Solanum lycopersicum Seedlings. Metabolic Responses Induced by the Alkamide Affinin
Abstract
:1. Introduction
2. Results
2.1. Developmental and Metabolic Response of Solanum lycopersicum to the Alkamide Affinin
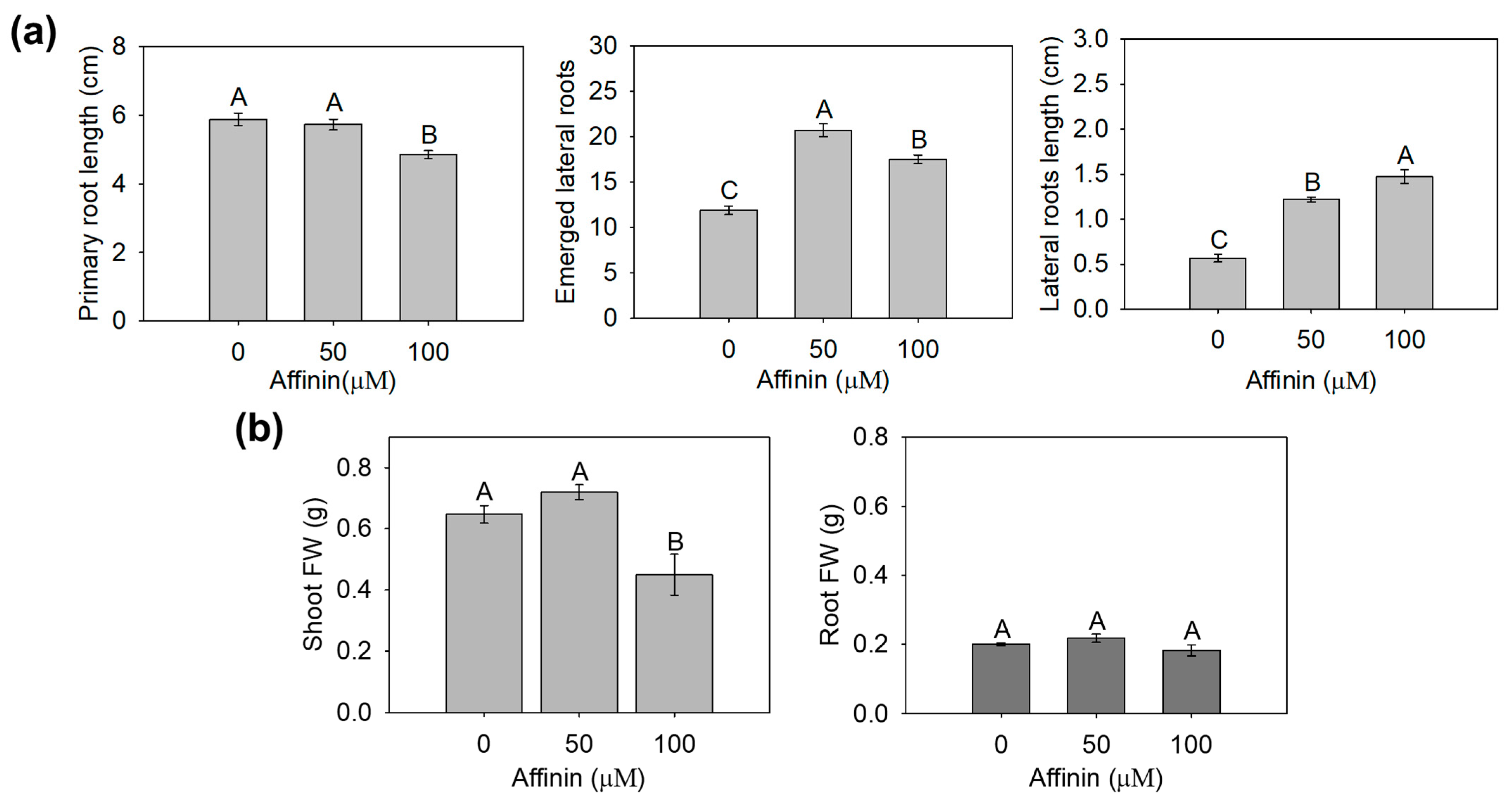
2.1.1. Effect of Affinin in Solanum lycopersicum Development
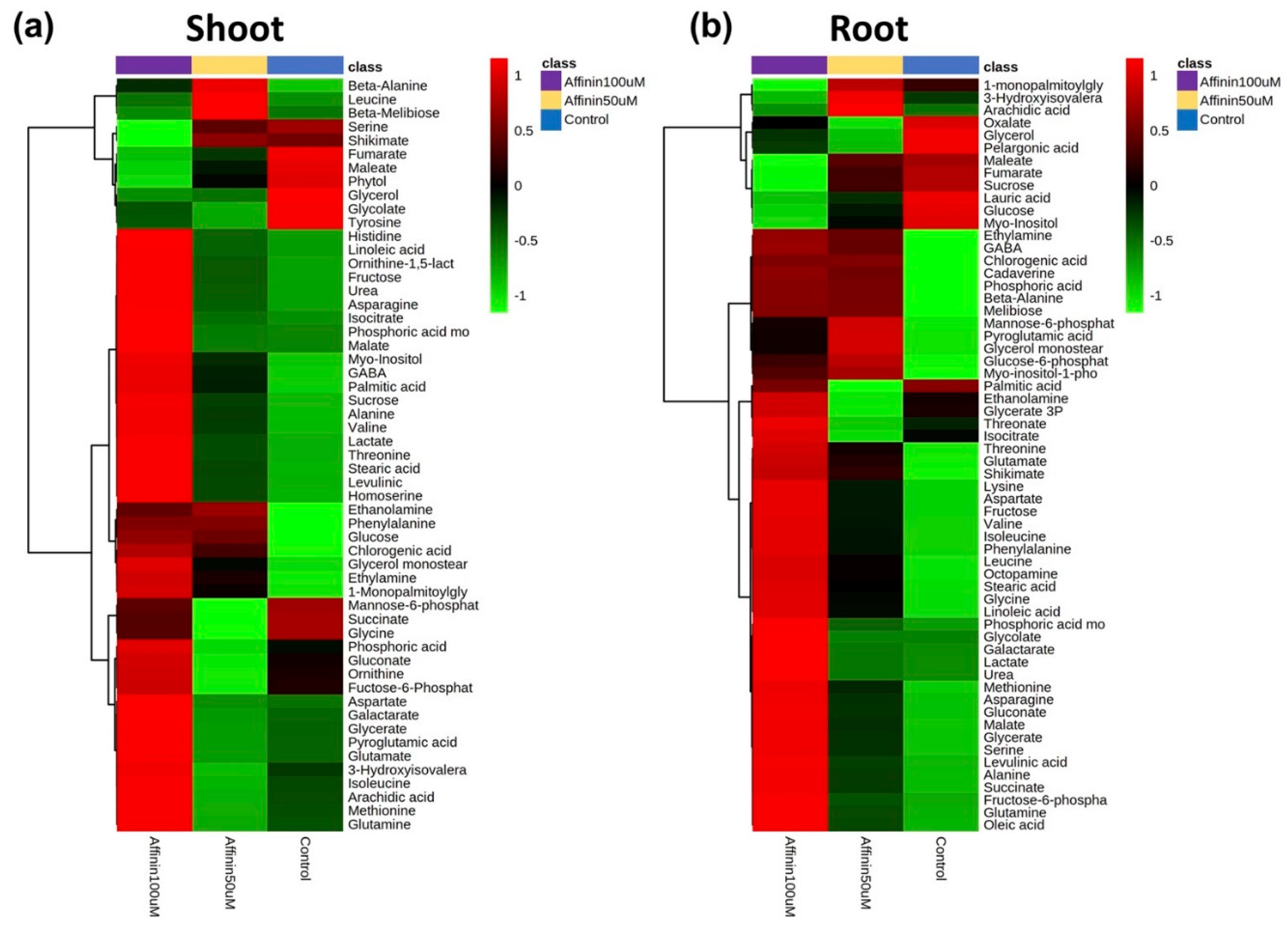
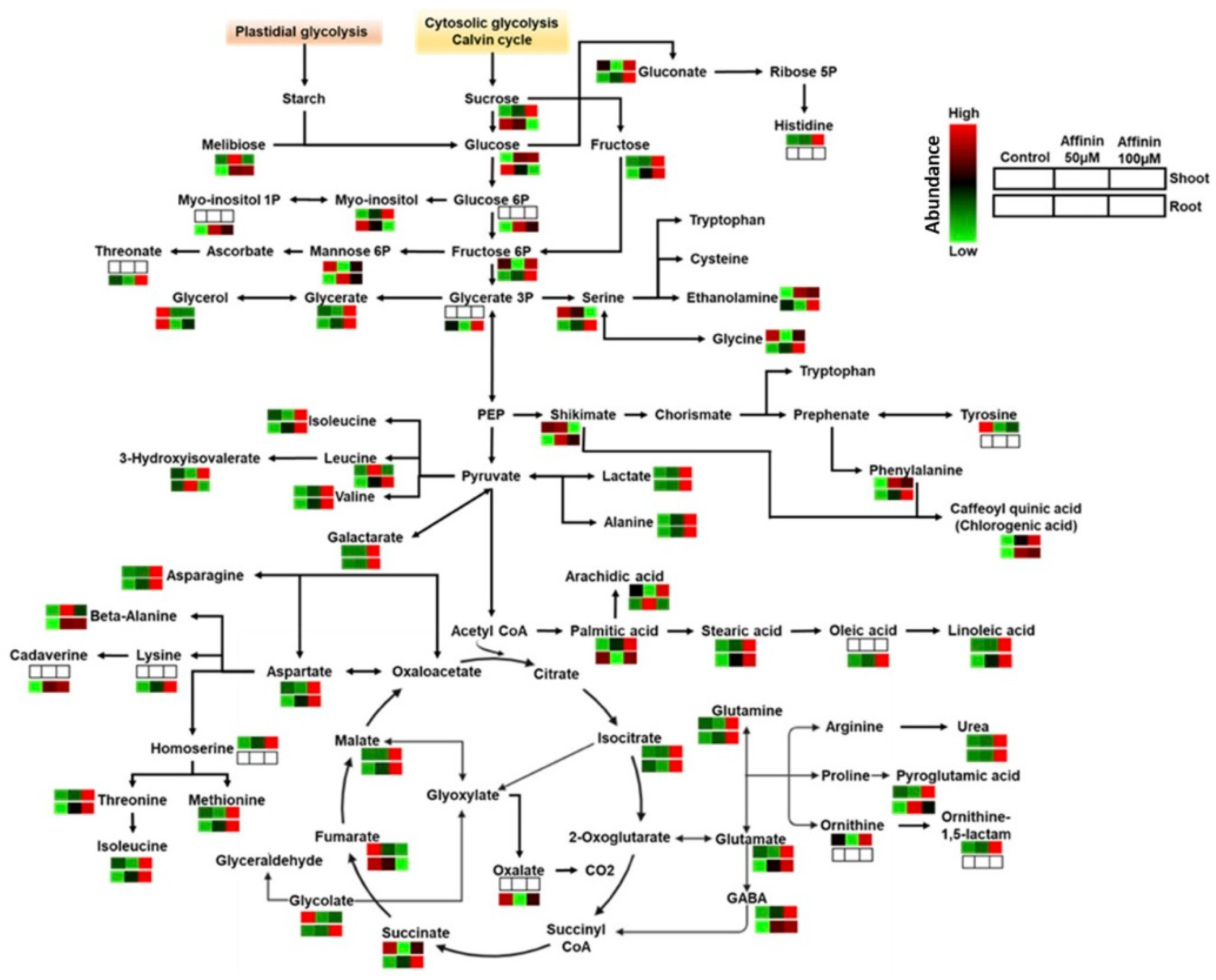
2.1.2. Metabolic Profiles Altered by Affinin
2.1.3. Metabolic Variations Induced by Affinin Treatments
