
- Rha, C.S.; Jung, Y.S.; Lee, J.D.; Jang, D.; Kim, M.S.; Lee, M.S.; Hong, Y.D.; Kim, D.O. Chemometric Analysis of Extracts and Fractions from Green, Oxidized, and Microbial Fermented Teas and Their Correlation to Potential Antioxidant and Anticancer Effects. Antioxidants 2020, 9, 1015. [Google Scholar] [CrossRef]
- Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. A Conversation on Data Mining Strategies in LC-MS Untargeted Metabolomics: Pre-Processing and Pre-Treatment Steps. Metabolites 2016, 6, 40. [Google Scholar] [CrossRef] [Green Version]
- Eriksson, L.; Byrne, T.; Johansson, E.; Trygg, J.; Vikström, C. Multi-and Megavariate Data Analysis Basic Principles and Applications; Umetrics Academy: Umeå, Sweden, 2013; Volume 1. [Google Scholar]
- Wiklund, S.; Johansson, E.; Sjöström, L.; Mellerowicz, E.J.; Edlund, U.; Shockcor, J.P.; Gottfries, J.; Moritz, T.; Trygg, J. Visualization of GC/TOF-MS-Based Metabolomics Data for Identification of Biochemically Interesting Compounds Using OPLS Class Models. Anal. Chem. 2008, 80, 115–122. [Google Scholar] [CrossRef]
- Cohen, J. Things I Have Learned (so far). In Proceedings of the 1990 98th Annual Convention of the American Psychological Association; Presented at the aforementioned conference; American Psychological Association: Boston, MA, USA, 1992. [Google Scholar]
- Toutenburg, H. Models for Categorical Response Variables. In Statistical Analysis of Designed Experiments, 3rd ed.; Toutenburg, H., Ed.; Springer Texts in Statistics; Springer: New York, NY, USA, 2010; pp. 329–393. [Google Scholar]
- Saia, S.; Fragasso, M.; De Vita, P.; Beleggia, R. Metabolomics Provides Valuable Insight for the Study of Durum Wheat: A Review. J. Agric. Food Chem. 2019, 67, 3069–3085. [Google Scholar] [CrossRef]
- Maria John, K.M.; Natarajan, S.; Luthria, D.L. Metabolite Changes in Nine Different Soybean Varieties Grown under Field and Greenhouse Conditions. Food Chem. 2016, 211, 347–355. [Google Scholar] [CrossRef] [Green Version]
- Yordi, E.G.; Koelig, R.; Matos, M.J.; Martínez, A.P.; Caballero, Y.; Santana, L.; Quintana, M.P.; Molina, E.; Uriarte, E. Artificial Intelligence Applied to Flavonoid Data in Food Matrices. Foods 2019, 8, 573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Kang, J.H.; Han, J.M.; Cho, M.H.; Chung, Y.J.; Park, K.H.; Shin, D.H.; Park, H.Y.; Choi, M.S.; Jeong, T.S. Anti-Obesity Effects of Soy Leaf Via Regulation of Adipogenic Transcription Factors and Fat Oxidation in Diet-Induced Obese Mice and 3T3-L1 Adipocytes. J. Med. Food 2015, 18, 899–908. [Google Scholar] [CrossRef]
- Ho, H.M.; Leung, L.K.; Chan, F.L.; Huang, Y.; Chen, Z.Y. Soy Leaf Lowers the Ratio of Non-HDL to HDL Cholesterol in Hamsters. J. Agric. Food Chem. 2003, 51, 4554–4558. [Google Scholar] [CrossRef]
