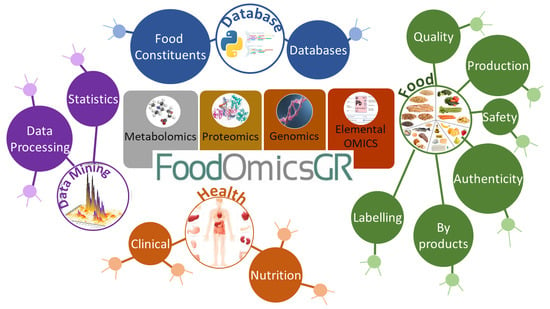
FoodOmicsGR_RI: A Consortium for Comprehensive Molecular Characterisation of Food Products
Abstract
:1. Introduction
2. FoodOmicsGR_RI, Structure
2.1. Governance
2.2. Access to Facility
- -
- Consultancy services and uptake of experimental intervention for foodomics studies (food product -oriented studies) or nutritional studies.
- -
- Big Data Handling and statistical analysis by various modes and special software including bespoke scripts and algorithms.
- -
- Genomic, proteomic and metabolomic analysis of foods, food products and biological samples from nutritional studies.
- -
- Quantitative Determination of key target molecules (nutrients, contaminants or other) in foods and biological samples.
3. FoodOmicsGR_RI Plan and Strategy
3.1. Literature Analysis of the Composition of Greek Foods and Compilation of Greek Food Composition Databases
3.2. Advancing Analytical Capability of the RI
3.3. Advancing and Harmonising the Analytical Portfolio
3.3.1. MS Based-Metabolomics
3.3.2. NMR Based-Metabolomics
3.3.3. Genetic and Genomic Analyses for Traceability of Animal Organisms
3.3.4. Proteomics Analysis
3.3.5. Elemental Metabolomics for Food Authentication
3.4. Sample Banking
3.5. Food Analysis, Traceability and Control of Geographical Origin
3.5.1. LC-MS and GC-MS Technology
3.5.2. NMR Technology
3.5.3. MALDI-TOF-MS Technology
3.5.4. Elemental Metabolomics Technology
3.5.5. Data Combination
3.6. Nutritional Studies, Highlighting the Value of Greek Foods
3.7. Valorization of Agro-Industry by-Products
3.8. Assessment of Food Safety
4. Impact on the National Research Environment
5. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cifuentes, A. Food Analysis: Present, Future, and Foodomics. ISRN Anal. Chem. 2012, 2012, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Valdés, A.; Cifuentes, A.; León, C. Foodomics evaluation of bioactive compounds in foods. Trac. Trends Anal. Chem. 2017, 96, 2–13. [Google Scholar] [CrossRef]
- Andjelković, U.; Šrajer Gajdošik, M.; Gašo-Sokač, D.; Martinović, T.; Josić, D. Foodomics and Food Safety: Where We Are. Food Technol. Biotechnol. 2017, 55, 290–307. [Google Scholar] [CrossRef] [PubMed]
- Putignani, L.; Dallapiccola, B. Foodomics as part of the host-microbiota-exposome interplay. J. Proteom. 2016, 147, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Lioupi, A.; Nenadis, N.; Theodoridis, G. Virgin olive oil metabolomics: A review. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2020, 1150, 122161. [Google Scholar] [CrossRef]
- Dasenaki, M.E.; Drakopoulou, S.K.; Katsianou, P.A.; Martakos, I.; Thomaidis, N.S. Authentication of Olive Products with Liquid Chromatographic techniques. In Chromatographic and Related Separation Techniques in Food Integrity and Authenticity, Volume B: Relevant Applications; Núñez, O., Campmajó, G., Eds.; World Scientific Publishing: London, UK, 2020; Volume B. [Google Scholar]
- Kalogiouri, N.P.; Aalizadeh, R.; Dasenaki, M.E.; Thomaidis, N.S. Application of High Resolution Mass Spectrometric methods coupled with chemometric techniques in olive oil authenticity studies—A review. Anal. Chim. Acta 2020, 1134, 150–173. [Google Scholar] [CrossRef]
- Diamantidou, D.; Zotou, A.; Theodoridis, G. Wine and grape marc spirits metabolomics. Metab. Off. J. Metab. Soc. 2018, 14, 159. [Google Scholar] [CrossRef]
- Dasenaki, M.E.; Drakopoulou, S.K.; Aalizadeh, R.; Thomaidis, N.S. Targeted and Untargeted Metabolomics as an Enhanced Tool for the Detection of Pomegranate Juice Adulteration. Foods 2019, 8, 212. [Google Scholar] [CrossRef] [Green Version]
- Amargianitaki, M.; Spyros, A. NMR-based metabolomics in wine quality control and authentication. Chem. Biol. Technol. Agric. 2017, 4, 9. [Google Scholar] [CrossRef] [Green Version]
- Spyros, A.; Dais, P. NMR Spectroscopy in Food Analysis; RSC food analysis monographs; RSC Publ: Cambridge, UK, 2012; Volume 10, ISBN 978-1-84973-533-9. [Google Scholar]
- Spyros, A. Application of NMR in food analysis. In Nuclear Magnetic Resonance; Royal Society of Chemistry: Cambridge, UK, 2016; pp. 269–308. [Google Scholar] [CrossRef]
- Tang, F.; Vasas, M.; Hatzakis, E.; Spyros, A. Magnetic resonance applications in food analysis. In Annual Reports on NMR Spectroscopy; Elsevier: Amsterdam, The Netherlands, 2019; pp. 239–306. [Google Scholar]
- Voidarou, C.; Tzora, A.; Malamou, O.; Akrida-Demertzi, K.; Demertzis, P.G.; Vassos, D.; Rozos, G.; Alexopoulos, A.; Plessas, S.; Stavropoulou, E.; et al. Chemical and microbiological characterization of artisan inoculants used for the fermentation of traditional dairy products in Epirus area (Greece). Anaerobe 2011, 17, 354–357. [Google Scholar] [CrossRef]
- Tzora, A.; Skoufos, I.; Fthenakis, C.G.; Tsangaris, G.; Bramis, G.; Karamoutsios, A.; Gelasakis, A.; Arsenos, G. Milk quality of indigenous Greek dairy sheep and goats raised under different production systems. In Book of Abstracts of the 65th Annual Meeting of the European Association for Animal Production: Copenhagen, Denmark, 25–28 August 2014; EAAP Book of Abstracts Series; EAAP scientific committee, Ed.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2014; Volume 20, p. 243. ISBN 978-90-8686-248-1. [Google Scholar]
- Skoufos, I.; Metsios, A.; Tzora, A.; Fthenakis, C.G.; Arsenos, G.; Tsangaris, G.; Papadopoulos, G.; Voidarou, C.; Karkabounas, S. In Vitro Studies on Milk Properties of the Indigenous Breed Capra Prisca in Nitric Oxide Stimulation. In Proceedings of the 65th Annual Meeting of the European Federation of Animal Science, Copenhagen, Denmark, 25–29 August 2014; p. 244. [Google Scholar]
- Skoufos, I.; Tzora, A.; Giannenas, I.; Karamoutsios, A.; Levic, J.; Ivanovic, D.; Tsangaris, G.; Fthenakis, G. Comparison of Milk Composition, Fatty Acid Profile and Conjugated Linoleic Acid (CLA) Content in Milk of Indigenous Breeds of Ewes and Goats Reared under a Similar Feeding System in the Region of Epirus. In Proceedings of the 32nd World Veterinary Congress (WVC), Greece, Istanbul, Turkey, 13–17 September 2015; pp. 357–358. [Google Scholar]