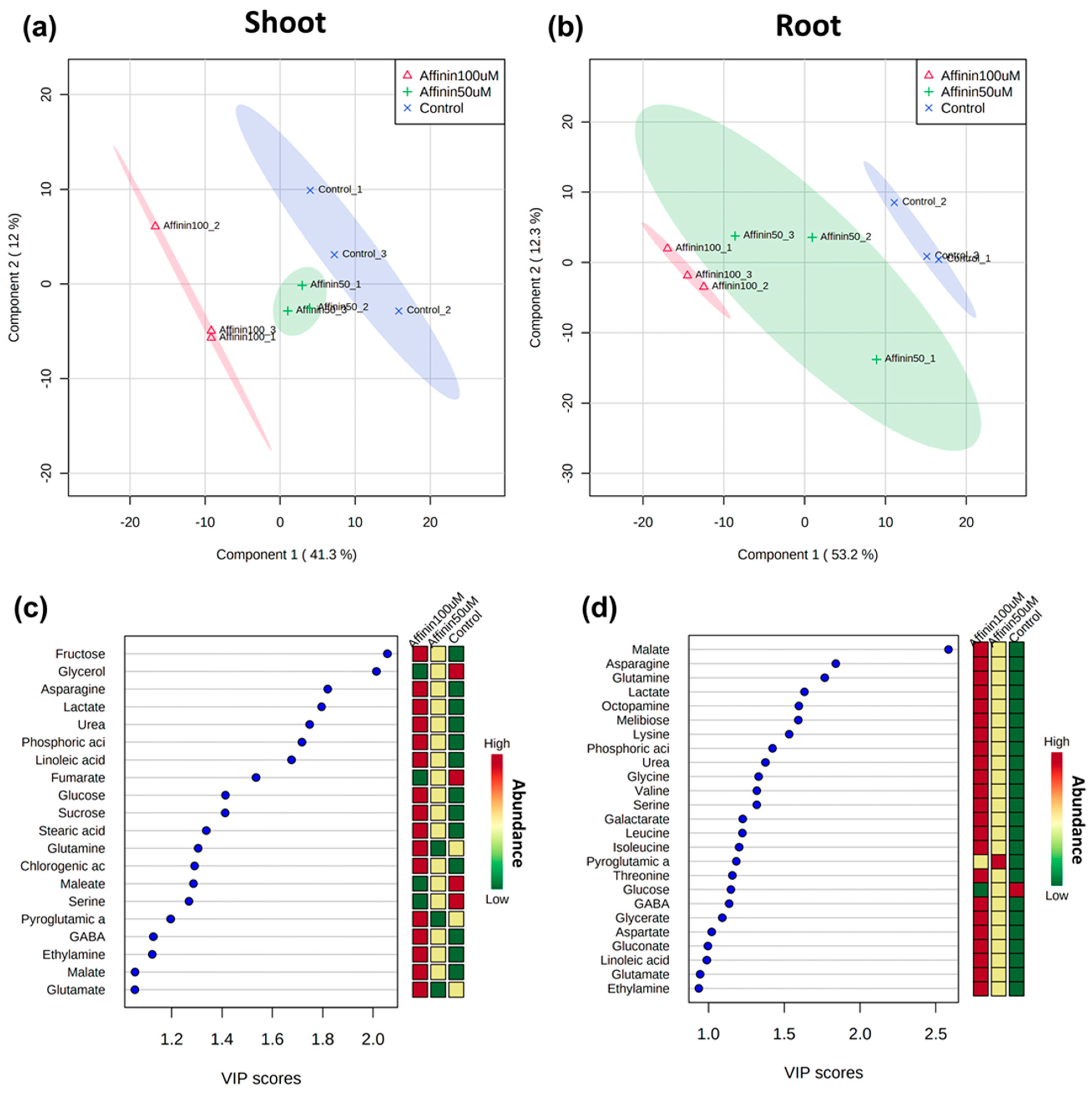
2.1.4. Global Metabolic Profile Changes Revealed by Partial Least Squares-Discriminant Analysis (PLS-DA) in Response to Affinin
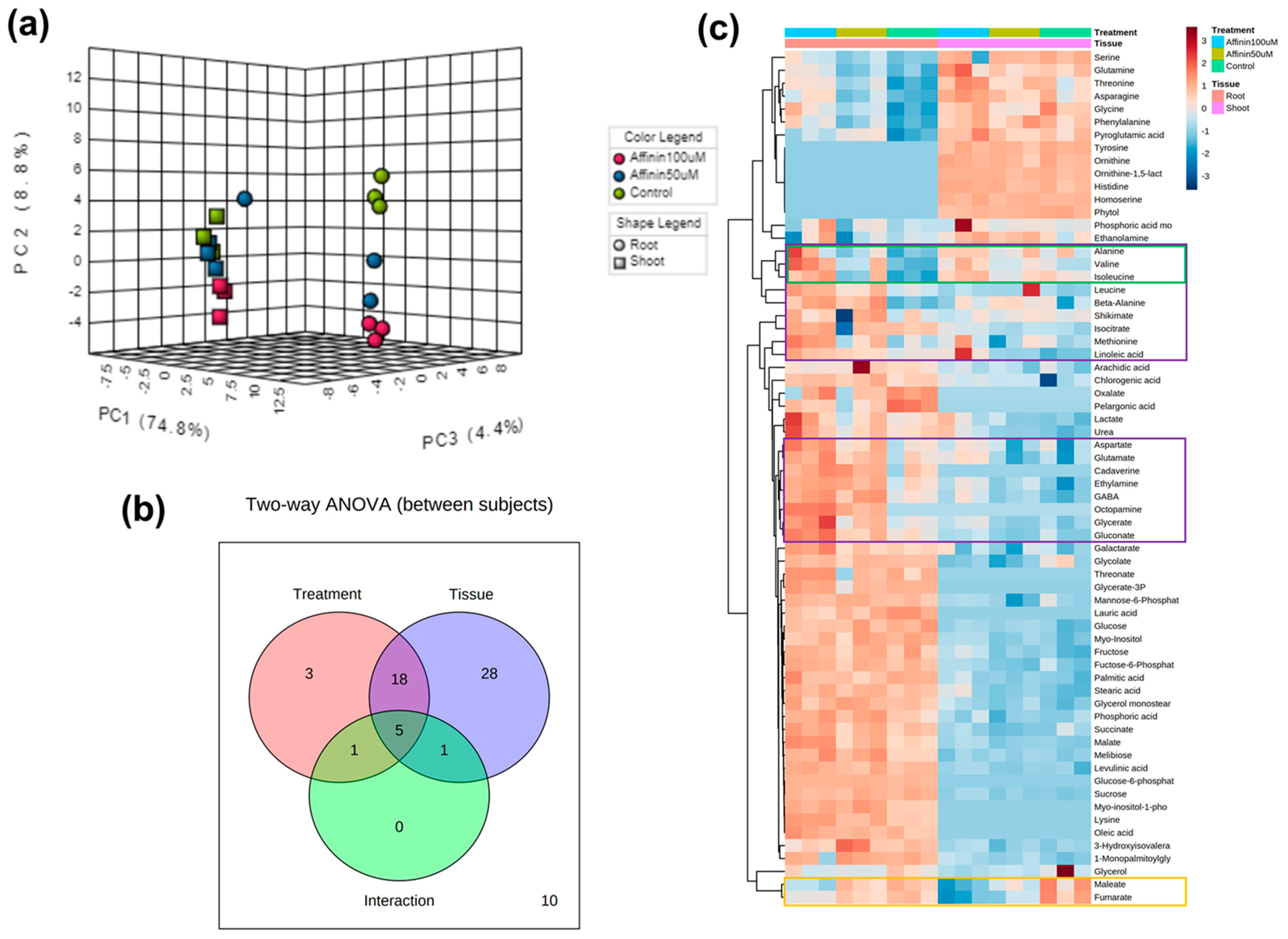
2.1.5. Independent Metabolic Response from Shoots and Roots Induced by Affinin
3. Discussion
3.1. Affinin Modulates Tomato Seedlings Development
3.2. Affinin Induces Metabolic Adjustment in Tomato Seedlings
3.2.1. Affinin Alters Sugar Metabolism
3.2.2. Affinin Alters Amino Acid Metabolism
3.2.3. Organic Acids and Sugar Acids Accumulated by Affinin Treatments
3.2.4. Fatty Acids Accumulated in Response to Affinin
3.2.5. Differences in Roots and Shoots Metabolic Profiles in Response to Affinin Treatment
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Plant Growth Analysis
4.3. Non-Targeted Metabolic Profiling by Gas Chromatography—Mass Spectrometry (GC/EIMS)
4.4. Data Processing and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Molinatorres, J.; Salgado-Garciglia, R.; Ramirez-Chavez, E.; del Rio, R.E. Purely olefinic alkamnides in Heliopsis longipes and Acmella (Spilanthes) oppositifolia. Biochem. Syst. Ecol. 1996, 24, 43–47. [Google Scholar] [CrossRef]
- Chávez, E.R.; Valdez, L.L.; Calleros, G.V.; Torres, J.M. Actividad fungicida de la afinina y del extracto crudo de raíces de Heliopsis longipes en dos especies de Sclerotium. Agrociencia 2000, 34, 207–215. Available online: www.redalyc.org/articulo.oa?id=30234210 (accessed on 23 October 2020).
