Pathway Tools Visualization of Organism-Scale Metabolic Networks
Abstract
:1. Introduction
1.1. Pathway Tools Background
1.2. Past Work on Pathway Tools Metabolic Network Diagrams
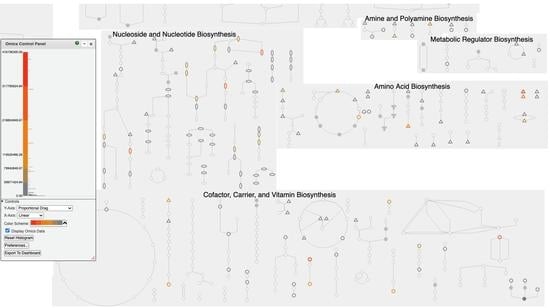
1.3. Organization of Pathway Tools Metabolic Charts
1.4. Creation of Pathway Tools Metabolic Charts
- Query the PGDB to determine the cellular architecture of the organism, meaning what membranes and cellular spaces are present
- Query the PGDB to determine the sets of reactions, proteins, and pathways present in different regions of the diagram, such as the sets of biosynthetic, catabolic, and energy-related pathways
- Lay out each individual pathway diagram using PTools pathway layout algorithms [14]
- Run a layout algorithm that arranges the pathways within each pathway block (such as Amino Acid Biosynthesis)
- Run a layout algorithm that arranges the pathway blocks relative to one another within the biosynthesis, catabolism, and energy-metabolism regions
- Lay out the grids of individual reactions in the cytosol, periplasm (if applicable), and extracellular space
- Arrange the transporters in the cellular membrane(s)
2. Results
2.1. New JavaScript Implementation of Metabolic Charts
2.2. Search and Highlight Operations
2.3. Cell-Type-Specific Metabolic Charts
2.4. Metabolic Charts for Pan-Genomes
2.5. Metabolic Charts for Organism Communities
2.6. Omics Data Analysis with Metabolic Charts
2.7. Integration with Other Modules of Pathway Tools
- SmartTables [25] enable users to generate or upload data such as a list of genes or metabolites to a “knowledge spreadsheet” and then manipulate it in various ways. Entities from one or more SmartTable columns can be painted as sets of highlights onto the metabolic chart for the appropriate organism. SmartTables containing omics data can be painted onto a metabolic chart. Alternatively, a set of highlighted objects in the metabolic chart can be exported to a new SmartTable.
- MetaFlux [26] enables construction of metabolic models using flux-balance analysis. Users can either create their own metabolic models using the desktop PTools or can run existing models within the BioCyc website (e.g., for E. coli). The steady state reaction fluxes computed by a model can be painted onto the metabolic chart for that organism, to visualize the activity levels of different portions of the metabolic network under the modeled conditions.
- A dead-end metabolite analysis tool generates lists of metabolites that are either consumed but not produced, or produced but not consumed by any reaction in an organism’s metabolic network. These metabolites represent either true cellular inputs and outputs, or gaps in the metabolic network corresponding to missing information. The metabolite lists generated by such an analysis can be highlighted on the metabolic chart.
- A chokepoint analysis tool identifies reactions that are potential drug targets because they represent chokepoints in the metabolic network, such that eliminating them would eliminate the only path to or from a particular metabolite. These reactions can be highlighted on a metabolic chart.
- The Pathway Collage Viewer [27] enables generation and editing of personalized multi-pathway diagrams called Pathway Collages. Pathway Collages bridge the gap between the low-level view of a single pathway and the high-level global view of the entire metabolic network by enabling users to select specific pathways of interest to combine into an intermediate-level diagram. Pathway Collages can be customized in various ways, and optionally overlaid with omics data. Users who have used the full metabolic chart to highlight one or more objects of interest can invoke the Pathway Collage Viewer to generate a collage of all pathways that either have themselves been highlighted or for which some component gene, reaction, or metabolite has been highlighted. In addition, when uploading omics data to a metabolic chart, users can choose between displaying data on the full metabolic chart or on a newly generated Pathway Collage consisting of only the N most perturbed pathways.
- The Omics Dashboard [28] is a novel tool for interactively exploring omics data through a hierarchical set of graphs organized around cellular systems. At its highest level, the Omics Dashboard contains panels for high-level systems such as Biosynthesis and Regulation, with plots for each of their subsystems, such as Amino Acid Biosynthesis and Cofactor Biosynthesis, that amalgamate the omics data values for all genes or metabolites in that subsystem. Drilling down on an individual subsystem such as Amino Acid Biosynthesis shows plots for the biosynthesis pathways of each individual amino acid. Users can continue to drill down on subsystems of interest until they reach the level of individual genes or metabolites. The Omics Dashboard offers a different, complementary view of omics data than do metabolic charts. From a metabolic chart painted with omics data, users can click a button to open a new tab with the data displayed on the Omics Dashboard and thus have access to both views simultaneously.