- Malissiova, E.; Tzora, A.; Katsioulis, A.; Hatzinikou, M.; Tsakalof, A.; Arvanitoyannis, I.S.; Govaris, A.; Hadjichristodoulou, C. Relationship between production conditions and milk gross composition in ewe’s and goat’s organic and conventional farms in central Greece. Dairy Sci. Technol. 2015, 95, 437–450. [Google Scholar] [CrossRef] [Green Version]
- Anagnostopoulos, A.K.; Katsafadou, A.I.; Pierros, V.; Kontopodis, E.; Fthenakis, G.C.; Arsenos, G.; Karkabounas, S.C.; Tzora, A.; Skoufos, I.; Tsangaris, G.T. Milk of Greek sheep and goat breeds; characterization by means of proteomics. J. Proteom. 2016, 147, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Anagnostopoulos, A.K.; Tsangaris, G.T.; Tzora, A.; Skoufos, I. Molecular Proteomic Characterization of Greek Single–Breed Dairy Products. In Proceedings of the 16th Human Proteome Organisation Word Congress, Dublin, Ireland, 17 September 2017; p. 127. [Google Scholar]
- Skoufos, I.; Tzora, A.; Giannenas, I.; Karamoutsios, A.; Tsangaris, G.; Fthenakis, G.C. Milk quality characteristics of Boutsiko, Frisarta and Karagouniko sheep breeds reared in the mountainous and semimountainous areas of Western and Central Greece. Int. J. Dairy Technol. 2017, 70, 345–353. [Google Scholar] [CrossRef]
- Michailidou, S.; Tsangaris, G.; Fthenakis, G.C.; Tzora, A.; Skoufos, I.; Karkabounas, S.C.; Banos, G.; Argiriou, A.; Arsenos, G. Genomic diversity and population structure of three autochthonous Greek sheep breeds assessed with genome-wide DNA arrays. Mol. Genet. Genom. 2018, 293, 753–768. [Google Scholar] [CrossRef]
- Michailidou, S.; Tsangaris, G.T.; Tzora, A.; Skoufos, I.; Banos, G.; Argiriou, A.; Arsenos, G. Analysis of genome-wide DNA arrays reveals the genomic population structure and diversity in autochthonous Greek goat breeds. PLoS ONE 2019, 14, e0226179. [Google Scholar] [CrossRef] [Green Version]
- Imsiridou, A.; Papapetrou, M.; Tilikidis, A.; Loukovitis, D.; Minos, G.; Gouva, E.; Chatzopoulos, A.; Skoufos, I.; Paschos, I. Can the Population Structure of Three Greek Marine Species (Sardina pilchardus, Penaeus kerathurus, Mullus barbatus) Become a Tool for Their Future Characterization as PGI Products? J. Nutr. Food Lipid Sci. 2019, 2019. [Google Scholar] [CrossRef]
- Gika, H.; Virgiliou, C.; Theodoridis, G.; Plumb, R.S.; Wilson, I.D. Untargeted LC/MS-based metabolic phenotyping (metabonomics/metabolomics): The state of the art. J. Chromatogr. B 2019, 1117, 136–147. [Google Scholar] [CrossRef]
- Gika, H.G.; Wilson, I.D.; Theodoridis, G.A. LC–MS-based holistic metabolic profiling. Problems, limitations, advantages, and future perspectives. J. Chromatogr. B 2014, 966, 1–6. [Google Scholar] [CrossRef]
- Kalogiouri, N.P.; Alygizakis, N.A.; Aalizadeh, R.; Thomaidis, N.S. Olive oil authenticity studies by target and nontarget LC-QTOF-MS combined with advanced chemometric techniques. Anal. Bioanal. Chem. 2016, 408, 7955–7970. [Google Scholar] [CrossRef]
- Kalogiouri, N.P.; Aalizadeh, R.; Thomaidis, N.S. Investigating the organic and conventional production type of olive oil with target and suspect screening by LC-QTOF-MS, a novel semi-quantification method using chemical similarity and advanced chemometrics. Anal. Bioanal. Chem. 2017, 409, 5413–5426. [Google Scholar] [CrossRef]
- Kalogiouri, N.P.; Aalizadeh, R.; Thomaidis, N.S. Application of an advanced and wide scope non-target screening workflow with LC-ESI-QTOF-MS and chemometrics for the classification of the Greek olive oil varieties. Food Chem. 2018, 256, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Kalogiouri, N.P.; Aalizadeh, R.; Dasenaki, M.E.; Thomaidis, N.S. Authentication of Greek PDO Kalamata Table Olives: A Novel Non-Target High Resolution Mass Spectrometric Approach. Molecules 2020, 25, 2919. [Google Scholar] [CrossRef] [PubMed]
- Aalizadeh, R.; Thomaidis, N.S.; Schymanski, E.L. AutoSuspect: An R package to Perform Automatic Suspect Screening based on Regulatory Databases. In Proceedings of the 15th International Conference on Environmental Science and Technology, Rhodes, Greece, 31 August–2 September 2017; Lekkas, D.F., Ed.; Global NEST Journal: Athens, Greece, 2017. [Google Scholar]
- Aalizadeh, R.; Nika, M.-C.; Thomaidis, N.S. Development and application of retention time prediction models in the suspect and non-target screening of emerging contaminants. J. Hazard. Mater. 2019, 363, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Nika, M.C.; Aalizadeh, R.; Thomaidis, N.S. Removal and Transformation of Citalopram and Four of its Biotransformation Products during Ozonation Experiments. In Proceedings of 15th International Conference on Environmental Science and Technology, Rhodes, Greece, 31 August–2 September 2017; Lekkas, D.F., Ed.; Global NEST Journal: Athens, Greece, 2017. [Google Scholar]
- Alygizakis, N.A.; Gago-Ferrero, P.; Hollender, J.; Thomaidis, N.S. Untargeted time-pattern analysis of LC-HRMS data to detect spills and compounds with high fluctuation in influent wastewater. J. Hazard. Mater. 2019, 361, 19–29. [Google Scholar] [CrossRef]
- Aalizadeh, R.; Thomaidis, N.S.; Bletsou, A.A.; Gago-Ferrero, P. Quantitative Structure-Retention Relationship Models To Support Nontarget High-Resolution Mass Spectrometric Screening of Emerging Contaminants in Environmental Samples. J. Chem. Inf. Modeling 2016, 56, 1384–1398. [Google Scholar] [CrossRef]
- Begou, O.; Gika, H.G.; Theodoridis, G.A.; Wilson, I.D. Quality Control and Validation Issues in LC-MS Metabolomics. Methods Mol. Biol. 2018, 1738, 15–26. [Google Scholar] [CrossRef]
- Gika, H.G.; Theodoridis, G.A.; Wingate, J.E.; Wilson, I.D. Within-day reproducibility of an HPLC-MS-based method for metabonomic analysis: Application to human urine. J. Proteome Res. 2007, 6, 3291–3303. [Google Scholar] [CrossRef]