- Ramírez-Chávez, E.; López-Bucio, J.; Herrera-Estrella, L.; Molina-Torres, J. Alkamides Isolated from Plants Promote Growth and Alter root Development in Arabidopsis. Plant Physiol. 2004, 134, 1058–1068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Morales, S.; Flores López, M.L.; Benavides Mendoza, A.; Flores Olivas, A. Actividad Inhibitoria del Extracto de Heliopsis longipes Sobre Fusarium oxysporum f. sp lycopersici. Rev. Mex. Fitopatol. 2011, 29, 146–153. [Google Scholar]
- Castro-Ruiz, J.E.; Rojas-Molina, A.; Luna-Vázquez, F.J.; Rivero-Cruz, F.; García-Gasca, T.; Ibarra-Alvarado, C. Affinin (Spilanthol), Isolated from Heliopsis longipes, Induces Vasodilation via Activation of Gasotransmitters and Prostacyclin Signaling Pathways. Int. J. Mol. Sci. 2017, 18, 218. [Google Scholar] [CrossRef]
- López-Bucio, J.; Millán-Godínez, M.; Méndez-Bravo, A.; Morquecho-Contreras, A.; Ramírez-Chávez, E.; Molina-Torres, J.; Pérez-Torres, A.; Higuchi, M.; Kakimoto, T.; Herrera-Estrella, L. Cytokinin Receptors Are Involved in Alkamide Regulation of Root and Shoot Development in Arabidopsis. Plant Physiol. 2007, 145, 1703–1713. [Google Scholar] [CrossRef] [Green Version]
- Méndez-Bravo, A.; Calderón-Vázquez, C.; Ibarra-Laclette, E.; Raya-González, J.; Ramírez-Chávez, E.; Molina-Torres, J.; Guevara-García, A.A.; López-Bucio, J.; Herrera-Estrella, L. Alkamides Activate Jasmonic Acid Biosynthesis and Signaling Pathways and Confer Resistance to Botrytis cinerea in Arabidopsis thaliana. PLoS ONE 2011, 6, e27251. [Google Scholar] [CrossRef]
- Blancaflor, E.B.; Hou, G.; Chapman, K.D. Elevated levels of N-lauroylethanolamine, an endogenous constituent of desiccated seeds, disrupt normal root development in Arabidopsis thaliana seedlings. Planta 2003, 217, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Coulon, D.; Faure, L.; Salmon, M.; Wattelet, V.; Bessoule, J.-J. N-Acylethanolamines and related compounds: Aspects of metabolism and functions. Plant Sci. 2012, 184, 129–140. [Google Scholar] [CrossRef]
- Blancaflor, E.B.; Kilaru, A.; Keereetaweep, J.; Khan, B.R.; Faure, L.; Chapman, K.D. N-Acylethanolamines: Lipid metabolites with functions in plant growth and development. Plant J. 2014, 79, 568–583. [Google Scholar] [CrossRef]
- Kunos, G.; Járai, Z.; Bátkai, S.; Goparaju, S.K.; Ishac, E.J.; Liu, J.; Wang, L.; A Wagner, J. Endocannabinoids as cardiovascular modulators. Chem. Phys. Lipids 2000, 108, 159–168. [Google Scholar] [CrossRef]
- Wilson, R.I. Endocannabinoid Signaling in the Brain. Science 2002, 296, 678–682. [Google Scholar] [CrossRef] [Green Version]
- Mathesius, U.; Mulders, S.; Gao, M.; Teplitski, M.; Caetano-Anollés, G.; Rolfe, B.G.; Bauer, W.D. Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc. Natl. Acad. Sci. USA 2003, 100, 1444–1449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortíz-Castro, R.; Martínez-Trujillo, M.; López-Bucio, J. N-acyl-L-homoserine lactones: A class of bacterial quorum-sensing signals alter post-embryonic root development in Arabidopsis thaliana. Plant Cell Environ. 2008, 31, 1497–1509. [Google Scholar] [CrossRef]
- Schenk, S.T.; Hernández-Reyes, C.; Samans, B.; Stein, E.; Neumann, C.; Schikora, M.; Reichelt, M.; Mithöfer, A.; Becker, A.; Kogel, K.-H.; et al. N-Acyl-Homoserine Lactone Primes Plants for Cell Wall Reinforcement and Induces Resistance to Bacterial Pathogens via the Salicylic Acid/Oxylipin Pathway. Plant Cell 2014, 26, 2708–2723. [Google Scholar] [CrossRef] [Green Version]