3. Discussion
4. Conclusions
5. Availability and Requirements
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Karp, P.D.; Midford, P.; Billington, R.; Kothari, A.; Krummenacker, M.; Ong, W.; Subhraveti, P.; Caspi, R.; Keseler, I.; Paley, S.M. Pathway Tools version 23.0 update: Software for pathway/genome informatics and systems biology. Brief. Bioinform. 2019, bbz104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karp, P.D.; Midford, P.; Paley, S.; Krummenacker, M.; Billington, R.; Kothari, A.; Ong, W.; Subhraveti, P.; Keseler, I.; Caspi, R. Pathway Tools version 24.0: Integrated software for pathway/genome informatics and systems biology [v4]. arXiv 2020, arXiv:1510.03964v4. [Google Scholar]
- Pathway/Genome Database Websites. Available online: https://BioCyc.org/otherpgdbs.shtml (accessed on 19 January 2021).
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midford, P.E.; Ong, Q.; et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 2017, 20, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Karp, P.; Riley, M.; Paley, S.; Pellegrini-Toole, A.; Krummenacker, M. EcoCyc: Electronic Encyclopedia of E. coli Genes and Metabolism. Nucleic Acids Res. 1997, 25, 43–50. [Google Scholar] [CrossRef] [Green Version]
- Karp, P.; Krummenacker, M.; Paley, S.; Wagg, J. Integrated pathway/genome databases and their role in drug discovery. Trends Biotechnol. 1999, 17, 275–281. [Google Scholar] [CrossRef]
- Karp, P.D. Pathway Databases: A Case Study in Computational Symbolic Theories. Science 2001, 293, 2040–2044. [Google Scholar] [CrossRef] [Green Version]
- Paley, S.M.; Karp, P.D. The Pathway Tools Cellular Overview Diagram and Omics Viewer. Nucleic Acids Res. 2006, 34, 3771–3778. Available online: http://nar.oxfordjournals.org/cgi/content/full/34/13/3771 (accessed on 19 January 2021). [CrossRef] [Green Version]
- Karp, P.D.; Paley, S.M.; Krummenacker, M.; Latendresse, M.; Dale, J.; Lee, T.; Kaipa, P.; Gilham, F.; Spaulding, A.; Popescu, L.; et al. Pathway Tools version 13.0: Integrated Software for Pathway/Genome Informatics and Systems Biology. Brief. Bioinform. 2010, 11, 40–79. [Google Scholar] [CrossRef] [Green Version]
- Karp, P.D.; Latendresse, M.; Paley, S.M. Using Cellular Network Diagrams to Interpret Large-Scale Datasets: Past Progress and Future Challenges. Proc. SPIE Int. Soc. Opt. Eng. 2011, 7865, 9. [Google Scholar] [CrossRef]
- Latendresse, M.; Karp, P.D. Web-Based Metabolic Network Visualization with a Zooming User Interface. BMC Bioinform. 2011, 12, 176–184. Available online: http://www.biomedcentral.com/1471-2105/12/176/abstract (accessed on 19 January 2021). [CrossRef] [Green Version]
- Latendresse, M.; Paley, S.M.; Karp, P.D. Browsing Metabolic and Regulatory Networks with BioCyc. Methods Mol. Biol. 2012, 804, 197–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- EcoCyc Cellular Overview. Available online: https://ecocyc.org/overviewsWeb/celOv.shtml (accessed on 1 December 2020).
- Karp, P.D.; Paley, S.M. Automated Drawing of Metabolic Pathways. In Proceedings Third International Conference on Bioinformatics and Genome Research; Lim, H., Cantor, C., Robbins, R., Eds.; World Scientific Publishing Co.: Singapore, 1994; pp. 225–238. Available online: http://www.ai.sri.com/pub_list/234 (accessed on 19 January 2021).
- HumanCyc Cellular Overview. Available online: https://humancyc.org/overviewsWeb/celOv.shtml (accessed on 1 December 2020).
- Bastian, F.; Parmentier, G.; Roux, J.; Moretti, S.; Laudet, V.; Robinson-Rechavi, M. Bgee: Integrating and Comparing Heterogeneous Transcriptome Data Among Species. In Data Integration in the Life Sciences; Bairoch, A., Cohen-Boulakia, S., Froidevaux, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 124–131. [Google Scholar]
- Pseudomonas Pan-Genome Cellular Overview. Available online: https://biocyc.org/overviewsWeb/celOv.shtml?orgid=PSEUDOMONAS-PANGENOME (accessed on 1 December 2020).