- Gika, H.G.; Theodoridis, G.A.; Earll, M.; Wilson, I.D. A QC approach to the determination of day-to-day reproducibility and robustness of LC-MS methods for global metabolite profiling in metabonomics/metabolomics. Bioanalysis 2012, 4, 2239–2247. [Google Scholar] [CrossRef]
- Gika, H.G.; Zisi, C.; Theodoridis, G.; Wilson, I.D. Protocol for quality control in metabolic profiling of biological fluids by U(H)PLC-MS. J. Chromatogr. B 2016, 1008, 15–25. [Google Scholar] [CrossRef]
- Lai, L.; Michopoulos, F.; Gika, H.; Theodoridis, G.; Wilkinson, R.W.; Odedra, R.; Wingate, J.; Bonner, R.; Tate, S.; Wilson, I.D. Methodological considerations in the development of HPLC-MS methods for the analysis of rodent plasma for metabonomic studies. Mol. Biosyst. 2009, 6, 108–120. [Google Scholar] [CrossRef]
- Want, E.J.; Wilson, I.D.; Gika, H.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Holmes, E.; Nicholson, J.K. Global metabolic profiling procedures for urine using UPLC-MS. Nat. Protoc. 2010, 5, 1005–1018. [Google Scholar] [CrossRef] [PubMed]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2013, 8, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Deda, O.; Gika, H.G.; Wilson, I.D.; Theodoridis, G.A. An overview of fecal sample preparation for global metabolic profiling. J. Pharm. Biomed. Anal. 2015, 113, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Deda, O.; Chatziioannou, A.C.; Fasoula, S.; Palachanis, D.; Raikos, Ν.; Theodoridis, G.A.; Gika, H.G. Sample preparation optimization in fecal metabolic profiling. J. Chromatogr. B 2017, 1047, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Gika, H.; Theodoridis, G. Sample preparation prior to the LC–MS-based metabolomics/metabonomics of blood-derived samples. Bioanalysis 2011, 3, 1647–1661. [Google Scholar] [CrossRef]
- Tsakelidou, E.; Virgiliou, C.; Valianou, L.; Gika, H.; Raikos, N.; Theodoridis, G. Sample Preparation Strategies for the Effective Quantitation of Hydrophilic Metabolites in Serum by Multi-Targeted HILIC-MS/MS. Metabolites 2017, 7, 13. [Google Scholar] [CrossRef] [Green Version]
- Moros, G.; Chatziioannou, A.C.; Gika, H.G.; Raikos, N.; Theodoridis, G. Investigation of the derivatization conditions for GC–MS metabolomics of biological samples. Bioanalysis 2017, 9, 53–65. [Google Scholar] [CrossRef]
- Theodoridis, G.; Gika, H.; Franceschi, P.; Caputi, L.; Arapitsas, P.; Scholz, M.; Masuero, D.; Wehrens, R.; Vrhovsek, U.; Mattivi, F. LC-MS based global metabolite profiling of grapes: Solvent extraction protocol optimisation. Metabolomics 2012, 8, 175–185. [Google Scholar] [CrossRef]
- Begou, O.; Gika, H.G.; Wilson, I.D.; Theodoridis, G. Hyphenated MS-based targeted approaches in metabolomics. Analyst 2017, 142, 3079–3100. [Google Scholar] [CrossRef]
- Tsochatzis, E.D.; Begou, O.; Gika, H.G.; Karayannakidis, P.D.; Kalogiannis, S. A hydrophilic interaction chromatography-tandem mass spectrometry method for amino acid profiling in mussels. J. Chromatogr. B 2017, 1047, 197–206. [Google Scholar] [CrossRef]
- Sarafian, M.H.; Lewis, M.R.; Pechlivanis, A.; Ralphs, S.; McPhail, M.J.W.; Patel, V.C.; Dumas, M.-E.; Holmes, E.; Nicholson, J.K. Bile Acid Profiling and Quantification in Biofluids Using Ultra-Performance Liquid Chromatography Tandem Mass Spectrometry. Anal. Chem. 2015, 87, 9662–9670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Virgiliou, C.; Sampsonidis, I.; Gika, H.G.; Raikos, N.; Theodoridis, G.A. Development and validation of a HILIC-MS/MS multitargeted method for metabolomics applications. Electrophoresis 2015, 36, 2215–2225. [Google Scholar] [CrossRef] [PubMed]
- Pina, A.; Begou, O.; Kanelis, D.; Gika, H.; Kalogiannis, S.; Tananaki, C.; Theodoridis, G.; Zotou, A. Targeted profiling of hydrophilic constituents of royal jelly by hydrophilic interaction liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 2018, 1531, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Virgiliou, C.; Kanelis, D.; Pina, A.; Gika, H.; Tananaki, C.; Zotou, A.; Theodoridis, G. A targeted approach for studying the effect of sugar bee feeding on the metabolic profile of Royal Jelly. J. Chromatogr. A 2020, 1616, 460783. [Google Scholar] [CrossRef]
- Arapitsas, P.; Della Corte, A.; Gika, H.; Narduzzi, L.; Mattivi, F.; Theodoridis, G. Studying the effect of storage conditions on the metabolite content of red wine using HILIC LC-MS based metabolomics. Food Chem. 2016, 197 Pt B, 1331–1340. [Google Scholar] [CrossRef]
- Gika, H.; Mattivi, F.; Vrhovsek, U.; Theodoridis, G. Hydrophilic interaction ultra performance liquid chromatography retention prediction under gradient elution. Anal. Bioanal. Chem. 2012, 404, 701–709. [Google Scholar] [CrossRef]
- Gika, H.G.; Theodoridis, G.A.; Vrhovsek, U.; Mattivi, F. Quantitative profiling of polar primary metabolites using hydrophilic interaction ultrahigh performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 2012, 1259, 121–127. [Google Scholar] [CrossRef]
- Spyros, A.; Dais, P. Application of (31)P NMR spectroscopy in food analysis. 1. Quantitative determination of the mono- and diglyceride composition of olive oils. J. Agric. Food Chem. 2000, 48, 802–805. [Google Scholar] [CrossRef]
- Fronimaki, P.; Spyros, A.; Christophoridou, S.; Dais, P. Determination of the diglyceride content in greek virgin olive oils and some commercial olive oils by employing (31)P NMR spectroscopy. J. Agric. Food Chem. 2002, 50, 2207–2213. [Google Scholar] [CrossRef]
- Vigli, G.; Philippidis, A.; Spyros, A.; Dais, P. Classification of edible oils by employing 31P and 1H NMR spectroscopy in combination with multivariate statistical analysis. A proposal for the detection of seed oil adulteration in virgin olive oils. J. Agric. Food Chem. 2003, 51, 5715–5722. [Google Scholar] [CrossRef]
- Fragaki, G.; Spyros, A.; Siragakis, G.; Salivaras, E.; Dais, P. Detection of extra virgin olive oil adulteration with lampante olive oil and refined olive oil using nuclear magnetic resonance spectroscopy and multivariate statistical analysis. J. Agric. Food Chem. 2005, 53, 2810–2816. [Google Scholar] [CrossRef] [PubMed]