- Méndez-Bravo, A.; Raya-González, J.; Herrera-Estrella, L.; López-Bucio, J. Nitric Oxide is Involved in Alkamide-Induced Lateral Root Development in Arabidopsis. Plant Cell Physiol. 2010, 51, 1612–1626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, L.; Wang, Y.-S.; Uppalapati, S.R.; Wang, K.; Tang, Y.; Vadapalli, V.; Venables, B.J.; Chapman, K.D.; Blancaflor, E.B.; Mysore, K.S. Overexpression of a fatty acid amide hydrolase compromises innate immunity in Arabidopsis. Plant J. 2008, 56, 336–349. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Ron, M.; Dorrity, M.W.; De Lucas, M.; Toal, T.; Hernandez, R.I.; Little, S.A.; Maloof, J.N.; Kliebenstein, D.J.; Brady, S.M. Identification of Novel Loci Regulating Interspecific Variation in Root Morphology and Cellular Development in Tomato. Plant Physiol. 2013, 162, 755–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sade, D.; Shriki, O.; Cuadros-Inostroza, A.; Tohge, T.; Semel, Y.; Haviv, Y.; Willmitzer, L.; Fernie, A.R.; Czosnek, H.; Brotman, Y. Comparative metabolomics and transcriptomics of plant response to Tomato yellow leaf curl virus infection in resistant and susceptible tomato cultivars. Metabolomics 2014, 11, 81–97. [Google Scholar] [CrossRef]
- Eloh, K.; Sasanelli, N.; Maxia, A.; Caboni, P. Untargeted Metabolomics of Tomato Plants after Root-Knot Nematode Infestation. J. Agric. Food Chem. 2016, 64, 5963–5968. [Google Scholar] [CrossRef]
- Li, Z.; Yu, J.; Peng, Y.; Huang, B. Metabolic pathways regulated by abscisic acid, salicylic acid and γ-aminobutyric acid in association with improved drought tolerance in creeping bentgrass (Agrostis stolonifera). Physiol. Plant. 2017, 159, 42–58. [Google Scholar] [CrossRef] [PubMed]
- Luo, B.; Ma, P.; Nie, Z.; Zhang, X.; He, X.; Ding, X.; Feng, X.; Lu, Q.; Ren, Z.; Lin, H.; et al. Metabolite profiling and genome-wide association studies reveal response mechanisms of phosphorus deficiency in maize seedling. Plant J. 2019, 97, 947–969. [Google Scholar] [CrossRef]
- Rojas, C.M.; Esenthil-Kumar, M.; Etzin, V.; Mysore, K.S. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 2014, 5, 17. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, H.; Li, S.C.; Li, Y.T. α-Galactosidase from Mortierella vinacea crystallization and properties. J. Biol. Chem. 1970, 245, 781–786. [Google Scholar] [CrossRef]
- Meyer, T.; Vigouroux, A.; Aumont-Nicaise, M.; Comte, G.; Vial, L.; Lavire, C.; Moréra, S. The plant defense signal galactinol is specifically used as a nutrient by the bacterial pathogen Agrobacterium fabrum. J. Biol. Chem. 2018, 293, 7930–7941. [Google Scholar] [CrossRef] [Green Version]
- Rai, V. Role of Amino Acids in Plant Responses to Stresses. Biol. Plant. 2002, 45, 481–487. [Google Scholar] [CrossRef]
- Ali, Q.; Habib-ur-Rehman Athar, M.Z.; Haider, S.S.; Aslam, N.; Shehzad, F.; Naseem, J.; Ashraf, R.; Ali, A.; Hussain, S.M. 12 Role of Amino Acids in Improving Abiotic Stress Tolerance to Plants. In Plant Tolerance to Environmental Stress: Role of Phytoprotectants; Hasanuzzaman, M., Fujita, M., Oku, H., Islam, M.T., Eds.; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Seifi, H.S.; van Bockhaven, J.; Angenon, G.; Höfte, M. Glutamate Metabolism in Plant Disease and Defense: Friend or Foe? Mol. Plant Microbe Interact. 2013, 26, 475–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Sousa, C.; Sodek, L. Alanine metabolism and alanine aminotransferase activity in soybean (Glycine max) during hypoxia of the root system and subsequent return to normoxia. Environ. Exp. Bot. 2003, 50, 1–8. [Google Scholar] [CrossRef]
- Subbarayan, K.; Rolletschek, H.; Senula, A.D.; Ulagappan, K.; Hajirezaei, M.-R.; Keller, E.J. Influence of oxygen deficiency and the role of specific amino acids in cryopreservation of garlic shoot tips. BMC Biotechnol. 2015, 15, 40. [Google Scholar] [CrossRef] [Green Version]
- Ros, R.; Muñoz-Bertomeu, J.; Krueger, S. Serine in plants: Biosynthesis, metabolism, and functions. Trends Plant Sci. 2014, 19, 564–569. [Google Scholar] [CrossRef] [PubMed]
- Igamberdiev, A.U.; Kleczkowski, L.A. The Glycerate and Phosphorylated Pathways of Serine Synthesis in Plants: The Branches of Plant Glycolysis Linking Carbon and Nitrogen Metabolism. Front. Plant Sci. 2018, 9, 318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, H.; Widhalm, J.R.; Qian, Y.; Maeda, H.; Cooper, B.R.; Jannasch, A.S.; Gonda, I.; Lewinsohn, E.; Rhodes, D.; Dudareva, N. An alternative pathway contributes to phenylalanine biosynthesis in plants via a cytosolic tyrosine:phenylpyruvate aminotransferase. Nat. Commun. 2013, 4, 2833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galili, G. The aspartate-family pathway of plants. Plant Signal. Behav. 2011, 6, 192–195. [Google Scholar] [CrossRef] [Green Version]