- Han, J.M.; Li, H.; Cho, M.H.; Baek, S.H.; Lee, C.H.; Park, H.Y.; Jeong, T.S. Soy-Leaf Extract Exerts Atheroprotective Effects Via Modulation of Kruppel-Like Factor 2 and Adhesion Molecules. Int. J. Mol. Sci. 2017, 18, 373. [Google Scholar] [CrossRef] [Green Version]




| Compound † | Positive | Negative | Tentative Identification | |||||
|---|---|---|---|---|---|---|---|---|
| MS1 a | Error ppm | MS2 b | MS1 a | Error ppm | MS2 b | Formula, (M) | ||
| CJ1 | 773.2120 | −0.2 | 303.0489, 465.1016, 611.1591, 627.1523 | 771.2016 | −0.1 | 301.0372 | C33H40O21 | quercetin 3-O-tri-glycoside A |
| CJ2 | 773.2133 | 0.1 | 303.0497, 465.1022, 611.1601, 627.1552 | 771.2041 | 0.5 | 301.0374 | C33H40O21 | quercetin 3-O-tri-glycoside B |
| DW1 | 757.2162 | −0.4 | 287.0542, 449.1076, 595.1634 611.1558 | 755.2048 | −0.3 | 285.0400 | C33H40O20 | kaempferol 3-O-tri-glycoside A |
| DW2 | 757.2173 | 0.1 | 287.0539, 449.1071, 595.1638 611.1597 | 755.2040 | −0.5 | 285.0408 | C33H40O20 | kaempferol 3-O-tri-glycoside B |
| NP1 | 627.1535 | 0.3 | 303.0494, 465.1018 | 625.1466 | −0.4 | 301.0372 | C27H30O17 | quercetin 3-O-di-glycoside A |
| NP2 | 627.1539 | −0.6 | 303.0489, 465.1012 | 625.1473 | −0.1 | 301.0365 | C27H30O17 | quercetin 3-O-di-glycoside B |
| WR1 | 741.2218 | −0.4 | 287.0541, 449.1070, 595.1646 | 739.2132 | 0.0 | 285.0331 | C33H40O19 | kaempferol 3-O-tri-glycoside C |
| WR2 | 595.1634 | −0.1 | 287.0553, 449.1067 | 593.1540 | 0.2 | 285.0414 | C27H30O15 | kaempferol 3-O-di-glycoside A |
| WR3 | 595.1628 | −0.3 | 287.0549, 449.1060 | 593.1545 | 0.2 | 285.0416 | C27H30O15 | kaempferol 3-O-di-glycoside B |
| No. a | CL b | Compound | RT c (min) | LOD d (μg/L) | LOQ e (μg/L) | Linearity (R f) | Accuracy (Recovery % ± SEM) g | Precision (RSD; %) h | |
|---|---|---|---|---|---|---|---|---|---|
| Intraday i | Interday j | ||||||||
| 1 | FL | quercetin 3-O-tri-glycoside A | 1.04 ± 0.002 | 8.21 ± 0.71 | 27.35 ± 2.35 | 0.9997 | 102.14 ± 1.24 | 3.84 | 3.17 |
| 2 | FL | quercetin 3-O-tri-glycoside B | 1.09 ± 0.001 | 1.23 ± 0.23 | 4.11 ± 0.77 | 0.9971 | 95.70 ± 2.66 | 8.32 | 5.35 |
| 3 | FL | quercetin 3-O-di-glycoside A | 1.34 ± 0.002 | 5.48 ± 0.57 | 18.26 ± 1.89 | 0.9998 | 93.96 ± 1.59 | 5.35 | 4.83 |
| 4 | FL | quercetin 3-O-di-glycoside B | 1.40 ± 0.002 | 8.12 ± 1.58 | 27.07 ± 5.27 | 0.9995 | 100.92 ± 1.52 | 4.76 | 2.89 |
| 5 | IS | daidzin | 1.45 ± 0.002 | 4.98 ± 0.32 | 16.61 ± 1.07 | 0.9991 | 108.81 ± 0.45 | 1.31 | 3.53 |
| 6 | FL | kaempferol 3-O-tri-glycoside A | 1.48 ± 0.002 | 38.09 ± 1.64 | 126.97 ± 5.46 | 0.9997 | 105.62 ± 0.62 | 1.87 | 6.50 |