- Beteinakis, S.; Papachristodoulou, A.; Gogou, G.; Katsikis, S.; Mikros, E.; Halabalaki, M. NMR-Based Metabolic Profiling of Edible Olives—Determination of Quality Parameters. Molecules 2020, 25, 3339. [Google Scholar] [CrossRef] [PubMed]
- Ralli, E.; Amargianitaki, M.; Manolopoulou, E.; Misiak, M.; Markakis, G.; Tachtalidou, S.; Kolesnikova, A.; Dais, P.; Spyros, A. NMR Spectroscopy Protocols for Food Metabolomics Applications. Methods Mol. Biol. 2018, 1738, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Halabalaki, M.; Vougogiannopoulou, K.; Mikros, E.; Skaltsounis, A.L. Recent advances and new strategies in the NMR-based identification of natural products. Curr. Opin. Biotechnol. 2014, 25, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Angelis, A.; Antoniadi, L.; Stathopoulos, P.; Halabalaki, M.; Skaltsounis, L.A. Oleocanthalic and Oleaceinic acids: New compounds from Extra Virgin Olive Oil (EVOO). Phytochem. Lett. 2018, 26, 190–194. [Google Scholar] [CrossRef]
- Karkoula, E.; Angelis, A.; Koulakiotis, N.-S.; Gikas, E.; Halabalaki, M.; Tsarbopoulos, A.; Skaltsounis, A.-L. Rapid isolation and characterization of crocins, picrocrocin, and crocetin from saffron using centrifugal partition chromatography and LC-MS. J. Sep. Sci. 2018, 41, 4105–4114. [Google Scholar] [CrossRef]
- Angelis, A.; Urbain, A.; Halabalaki, M.; Aligiannis, N.; Skaltsounis, A.-L. One-step isolation of oryzanol from rice bran oil by non-aqueous hydrostatic countercurrent chromatography. J. Sep. Sci. 2011, 34, 2528–2537. [Google Scholar] [CrossRef]
- Michailidis, D.; Angelis, A.; Aligiannis, N.; Mitakou, S.; Skaltsounis, L. Recovery of Sesamin, Sesamolin, and Minor Lignans From Sesame Oil Using Solid Support-Free Liquid–Liquid Extraction and Chromatography Techniques and Evaluation of Their Enzymatic Inhibition Properties. Front. Pharmacol. 2019, 10. [Google Scholar] [CrossRef]
- Brieudes, V.; Angelis, A.; Vougogiannopoulou, K.; Pratsinis, H.; Kletsas, D.; Mitakou, S.; Halabalaki, M.; Skaltsounis, L. Phytochemical Analysis and Antioxidant Potential of the Phytonutrient-Rich Decoction of Cichorium spinosum and C. intybus. Planta Med. 2016, 82, 1070–1078. [Google Scholar] [CrossRef] [Green Version]
- Mackie, I.M.; Pryde, S.E.; Gonzales-Sotelo, C.; Medina, I.; Peréz-Martín, R.; Quinteiro, J.; Rey-Mendez, M.; Rehbein, H. Challenges in the identification of species of canned fish. Trends Food Sci. Technol. 1999, 10, 9–14. [Google Scholar] [CrossRef]
- Cunningham, E.P.; Meghen, C.M. Biological identification systems: Genetic markers. Rev. Sci. Et Tech. (Int. Off. Epizoot.) 2001, 20, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Mariani, P.; Panzitta, F.; Nardelli Costa, J.; Lazzari, B.; Crepaldi, P.; Marilli, M.; Fornarelli, F.; Fusi, M.; Milanesi, E.; Negrini, R.; et al. Metodi molecolari per la tracciabilità dei prodotti di origine animale. In Proceedings of the 4th World Italian Beef Cattle Congress, Perugia, Italy, 29 April–1 May 2005; pp. 297–302. [Google Scholar]
- Minoudi, S.; Karaiskou, N.; Avgeris, M.; Gkagkavouzis, K.; Tarantili, P.; Triantafyllidou, D.; Palilis, L.; Avramopoulou, V.; Tsikliras, A.; Barmperis, K.; et al. Seafood mislabeling in Greek market using DNA barcoding. Food Control 2020, 113, 107213. [Google Scholar] [CrossRef]
- Siametis, A.; Gkagkavouzis, A.; Kavakiotis, I.; Karaiskou, N.; Triantafyllidis, A. Genetic Discrimination of Wild and Farmed EUROPEAN Sea Bass (Dicentrarchus labrax) in Greek Waters. In Proceedings of the 16th Hellenic Conference of Ichthyologists, Kavala, Greece, 6–9 October 2016; pp. 453–457. [Google Scholar]
- Gkagkavouzis, K.; Ogden, R.; Murray-Dickson, G.; Maroso, F.; Karaiskou, N.; Triantafyllidis, A.; Taylor, M. Targeted SNP Genotyping Enables Identification of Wild and Farmed Greek Sea Bream (Sparus aurata). In Proceedings of the International Symposium on Genomics in Aquaculture, Athens, Greece, 20–22 April 2016; p. 54. [Google Scholar]
- Tsartsianidou, V.; Banos, G.; Basdagianni, Z.; Chatziplis, D.; Kapsona, V.; Sánchez-Molano, E.; Gkagkavouzis, K.; Karaiskou, N.; Arsenos, G.; Triantafyllidis, A. Phenotypic and Genetic Characterization of Dairy Sheep Production Resilience to Climate Fluctuations. In Proceedings of the 71st Annual Meeting of the European Federation of Animal Science, 1–4 December 2020; p. 572. [Google Scholar]
- Tomazou, M.; Oulas, A.; Anagnostopoulos, A.K.; Tsangaris, G.T.; Spyrou, G.M. In Silico Identification of Antimicrobial Peptides in the Proteomes of Goat and Sheep Milk and Feta Cheese. Proteomes 2019, 7, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anagnostopoulos, A.K.; Stravopodis, D.J.; Tsangaris, G.T. Yield of 6,000 proteins by 1D nLC-MS/MS without pre-fractionation. J. Chromatogr. BAnal. Technol. Biomed. Life Sci. 2017, 1047, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Zdolec, N.; Lorenzo, J.M.; Ray, R.C. Use of Microbes for Improving Food Safety and Quality. Biomed Res. Int. 2018, 2018, 3902698. [Google Scholar] [CrossRef] [Green Version]
- Rozos, G.; Voidarou, C.; Stavropoulou, E.; Skoufos, I.; Tzora, A.; Alexopoulos, A.; Bezirtzoglou, E. Biodiversity and Microbial Resistance of Lactobacilli Isolated From the Traditional Greek Cheese Kopanisti. Front. Microbiol. 2018, 9, 517. [Google Scholar] [CrossRef]
- Zhang, P.; Georgiou, C.A.; Brusic, V. Elemental metabolomics. Brief. Bioinform. 2017, bbw131. [Google Scholar] [CrossRef]
- Georgiou, C.A.; Danezis, G.P. Elemental and Isotopic Mass Spectrometry. In Advanced Mass Spectrometry for Food Safety and Quality, Comprehensive analytical chemistry; Picó, Y., Ed.; Elsevier: Amsterdam, The Netherlands, 2015; Volume 68, ISBN 978-0-444-63340-8. [Google Scholar]
- Danezis, G.P.; Georgiou, C.A. Elemental Metabolomics for food authentication. In Food Quality, Traceability and Foodomics; Cozzolino, D., Cifuentes, A., Eds.; Comprehensive Foodomics; Elsevier: Amsterdam, The Netherlands, 2020; ISBN 9780128163962. (In Press). [Google Scholar]