- Tomar, P.C.; Lakra, N.; Mishra, S.N. Cadaverine. Plant Signal. Behav. 2013, 8, e25850. [Google Scholar] [CrossRef] [Green Version]
- Jancewicz, A.L.; Gibbs, N.M.; Masson, P.H. Cadaverine’s Functional Role in Plant Development and Environmental Response. Front. Plant Sci. 2016, 7, 870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, M.D.; Che, P.; Song, J.; Nikolau, B.J.; Wurtele, E.S. 3-Methylcrotonyl-Coenzyme A Carboxylase Is a Component of the Mitochondrial Leucine Catabolic Pathway in Plants. Plant Physiol. 1998, 118, 1127–1138. [Google Scholar] [CrossRef] [Green Version]
- Binder, S. Branched-Chain Amino Acid Metabolism in Arabidopsis thaliana. Arab. Book Am. Soc. Plant Biol. 2010, 8, e0137. [Google Scholar] [CrossRef] [Green Version]
- López-Bucio, J.; Nieto-Jacobo, M.F.; Ramírez-Rodríguez, V.; Herrera-Estrella, L. Organic acid metabolism in plants: From adaptive physiology to transgenic varieties for cultivation in extreme soils. Plant Sci. 2000, 160, 1–13. [Google Scholar] [CrossRef]
- Corkins, M.E.; Wilson, S.; Cocuron, J.-C.; Alonso, A.P.; Bird, A.J. The gluconate shunt is an alternative route for directing glucose into the pentose phosphate pathway in fission yeast. J. Biol. Chem. 2017, 292, 13823–13832. [Google Scholar] [CrossRef] [Green Version]
- Wushensky, J.A.; Youngster, T.; Mendonca, C.M.; Aristilde, L. Flux Connections between Gluconate Pathway, Glycolysis, and Pentose–Phosphate Pathway During Carbohydrate Metabolism in Bacillus megaterium QM B1551. Front. Microbiol. 2018, 9, 2789. [Google Scholar] [CrossRef] [Green Version]
- Degenkolbe, T.; Do, P.T.; Kopka, J.; Zuther, E.; Hincha, D.K.; Köhl, K.I. Identification of Drought Tolerance Markers in a Diverse Population of Rice Cultivars by Expression and Metabolite Profiling. PLoS ONE 2013, 8, e63637. [Google Scholar] [CrossRef]
- Gupta, P.; De, B. Metabolomics analysis of rice responses to salinity stress revealed elevation of serotonin, and gentisic acid levels in leaves of tolerant varieties. Plant Signal. Behav. 2017, 12, e1335845. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, N.O.; Olivares, F.L.; Novotny, E.H.; Canellas, L.P. Changes in metabolic profiling of sugarcane leaves induced by endophytic diazotrophic bacteria and humic acids. PeerJ 2018, 6, e5445. [Google Scholar] [CrossRef]
- Truffault, V.; Fry, S.C.; Stevens, R.G.; Gautier, H. Ascorbate degradation in tomato leads to accumulation of oxalate, threonate and oxalyl threonate. Plant J. 2017, 89, 996–1008. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Braun, H.; Gawryluk, R.M.R.; Millar, A.H. Mitochondrial complexIIof plants: Subunit composition, assembly, and function in respiration and signaling. Plant J. 2019, 98, 405–417. [Google Scholar] [CrossRef] [Green Version]
- António, C.; Päpke, C.; Rocha, M.; Diab, H.; Limami, A.M.; Obata, T.; Fernie, A.R.; Van Dongen, J.T. Regulation of Primary Metabolism in Response to Low Oxygen Availability as Revealed by Carbon and Nitrogen Isotope Redistribution. Plant Physiol. 2016, 170, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Simonin, V.; Galina, A. Nitric oxide inhibits succinate dehydrogenase-driven oxygen consumption in potato tuber mitochondria in an oxygen tension-independent manner. Biochem. J. 2012, 449, 263–273. [Google Scholar] [CrossRef] [Green Version]
- Millar, A.A.; Smith, M.A.; Kunst, L. All fatty acids are not equal: Discrimination in plant membrane lipids. Trends Plant Sci. 2000, 5, 95–101. [Google Scholar] [CrossRef]
- Xue, H.Q.; Upchurch, R.G.; Kwanyuen, P. Ergosterol as a Quantifiable Biomass Marker for Diaporthe haseolorum and Cercospora kikuchi. Plant Dis. 2006, 90, 1395–1398. [Google Scholar] [CrossRef]
- Kachroo, A.; Venugopal, S.C.; Lapchyk, L.; Falcone, D.; Hildebrand, D.; Kachroo, P. Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 5152–5157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kishimoto, K.; Matsui, K.; Ozawa, R.; Takabayashi, J. Volatile C6-aldehydes and Allo-ocimene Activate Defense Genes and Induce Resistance against Botrytis cinerea in Arabidopsis thaliana. Plant Cell Physiol. 2005, 46, 1093–1102. [Google Scholar] [CrossRef] [Green Version]