| 7 | FL | kaempferol 3-O-tri-glycoside B | 1.62 ± 0.002 | 57.03 ± 2.53 | 190.10 ± 8.44 | 0.9991 | 102.07 ± 2.61 | 8.09 | 8.97 |
| 8 | IS | glycitin | 1.67 ± 0.002 | 17.53 ± 1.41 | 58.43 ± 4.71 | 0.9992 | 104.96 ± 1.40 | 4.23 | 3.44 |
| 9 | FL | kaempferol 3-O-tri-glycoside C | 2.01 ± 0.002 | 8.26 ± 0.97 | 27.55 ± 3.25 | 0.9996 | 103.88 ± 1.07 | 3.26 | 2.50 |
| 10 | FL | rutin | 2.45 ± 0.001 | 5.81 ± 0.40 | 19.38 ± 1.33 | 0.9998 | 101.57 ± 1.00 | 3.11 | 3.35 |
| 11 | FL | isoquercitrin | 2.61 ± 0.001 | 3.58 ± 0.16 | 11.94 ± 0.52 | 0.9997 | 104.43 ± 0.81 | 2.45 | 3.43 |
| 12 | IS | genistin | 2.70 ± 0.000 | 14.76 ± 0.81 | 49.19 ± 2.71 | 0.9995 | 102.51 ± 1.03 | 3.18 | 2.97 |
| 13 | FL | kaempferol 3-O-di-glycoside A | 2.72 ± 0.000 | 2.64 ± 0.13 | 8.80 ± 0.45 | 0.9997 | 100.51 ± 1.12 | 3.51 | 1.49 |
| 14 | FL | kaempferol 3-O-di-glycoside B | 2.91 ± 0.000 | 7.09 ± 0.10 | 23.65 ± 0.33 | 0.9992 | 96.73 ± 1.07 | 3.48 | 4.42 |
| 15 | IS | malonyl daidzin | 2.92 ± 0.000 | 13.12 ± 0.80 | 43.72 ± 2.66 | 0.9997 | 102.90 ± 1.25 | 3.85 | 3.70 |
| 16 | IS | malonyl glycitin | 2.98 ± 0.000 | 5.55 ± 0.52 | 18.49 ± 1.72 | 0.9998 | 100.27 ± 1.18 | 3.71 | 4.65 |
| 17 | FL | astragalin | 3.03 ± 0.000 | 1.57 ± 0.36 | 5.23 ± 1.21 | 0.9995 | 103.24 ± 0.61 | 1.86 | 3.62 |
| 18 | FE | apigenin 7-O-glucoside | 3.15 ± 0.000 | 0.38 ± 0.07 | 1.27 ± 0.23 | 0.9981 | 106.70 ± 1.36 | 4.04 | 2.38 |
| 19 | IS | acetyl daidzin | 3.29 ± 0.000 | 5.45 ± 0.25 | 18.16 ± 0.84 | 0.9993 | 101.76 ± 0.86 | 2.68 | 2.45 |
| 20 | IS | acetyl glycitin | 3.38 ± 0.000 | 2.46 ± 0.03 | 8.20 ± 0.09 | 0.9997 | 102.34 ± 0.51 | 1.56 | 2.15 |
| 21 | IS | malonyl genistin | 3.45 ± 0.000 | 8.52 ± 0.03 | 28.39 ± 0.09 | 1.0000 | 99.12 ± 0.44 | 1.40 | 2.14 |
| 22 | IS | daidzein | 3.79 ± 0.000 | 7.87 ± 0.15 | 26.22 ± 0.50 | 0.9995 | 100.31 ± 0.94 | 2.98 | 2.61 |
| 23 | IS | acetyl genistin | 3.97 ± 0.000 | 12.03 ± 0.18 | 40.10 ± 0.61 | 0.9995 | 100.90 ± 0.95 | 2.99 | 2.76 |
| 24 | IS | glycitein | 4.01 ± 0.000 | 18.12 ± 0.35 | 60.40 ± 1.18 | 0.9995 | 100.45 ± 0.71 | 2.24 | 1.52 |
| 25 | FE | luteolin | 4.19 ± 0.001 | 7.76 ± 0.22 | 25.88 ± 0.75 | 0.9998 | 101.01 ± 0.81 | 2.53 | 2.27 |
| 26 | FL | quercetin | 4.21 ± 0.000 | 9.94 ± 0.38 | 33.14 ± 1.26 | 0.9995 | 101.08 ± 0.67 | 2.09 | 2.30 |
| 27 | IS | genistein | 5.40 ± 0.001 | 45.18 ± 2.02 | 150.59 ± 6.73 | 0.9998 | 100.92 ± 0.43 | 1.35 | 0.70 |
| 28 | FE | apigenin | 5.52 ± 0.001 | 15.20 ± 0.87 | 50.65 ± 2.90 | 0.9958 | 101.30 ± 1.21 | 3.79 | 3.90 |
| 29 | CM | coumestrol | 5.58 ± 0.000 | 2.16 ± 0.19 | 7.19 ± 0.63 | 0.9965 | 105.20 ± 0.60 | 1.80 | 2.24 |
| 30 | FL | kaempferol | 5.65 ± 0.000 | 92.49 ± 10.43 | 308.29 ± 34.78 | 0.9992 | 104.37 ± 0.68 | 2.05 | 3.73 |