- Drivelos, S.A.; Danezis, G.P.; Halagarda, M.; Popek, S.; Georgiou, C.A. Geographical origin and botanical type honey authentication through elemental metabolomics via chemometrics. Food Chem. 2020, 338, 127936. [Google Scholar] [CrossRef]
- Danezis, G.P.; Pappas, A.C.; Tsiplakou, E.; Pappa, E.C.; Zacharioudaki, M.; Tsagkaris, A.S.; Papachristidis, C.A.; Sotirakoglou, K.; Zervas, G.; Georgiou, C.A. Authentication of Greek Protected Designation of Origin cheeses through elemental metabolomics. Int. Dairy J. 2020, 104, 104599. [Google Scholar] [CrossRef]
- Pappas, A.C.; Zoidis, E.; Goliomytis, M.; Simitzis, P.E.; Sotirakoglou, K.; Charismiadou, M.A.; Nikitas, C.; Danezis, G.; Deligeorgis, S.G.; Georgiou, C.A. Elemental Metabolomics: Modulation of Egg Metallome with Flavonoids, an Exploratory Study. Antioxidants 2019, 8, 361. [Google Scholar] [CrossRef] [Green Version]
- Danezis, G.; Theodorou, C.; Massouras, T.; Zoidis, E.; Hadjigeorgiou, I.; Georgiou, C.A. Greek Graviera Cheese Assessment through Elemental Metabolomics-Implications for Authentication, Safety and Nutrition. Molecules 2019, 24, 670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danezis, G.P.; Pappas, A.C.; Zoidis, E.; Papadomichelakis, G.; Hadjigeorgiou, I.; Zhang, P.; Brusic, V.; Georgiou, C.A. Game meat authentication through rare earth elements fingerprinting. Anal. Chim. Acta 2017, 991, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Drivelos, S.A.; Danezis, G.P.; Haroutounian, S.A.; Georgiou, C.A. Rare earth elements minimal harvest year variation facilitates robust geographical origin discrimination: The case of PDO “Fava Santorinis”. Food Chem. 2016, 213, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Drivelos, S.A.; Higgins, K.; Kalivas, J.H.; Haroutounian, S.A.; Georgiou, C.A. Data fusion for food authentication. Combining rare earth elements and trace metals to discriminate “Fava Santorinis” from other yellow split peas using chemometric tools. Food Chem. 2014, 165, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Farmaki, E.G.; Thomaidis, N.S.; Minioti, K.S.; Ioannou, E.; Georgiou, C.A.; Efstathiou, C.E. Geographical Characterization of Greek Olive Oils Using Rare Earth Elements Content and Supervised Chemometric Techniques. Anal. Lett. 2012, 45, 920–932. [Google Scholar] [CrossRef]
- Voidarou, C.; Alexopoulos, A.; Tsinas, A.; Rozos, G.; Tzora, A.; Skoufos, I.; Vartzakas, T.; Bezirtzoglou, E. Lactic acid bacteria and Bifidobacteria isolated from honeycombs, as bioprotective agents against pathogens and spoilage microorganisms in fresh fruits and vegetables. Appl. Sci. 2020, 10, 7309. [Google Scholar] [CrossRef]
- Tzora, A.; Bonos, E.; Fotou, K.; Karamoutsios, A.; Barka, E.; Gouva, E.; Skoufos, I. Determination of Bacterial Communities of Four Greek Feta Cheeses Using 16S rRNA Genome Sequencing, Presented at 34th International EFFoST 2020, Israel (Online). 2020. Available online: http://www.effostconference.com/resources/updateable/pdf/EFFOST2020%20programme_06112020.pdf (accessed on 15 November 2020).
- Tzora, A.; Bonos, E.; Sidiropoulou, E.; Grigoriadou, K.; Fotou, K.; Karamoutsios, A.; Basdagianni, Z.; Karaiskou, C.; Giannenas, I.; Skoufos, I. Effect of Dietary Polyunsaturated Fatty Acids Enrichment on the Chemical and Microbiological Composition of Kefalograviera Cheese. Israel (Online), 2020. [Google Scholar]
- Paraschos, S.; Magiatis, P.; Gikas, E.; Smyrnioudis, I.; Skaltsounis, A.-L. Quality profile determination of Chios mastic gum essential oil and detection of adulteration in mastic oil products with the application of chiral and non-chiral GC-MS analysis. Fitoterapia 2016, 114, 12–17. [Google Scholar] [CrossRef]
- Nikou, T.; Witt, M.; Stathopoulos, P.; Barsch, A.; Halabalaki, M. Olive Oil Quality and Authenticity Assessment Aspects Employing FIA-MRMS and LC-Orbitrap MS Metabolomic Approaches. Front. Public Health 2020, 8. [Google Scholar] [CrossRef]
- Aprea, E.; Gika, H.; Carlin, S.; Theodoridis, G.; Vrhovsek, U.; Mattivi, F. Metabolite profiling on apple volatile content based on solid phase microextraction and gas-chromatography time of flight mass spectrometry. J. Chromatogr. A 2011, 1218, 4517–4524. [Google Scholar] [CrossRef]
- Anagnostopoulos, A.K.; Tsangaris, G.T. Feta cheese proteins: Manifesting the identity of Greece’s National Treasure. Data Brief 2018, 19, 2037–2040. [Google Scholar] [CrossRef]
- Mikropoulou, E.; Vougogiannopoulou, K.; Kalpoutzakis, E.; Sklirou, A.; Skaperda, Z.; Houriet, J.; Wolfender, J.-L.; Trougakos, I.; Kouretas, D.; Halabalaki, M.; et al. Phytochemical Composition of the Decoctions of Greek Edible Greens (Chórta) and Evaluation of Antioxidant and Cytotoxic Properties. Molecules 2018, 23, 1541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xynos, N.; Termentzi, A.; Fokialakis, N.; Skaltsounis, L.A.; Aligiannis, N. Supercritical CO2 extraction of mastic gum and chemical characterization of bioactive fractions using LC-HRMS/MS and GC–MS. J. Supercrit. Fluids 2018, 133, 349–356. [Google Scholar] [CrossRef]
- Axiotis, E.; Petrakis, E.A.; Halabalaki, M.; Mitakou, S. Phytochemical Profile and Biological Activity of Endemic Sideritis sipylea Boiss. in North Aegean Greek Islands. Molecules 2020, 25, 2022. [Google Scholar] [CrossRef] [PubMed]
- Tuenter, E.; Sakavitsi, M.E.; Rivera-Mondragón, A.; Hermans, N.; Foubert, K.; Halabalaki, M.; Pieters, L. Ruby chocolate: A study of its phytochemical composition and quantitative comparison with dark, milk and white chocolate. Food Chem. 2020, 128446. [Google Scholar] [CrossRef]
- Termentzi, A.; Halabalaki, M.; Skaltsounis, A.L. From Drupes to Olive Oil: An Exploration of Olive Key Metabolites. In Olive and Olive Oil Bioactive Constituents; Elsevier: Amsterdam, The Netherlands, 2015; pp. 147–177. [Google Scholar]