- Shanab, S.M.; Hafez, R.M.; Fouad, A.S. A review on algae and plants as potential source of arachidonic acid. J. Adv. Res. 2018, 11, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Bais, H.P.; Loyola-Vargas, V.M.; Flores, H.E.; Vivanco, J.M. Root-specific metabolism: The biology and biochemistry of underground organs. Vitr. Cell. Dev. Biol. Anim. 2001, 37, 730–741. [Google Scholar] [CrossRef]
- Walch-Liu, P.; Liu, L.-H.; Remans, T.; Tester, M.; Forde, B.G. Evidence that l -Glutamate Can Act as an Exogenous Signal to Modulate Root Growth and Branching in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 1045–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Weckwerth, W.; Loureiro, M.E.; Wenzel, K.; Fiehn, O. Differential metabolic networks unravel the effects of silent plant phenotypes. Proc. Natl. Acad. Sci. USA 2004, 101, 7809–7814. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Wishart, D.S. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 2011, 6, 743–760. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. MetPA: A web-based metabolomics tool for pathway analysis and visualization. Bioinformatics 2010, 26, 2342–2344. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Metabolite | ChEBI ID | StDev |
|---|---|---|
| SHOOTS | ||
| Asparagine | 17,196 | 6.4627 |
| Chlorogenic acid | 16,112 | 4.0187 |
| Fructose | 28,757 | 2.9712 |
| Glucose | 17,634 | 3.9011 |
| Glutamate | 16,015 | 1.61 |
| Glutamine | 18,050 | 13.8 |
| Lactate | 24,996 | 6.1811 |
| Linoleic acid | 17,351 | 5.1357 |
| Phosphoric acid monomethyl ester | 340,824 | 23.675 |
| Pyroglutamic acid | 16,010 | 8.5086 |
| Stearic acid | 28,842 | 1.8317 |
| Sucrose | 17,992 | 3.7142 |
| Urea | 16,199 | 3.2501 |
| ROOTS | ||
| Alanine | 15,570 | 1.2658 |
| Asparagine | 17,196 | 3.1375 |
| Aspartate | 22,660 | 1.0977 |
| Beta-alanine | 16,958 | 0.19073 |
| Cadaverine | 18,127 | 1.3855 |
| Chlorogenic acid | 16,112 | 3.8009 |
| Ethylamine | 15,862 | 0.84901 |
| GABA | 16,865 | 1.2353 |
| Galactarate | 30,852 | 1.375 |
| Gluconate | 86,359 | 0.41726 |
| Glucose-6-phosphate | 14,314 | 1.3352 |
| Glutamate | 16,015 | 1.5105 |
| Glutamine | 18,050 | 3.1626 |
| Glycerate | 16,659 | 1.2093 |
| Glycerate 3P | 17,050 | 1.8376 |
| Glycine | 15,428 | 1.6957 |
| Isocitrate | 30,887 | 6.5588 |
| Isoleucine | 27,730 | 1.6544 |
| Lactate | 24,996 | 13.919 |
| Leucine | 25,017 | 1.114 |
| Linoleic acid | 17,351 | 0.59333 |
| Lysine | 18,019 | 1.2269 |
| Malate | 6650 | 4.1396 |
| Melibiose | 28,053 | 7.8211 |
| Methionine | 16,811 | 0.67585 |
| Myo-inositol-1-phosphate | 18,297 | 0.89362 |
| Octopamine | 17,134 | 0.12966 |
| Oleic acid | 16,196 | 2.1569 |
| Phenylalanine | 17,295 | 0.8976 |
| Phosphoric acid | 26,078 | 5.5247 |
| Pyroglutamic acid | 16,010 | 1.5695 |
| Serine | 17,822 | 3.5762 |
| Succinate | 15,741 | 1.5881 |
| Threonate | 15,908 | 2.5509 |
| Threonine | 16,398 | 0.51273 |
| Urea | 16,199 | 9.7979 |
| Valine | 27,266 | 1.0954 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campos-García, T.; Molina-Torres, J. Solanum lycopersicum Seedlings. Metabolic Responses Induced by the Alkamide Affinin. Metabolites 2021, 11, 143. https://doi.org/10.3390/metabo11030143
Campos-García T, Molina-Torres J. Solanum lycopersicum Seedlings. Metabolic Responses Induced by the Alkamide Affinin. Metabolites. 2021; 11(3):143. https://doi.org/10.3390/metabo11030143
Chicago/Turabian StyleCampos-García, Tonatiu, and Jorge Molina-Torres. 2021. "Solanum lycopersicum Seedlings. Metabolic Responses Induced by the Alkamide Affinin" Metabolites 11, no. 3: 143. https://doi.org/10.3390/metabo11030143
APA StyleCampos-García, T., & Molina-Torres, J. (2021). Solanum lycopersicum Seedlings. Metabolic Responses Induced by the Alkamide Affinin. Metabolites, 11(3), 143. https://doi.org/10.3390/metabo11030143