- Wannemuehler, M.J.; Overstreet, A.M.; Ward, D.V.; Phillips, G.J. Draft genome sequences of the altered Schaedler flora, a defined bacterial community from gnotobiotic mice. Genome Announc. 2014, 2, e00287-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Presentation: New Omics Viewer Controls. Available online: https://www.youtube.com/watch?v=6DaDQwLlB38 (accessed on 19 January 2021).
- von Wulffen, J.; Ulmer, A.; Jager, G.; Sawodny, O.; Feuer, R. Rapid Sampling of Escherichia coli After Changing Oxygen Conditions Reveals Transcriptional Dynamics. Genes 2017, 8, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- E. coli Cellular Overview with Animated Display of Gene Expression Dataset. Available online: https://biocyc.org/overviewsWeb/celOv.shtml?omics=t&url=file://expr-examples/ecoli-feuer-toaerobic-significant.txt&orgid=ecoli&zoomlevel=0&column1=1-6&class=gene&expressiontype=absolute (accessed on 19 January 2021).
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets–update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sud, M.; Fahy, E.; Cotter, D.; Azam, K.; Vadivelu, I.; Burant, C.; Edison, A.; Fiehn, O.; Higashi, R.; Sumner, K.S.N.S.; et al. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 2015, 44, D463–D470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paley, S.M.; Latendresse, M.; Karp, P.D. Regulatory Network Operations in the Pathway Tools Software. BMC Bioinform. 2012, 13, 243–256. Available online: http://www.biomedcentral.com/1471-2105/13/243 (accessed on 19 January 2021). [CrossRef] [PubMed] [Green Version]
- Travers, M.; Paley, S.M.; Shrager, J.; Holland, T.; Karp, P.D. Groups: Knowledge spreadsheets for symbolic biocomputing. Database 2013, 1–12. Available online: http://database.oxfordjournals.org/content/2013/bat061.abstract (accessed on 19 January 2021). [CrossRef]
- Latendresse, M.; Krummenacker, M.; Trupp, M.; Karp, P.D. Construction and completion of flux balance models from pathway databases. Bioinformatics 2012, 28, 388–396. Available online: http://bioinformatics.oxfordjournals.org/content/28/3/388 (accessed on 19 January 2021). [CrossRef] [PubMed] [Green Version]
- Paley, S.; O’Maille, P.E.; Weaver, D.; Karp, P.D. Pathway collages: Personalized multi-pathway diagrams. BMC Bioinform. 2016, 17, 529–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paley, S.M.; Parker, K.; Spaulding, A.; Tomb, J.; O’Maille, P.; Karp, P.D. The Omics Dashboard for interactive exploration of gene-expression data. Nucleic Acids Res. 2017, 45, 12113–12124. Available online: https://academic.oup.com/nar/article/45/21/12113/4508872 (accessed on 19 January 2021). [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okuda, S.; Yamada, T.; Hamajima, M.; Itoh, M.; Katayama, T.; Bork, P.; Goto, S.; Kanehisa, M. KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 2008, 36, W423–W426. Available online: http://nar.oxfordjournals.org/content/36/suppl_2/W423.abstract (accessed on 19 January 2021). [CrossRef] [PubMed]
- Yamada, T.; Letunic, I.; Okuda, S.; Kanehisa, M.; Bork, P. iPath2.0: Interactive pathway explorer. Nucleic Acids Res. 2011, 39, W412–W415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, Z.A.; Drager, A.; Ebrahim, A.; Sonnenschein, N.; Lewis, N.E.; Palsson, B.O. Escher: A Web Application for Building, Sharing, and Embedding Data-Rich Visualizations of Biological Pathways. PLoS Comput. Biol. 2015, 11, e1004321. [Google Scholar] [CrossRef] [PubMed] [Green Version]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paley, S.; Billington, R.; Herson, J.; Krummenacker, M.; Karp, P.D. Pathway Tools Visualization of Organism-Scale Metabolic Networks. Metabolites 2021, 11, 64. https://doi.org/10.3390/metabo11020064
Paley S, Billington R, Herson J, Krummenacker M, Karp PD. Pathway Tools Visualization of Organism-Scale Metabolic Networks. Metabolites. 2021; 11(2):64. https://doi.org/10.3390/metabo11020064
Chicago/Turabian StylePaley, Suzanne, Richard Billington, James Herson, Markus Krummenacker, and Peter D. Karp. 2021. "Pathway Tools Visualization of Organism-Scale Metabolic Networks" Metabolites 11, no. 2: 64. https://doi.org/10.3390/metabo11020064
APA StylePaley, S., Billington, R., Herson, J., Krummenacker, M., & Karp, P. D. (2021). Pathway Tools Visualization of Organism-Scale Metabolic Networks. Metabolites, 11(2), 64. https://doi.org/10.3390/metabo11020064