- Kazalaki, A.; Misiak, M.; Spyros, A.; Dais, P. Identification and quantitative determination of carbohydrate molecules in Greek honey by employing 13C NMR spectroscopy. Anal. Methods 2015, 7, 5962–5972. [Google Scholar] [CrossRef]
- Spyros, A. Phosphorus Derivatization as a Tool to Enhance Specificity of Quantitative NMR Analysis of Foods. In Modern Magnetic Resonance; Webb, G., Ed.; Springer: Cham, Switzerland, 2017; pp. 1–9. [Google Scholar] [CrossRef]
- Petrakis, P.V.; Agiomyrgianaki, A.; Christophoridou, S.; Spyros, A.; Dais, P. Geographical characterization of greek virgin olive oils (cv. Koroneiki) using 1H and 31P NMR fingerprinting with canonical discriminant analysis and classification binary trees. J. Agric. Food Chem. 2008, 56, 3200–3207. [Google Scholar] [CrossRef]
- Anastasiadi, M.; Zira, A.; Magiatis, P.; Haroutounian, S.A.; Skaltsounis, A.L.; Mikros, E. 1H NMR-based metabonomics for the classification of Greek wines according to variety, region, and vintage. Comparison with HPLC data. J. Agric. Food Chem. 2009, 57, 11067–11074. [Google Scholar] [CrossRef]
- Fotou, K.; Tzora, A.; Voidarou, C.; Alexopoulos, A.; Plessas, S.; Avgeris, I.; Bezirtzoglou, E.; Akrida-Demertzi, K.; Demertzis, P.G. Isolation of microbial pathogens of subclinical mastitis from raw sheep’s milk of Epirus (Greece) and their role in its hygiene. Anaerobe 2011, 17, 315–319. [Google Scholar] [CrossRef]
- Alispahic, M.; Hummel, K.; Jandreski-Cvetkovic, D.; Nöbauer, K.; Razzazi-Fazeli, E.; Hess, M.; Hess, C. Species-specific identification and differentiation of Arcobacter, Helicobacter and Campylobacter by full-spectral matrix-associated laser desorption/ionization time of flight mass spectrometry analysis. J. Med. Microbiol. 2010, 59, 295–301. [Google Scholar] [CrossRef] [Green Version]
- Ndoye, B.; Rasolofo, E.A.; LaPointe, G.; Roy, D. A review of the molecular approaches to investigate the diversity and activity of cheese microbiota. Dairy Sci. Technol. 2011, 91, 495–524. [Google Scholar] [CrossRef] [Green Version]
- Foddai, A.C.G.; Grant, I.R. Methods for detection of viable foodborne pathogens: Current state-of-art and future prospects. Appl. Microbiol. Biotechnol. 2020, 104, 4281–4288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsangaris, G.T. Trophometry: A New Approach for Studying Food. In Proceedings of the XIII Annual Conference of Italian Proteomics Association, Como, Italy, 5–7 September 2018; p. 18. [Google Scholar]
- Gika, H.G.; Macpherson, E.; Theodoridis, G.A.; Wilson, I.D. Evaluation of the repeatability of ultra-performance liquid chromatography-TOF-MS for global metabolic profiling of human urine samples. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2008, 871, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Michopoulos, F.; Lai, L.; Gika, H.; Theodoridis, G.; Wilson, I. UPLC-MS-based analysis of human plasma for metabonomics using solvent precipitation or solid phase extraction. J. Proteome Res. 2009, 8, 2114–2121. [Google Scholar] [CrossRef] [PubMed]
- Gika, H.G.; Ji, C.; Theodoridis, G.A.; Michopoulos, F.; Kaplowitz, N.; Wilson, I.D. Investigation of chronic alcohol consumption in rodents via ultra-high-performance liquid chromatography–mass spectrometry based metabolite profiling. J. Chromatogr. A 2012, 1259, 128–137. [Google Scholar] [CrossRef] [Green Version]
- Loftus, N.; Barnes, A.; Ashton, S.; Michopoulos, F.; Theodoridis, G.; Wilson, I.; Ji, C.; Kaplowitz, N. Metabonomic investigation of liver profiles of nonpolar metabolites obtained from alcohol-dosed rats and mice using high mass accuracy MSn analysis. J. Proteome Res. 2011, 10, 705–713. [Google Scholar] [CrossRef] [Green Version]
- Begou, O.; Deda, O.; Agapiou, A.; Taitzoglou, I.; Gika, H.; Theodoridis, G. Urine and fecal samples targeted metabolomics of carobs treated rats. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2019, 1114–1115, 76–85. [Google Scholar] [CrossRef]
- Spagou, K.; Theodoridis, G.; Wilson, I.; Raikos, N.; Greaves, P.; Edwards, R.; Nolan, B.; Klapa, M.I. A GC-MS metabolic profiling study of plasma samples from mice on low- and high-fat diets. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2011, 879, 1467–1475. [Google Scholar] [CrossRef]
- Gika, H.G.; Theodoridis, G.A.; Wilson, I.D. Hydrophilic interaction and reversed-phase ultra-performance liquid chromatography TOF-MS for metabonomic analysis of Zucker rat urine. J. Sep. Sci. 2008, 31, 1598–1608. [Google Scholar] [CrossRef]
- Peroulis, N.; Androutsopoulos, V.P.; Notas, G.; Koinaki, S.; Giakoumaki, E.; Spyros, A.; Manolopoulou, E.; Kargaki, S.; Tzardi, M.; Moustou, E.; et al. Significant metabolic improvement by a water extract of olives: Animal and human evidence. Eur. J. Nutr. 2019, 58, 2545–2560. [Google Scholar] [CrossRef]
- Andreadou, I.; Iliodromitis, E.K.; Mikros, E.; Constantinou, M.; Agalias, A.; Magiatis, P.; Skaltsounis, A.L.; Kamber, E.; Tsantili-Kakoulidou, A.; Kremastinos, D.T. The olive constituent oleuropein exhibits anti-ischemic, antioxidative, and hypolipidemic effects in anesthetized rabbits. J. Nutr. 2006, 136, 2213–2219. [Google Scholar] [CrossRef] [Green Version]
- Andreadou, I.; Papaefthimiou, M.; Zira, A.; Constantinou, M.; Sigala, F.; Skaltsounis, A.-L.; Tsantili-Kakoulidou, A.; Iliodromitis, E.K.; Kremastinos, D.T.; Mikros, E. Metabonomic identification of novel biomarkers in doxorubicin cardiotoxicity and protective effect of the natural antioxidant oleuropein. Nmr Biomed. 2009, 22, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Andreadou, I.; Iliodromitis, E.K.; Souridis, V.; Prokovas, E.; Kostidis, S.; Zoga, A.; Dagres, N.; Tsantili-Kakoulidou, A.; Kremastinos, D.T.; Mikros, E.; et al. Investigating the effect of antioxidant treatment on the protective effect of preconditioning in anesthetized rabbits. J. Cardiovasc. Pharmacol. 2011, 58, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Andreadou, I.; Mikros, E.; Ioannidis, K.; Sigala, F.; Naka, K.; Kostidis, S.; Farmakis, D.; Tenta, R.; Kavantzas, N.; Bibli, S.-I.; et al. Oleuropein prevents doxorubicin-induced cardiomyopathy interfering with signaling molecules and cardiomyocyte metabolism. J. Mol. Cell. Cardiol. 2014, 69, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Andreadou, I.; Benaki, D.; Efentakis, P.; Bibli, S.-I.; Milioni, A.-I.; Papachristodoulou, A.; Zoga, A.; Skaltsounis, A.-L.; Mikros, E.; Iliodromitis, E.K. The natural olive constituent oleuropein induces nutritional cardioprotection in normal and cholesterol-fed rabbits: Comparison with preconditioning. Planta Med. 2015, 81, 655–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Efentakis, P.; Iliodromitis, E.K.; Mikros, E.; Papachristodoulou, A.; Dagres, N.; Skaltsounis, A.-L.; Andreadou, I. Effects of the olive tree leaf constituents on myocardial oxidative damage and atherosclerosis. Planta Med. 2015, 81, 648–654. [Google Scholar] [CrossRef] [Green Version]