| 31 | FL | isorhamnetin | 5.71 ± 0.000 | 35.14 ± 3.14 | 117.12 ± 10.47 | 0.9990 | 108.85 ± 1.19 | 3.45 | 5.41 |
| Class of Data Set a | Compound b | OPLS or OPLS-DA c | BF or BT g | |||
|---|---|---|---|---|---|---|
| p d | p (corr) e | VIP f | G2 h | Portion i | ||
| Day | daidzein | 0.345 | 0.636 | 1.51 | 5.00 | 0.14 |
| genistein | 0.203 | 0.512 | LL j | 4.82 | 0.13 | |
| glycitein | 0.217 | 0.696 | LL | 7.96 | 0.22 | |
| malonyl glycitin | 0.174 | 0.512 | 0.79 | 6.05 | 0.17 | |
| Variety | apigenin | 0.049 | 0.656 | LL | 581.90 | 0.27 |
| luteolin | −0.095 | −0.556 | LL | 959.18 | 0.44 | |
| isoquercitrin | −0.200 | −0.661 | 0.92 | LL | LL | |
| quercetin 3-O-di-glycoside A | −0.500 | −0.688 | 2.31 | LL | LL | |
| quercetin 3-O-di-glycoside B | −0.457 | −0.705 | 2.07 | LL | LL | |
| rutin | −0.386 | −0.703 | 1.77 | LL | LL | |
| quercetin 3-O-tri-glycoside A | LL | LL | 1.02 | 593.46 | 0.27 | |
| City | astragalin | 0.060 | 0.596 | LL | 17.81 | LL |
| kaempferol 3-O-di-glycoside A | 0.221 | 0.633 | 0.94 | 258.71 | 0.12 | |
| malonyl daidzin | −0.290 | −0.519 | 1.41 | LL | LL | |
| quercetin 3-O-tri-glycoside A | −0.340 | −0.597 | 1.37 | 68.08 | LL | |
| quercetin 3-O-tri-glycoside B | −0.448 | −0.705 | 1.88 | 183.69 | 0.08 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rha, C.-S.; Jang, E.K.; Hong, Y.D.; Park, W.S. Supervised Statistical Learning Prediction of Soybean Varieties and Cultivation Sites Using Rapid UPLC-MS Separation, Method Validation, and Targeted Metabolomic Analysis of 31 Phenolic Compounds in the Leaves. Metabolites 2021, 11, 884. https://doi.org/10.3390/metabo11120884
Rha C-S, Jang EK, Hong YD, Park WS. Supervised Statistical Learning Prediction of Soybean Varieties and Cultivation Sites Using Rapid UPLC-MS Separation, Method Validation, and Targeted Metabolomic Analysis of 31 Phenolic Compounds in the Leaves. Metabolites. 2021; 11(12):884. https://doi.org/10.3390/metabo11120884
Chicago/Turabian StyleRha, Chan-Su, Eun Kyu Jang, Yong Deog Hong, and Won Seok Park. 2021. "Supervised Statistical Learning Prediction of Soybean Varieties and Cultivation Sites Using Rapid UPLC-MS Separation, Method Validation, and Targeted Metabolomic Analysis of 31 Phenolic Compounds in the Leaves" Metabolites 11, no. 12: 884. https://doi.org/10.3390/metabo11120884
APA StyleRha, C. -S., Jang, E. K., Hong, Y. D., & Park, W. S. (2021). Supervised Statistical Learning Prediction of Soybean Varieties and Cultivation Sites Using Rapid UPLC-MS Separation, Method Validation, and Targeted Metabolomic Analysis of 31 Phenolic Compounds in the Leaves. Metabolites, 11(12), 884. https://doi.org/10.3390/metabo11120884