- Malliou, F.; Andreadou, I.; Gonzalez, F.J.; Lazou, A.; Xepapadaki, E.; Vallianou, I.; Lambrinidis, G.; Mikros, E.; Marselos, M.; Skaltsounis, A.-L.; et al. The olive constituent oleuropein, as a PPAR\textbackslashtextgreeka agonist, markedly reduces serum triglycerides. J. Nutr. Biochem. 2018, 59, 17–28. [Google Scholar] [CrossRef]
- Papachristodoulou, A.; Tsoukala, M.; Benaki, D.; Kostidis, S.; Gioti, K.; Aligiannis, N.; Pratsinis, H.; Kletsas, D.; Skaltsounis, A.-L.; Mikros, E.; et al. Oleuropein is a Powerful Sensitizer of Doxorubicin-mediated Killing of Prostate Cancer Cells and Exerts Its Action via Induction of Autophagy. J. Cancer Res. Treat. 2016, 4, 61–68. [Google Scholar] [CrossRef] [Green Version]
- Sklavos, S.; Gatidou, G.; Stasinakis, A.S.; Haralambopoulos, D. Use of solar distillation for olive mill wastewater drying and recovery of polyphenolic compounds. J. Environ. Manag. 2015, 162, 46–52. [Google Scholar] [CrossRef]
- Kotoula, D.; Iliopoulou, A.; Irakleous-Palaiologou, E.; Gatidou, G.; Aloupi, M.; Antonopoulou, P.; Fountoulakis, M.S.; Stasinakis, A.S. Municipal wastewater treatment by combining in series microalgae Chlorella sorokiniana and macrophyte Lemna minor: Preliminary results. J. Clean. Prod. 2020, 271, 122704. [Google Scholar] [CrossRef]
- Agalias, A.; Magiatis, P.; Skaltsounis, A.-L.; Mikros, E.; Tsarbopoulos, A.; Gikas, E.; Spanos, I.; Manios, T. A new process for the management of olive oil mill waste water and recovery of natural antioxidants. J. Agric. Food Chem. 2007, 55, 2671–2676. [Google Scholar] [CrossRef] [PubMed]
- Bazoti, F.N.; Gikas, E.; Skaltsounis, A.L.; Tsarbopoulos, A. Development of a liquid chromatography–electrospray ionization tandem mass spectrometry (LC–ESI MS/MS) method for the quantification of bioactive substances present in olive oil mill wastewaters. Anal. Chim. Acta 2006, 573–574, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadi, M.; Pratsinis, H.; Kletsas, D.; Skaltsounis, A.-L.; Haroutounian, S.A. Bioactive non-coloured polyphenols content of grapes, wines and vinification by-products: Evaluation of the antioxidant activities of their extracts. Food Res. Int. 2010, 43, 805–813. [Google Scholar] [CrossRef]
- Marinou, K.A.; Georgopoulou, K.; Agrogiannis, G.; Karatzas, T.; Iliopoulos, D.; Papalois, A.; Chatziioannou, A.; Magiatis, P.; Halabalaki, M.; Tsantila, N.; et al. Differential effect of Pistacia vera extracts on experimental atherosclerosis in the rabbit animal model: An experimental study. Lipids Health Dis. 2010, 9, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xynos, N.; Abatis, D.; Argyropoulou, A.; Polychronopoulos, P.; Aligiannis, N.; Skaltsounis, A.-L. Development of a Sustainable Procedure for the Recovery of Hydroxytyrosol from Table Olive Processing Wastewater Using Adsorption Resin Technology and Centrifugal Partition Chromatography. Planta Med. 2015, 81, 1621–1627. [Google Scholar] [CrossRef] [PubMed]
- Vougogiannopoulou, K.; Angelopoulou, M.; Pratsinis, H.; Grougnet, R.; Halabalaki, M.; Kletsas, D.; Deguin, B.; Skaltsounis, L. Chemical and Biological Investigation of Olive Mill Waste Water—OMWW Secoiridoid Lactones. Planta Med. 2015, 81, 1205–1212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadopoulou, A.; Petrotos, K.; Stagos, D.; Gerasopoulos, K.; Maimaris, A.; Makris, H.; Kafantaris, I.; Makri, S.; Kerasioti, E.; Halabalaki, M.; et al. Enhancement of Antioxidant Mechanisms and Reduction of Oxidative Stress in Chickens after the Administration of Drinking Water Enriched with Polyphenolic Powder from Olive Mill Waste Waters. Oxidative Med. Cell. Longev. 2017, 2017, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Haroutinian, S.; Skaltsounis, A.L.; Magiatis, P.; Kazantzoglou, G.; Evergetis, E. Products derived from the vinification residues of cultivated plants belonging to the Vitis viniferae species for producing cosmetic products, food supplements and bio-active foodstuffs. GR Patent (GR1005157), 09 September 2004. [Google Scholar]
- Lambropoulou, D.A.; Albanis, T.A. Methods of sample preparation for determination of pesticide residues in food matrices by chromatography-mass spectrometry-based techniques: A review. Anal. Bioanal. Chem. 2007, 389, 1663–1683. [Google Scholar] [CrossRef]
- Tsoutsi, C.S.; Konstantinou, I.K.; Hela, D.G. Organophosphorus pesticide residues in Greek virgin olive oil: Levels, dietary intake and risk assessment. Food Addit. Contam. Part AChem. Anal. ControlExpo. Risk Assess. 2008, 25, 1225–1236. [Google Scholar] [CrossRef]
- Zioris, I.V.; Lambropoulou, D.A.; Danis, T.G.; Karagiozoglou, D.T.; Albanis, T.A. Assessment of pesticide residues in fresh peach samples produced under integrated crop management in an agricultural region of northern Greece. Food Addit. Contam. Part A 2009, 26, 1256–1264. [Google Scholar] [CrossRef]
- Boti, V.I.; Sakkas, V.A.; Albanis, T.A. An experimental design approach employing artificial neural networks for the determination of potential endocrine disruptors in food using matrix solid-phase dispersion. J. Chromatogr. A 2009, 1216, 1296–1304. [Google Scholar] [CrossRef]
- Synaridou, M.-E.S.; Sakkas, V.A.; Stalikas, C.D.; Albanis, T.A. Evaluation of magnetic nanoparticles to serve as solid-phase extraction sorbents for the determination of endocrine disruptors in milk samples by gas chromatography mass spectrometry. J. Chromatogr. A 2014, 1348, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Garbi, A.; Sakkas, V.; Fiamegos, Y.C.; Stalikas, C.D.; Albanis, T. Sensitive determination of pesticides residues in wine samples with the aid of single-drop microextraction and response surface methodology. Talanta 2010, 82, 1286–1291. [Google Scholar] [CrossRef] [PubMed]
- Aznar-Alemany, Ò.; Aminot, Y.; Vilà-Cano, J.; Köck-Schulmeyer, M.; Readman, J.W.; Marques, A.; Godinho, L.; Botteon, E.; Ferrari, F.; Boti, V.; et al. Halogenated and organophosphorus flame retardants in European aquaculture samples. Sci. Total Environ. 2018, 612, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Maragou, N.C.; Makri, A.; Lampi, E.N.; Thomaidis, N.S.; Koupparis, M.A. Migration of bisphenol A from polycarbonate baby bottles under real use conditions. Food Addit. Contam. Part A 2008, 25, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Papaspyrou, S.D.; Thomaidis, N.S.; Lampi, E.N.; Lioupis, A. Determination of migration of n-butyltins and n-octyltins to food simulants by gas chromatography–mass spectrometry. Appl. Organomet. Chem. 2007, 21, 412–424. [Google Scholar] [CrossRef]
- Raptopoulou, K.G.; Pasias, I.N.; Thomaidis, N.S.; Proestos, C. Study of the migration phenomena of specific metals in canned tomato paste before and after opening. Validation of a new quality indicator for opened cans. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2014, 69, 25–31. [Google Scholar] [CrossRef]
- Petropoulos, G.; Raptopoulou, K.G.; Pasias, I.N.; Thomaidis, N.S.; Proestos, C. Chemometric determination of the shelf life of opened cans using the migration of specific metals as quality indicators. Food Chem. 2018, 267, 313–318. [Google Scholar] [CrossRef]
- Diamantidou, D.; Begou, O.; Theodoridis, G.; Gika, H.; Tsochatzis, E.; Kalogiannis, S.; Kataiftsi, N.; Soufleros, E.; Zotou, A. Development and validation of an ultra high performance liquid chromatography-tandem mass spectrometry method for the determination of phthalate esters in Greek grape marc spirits. J. Chromatogr. A 2019, 1603, 165–178. [Google Scholar] [CrossRef]
- Tsochatzis, E.D.; Gika, H.; Theodoridis, G. Development and validation of a fast gas chromatography mass spectrometry method for the quantification of selected non-intentionally added substances and polystyrene/polyurethane oligomers in liquid food simulants. Anal. Chim. Acta 2020, 1130, 49–59. [Google Scholar] [CrossRef]
- Dasenaki, M.E.; Thomaidis, N.S. Multi-residue determination of seventeen sulfonamides and five tetracyclines in fish tissue using a multi-stage LC–ESI–MS/MS approach based on advanced mass spectrometric techniques. Anal. Chim. Acta 2010, 672, 93–102. [Google Scholar] [CrossRef]
- Dasenaki, M.E.; Thomaidis, N.S. Multi-residue determination of 115 veterinary drugs and pharmaceutical residues in milk powder, butter, fish tissue and eggs using liquid chromatography–tandem mass spectrometry. Anal. Chim. Acta 2015, 880, 103–121. [Google Scholar] [CrossRef] [PubMed]
- Dasenaki, M.E.; Bletsou, A.A.; Koulis, G.A.; Thomaidis, N.S. Qualitative Multiresidue Screening Method for 143 Veterinary Drugs and Pharmaceuticals in Milk and Fish Tissue Using Liquid Chromatography Quadrupole-Time-of-Flight Mass Spectrometry. J. Agric. Food Chem. 2015, 63, 4493–4508. [Google Scholar] [CrossRef] [PubMed]
- Dasenaki, M.E.; Michali, M.S.; Thomaidis, N.S. Analysis of 76 veterinary pharmaceuticals from 13 classes including aminoglycosides in bovine muscle by hydrophilic interaction liquid chromatography–tandem mass spectrometry. J. Chromatogr. A 2016, 1452, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Dasenaki, M.E.; Thomaidis, N.S. Multi-residue methodology for the determination of 16 coccidiostats in animal tissues and eggs by hydrophilic interaction liquid chromatography—Tandem mass spectrometry. Food Chem. 2019, 275, 668–680. [Google Scholar] [CrossRef]
- Orfanidis, A.; Gika, H.G.; Theodoridis, G.; Mastrogianni, O.; Raikos, N. An UHPLC-MS-MS Method for the Determination of 84 Drugs of Abuse and Pharmaceuticals in Blood. J. Anal. Toxicol 2020, bkaa032. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Theodoridis, G.; Pechlivanis, A.; Thomaidis, N.S.; Spyros, A.; Georgiou, C.A.; Albanis, T.; Skoufos, I.; Kalogiannis, S.; Tsangaris, G.T.; Stasinakis, A.S.; et al. FoodOmicsGR_RI: A Consortium for Comprehensive Molecular Characterisation of Food Products. Metabolites 2021, 11, 74. https://doi.org/10.3390/metabo11020074
Theodoridis G, Pechlivanis A, Thomaidis NS, Spyros A, Georgiou CA, Albanis T, Skoufos I, Kalogiannis S, Tsangaris GT, Stasinakis AS, et al. FoodOmicsGR_RI: A Consortium for Comprehensive Molecular Characterisation of Food Products. Metabolites. 2021; 11(2):74. https://doi.org/10.3390/metabo11020074
Chicago/Turabian StyleTheodoridis, Georgios, Alexandros Pechlivanis, Nikolaos S. Thomaidis, Apostolos Spyros, Constantinos A. Georgiou, Triantafyllos Albanis, Ioannis Skoufos, Stavros Kalogiannis, George Th. Tsangaris, Athanasios S. Stasinakis, and et al. 2021. "FoodOmicsGR_RI: A Consortium for Comprehensive Molecular Characterisation of Food Products" Metabolites 11, no. 2: 74. https://doi.org/10.3390/metabo11020074
APA StyleTheodoridis, G., Pechlivanis, A., Thomaidis, N. S., Spyros, A., Georgiou, C. A., Albanis, T., Skoufos, I., Kalogiannis, S., Tsangaris, G. T., Stasinakis, A. S., Konstantinou, I., Triantafyllidis, A., Gkagkavouzis, K., Kritikou, A. S., Dasenaki, M. E., Gika, H., Virgiliou, C., Kodra, D., Nenadis, N., ... on behalf of the FoodOmicsGR_RI Consortium. (2021). FoodOmicsGR_RI: A Consortium for Comprehensive Molecular Characterisation of Food Products. Metabolites, 11(2), 74. https://doi.org/10.3390/metabo11020074