OS-PCA: Orthogonal Smoothed Principal Component Analysis Applied to Metabolome Data
Abstract
1. Introduction
2. Results and Discussion
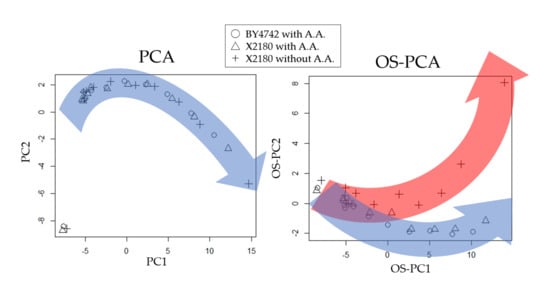
2.1. Case Study 1: Metabolic Turnover Analysis
2.2. Case Study 2: Metabolome Analysis for the Taste of Japanese Green Tea
3. Methods
3.1. Smoothed Principal Component Analysis (PCA)
3.2. Orthogonal Smoothed Principal Component Analysis (OS-PCA)
3.3. OS-PCA for Repeated Measurement Data
3.4. Statistical Property of OS-PC Loading for Autoscaled Data
3.5. Statistical Property of OS-PC Loading for Repeated Measurements and Autoscaled Data
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jolliffe, I.T. Principal Component Analysis, 2nd ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Wold, S.; Sjostrom, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Barker, M.; Rayens, W. Partial least squares for discrimination. J. Chemom. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Stanstrup, J.; Broeckling, C.D.; Helmus, R.; Hoffmann, N.; Mathe, E.; Naake, T.; Nicolotti, L.; Peters, K.; Rainer, J.; Salek, R.M.; et al. The metaRbolomics Toolbox in Bioconductor and beyond. Metabolites 2019, 9, 200. [Google Scholar] [CrossRef] [PubMed]
- Smilde, A.K.; Westerhuis, J.A.; Hoefsloot, H.C.J.; Bijlsma, S.; Rubingh, C.M.; Vis, D.J.; Jellema, R.H.; Pijl, H.; Roelfsema, F.; van der Greef, J. Dynamic metabolomic data analysis: A tutorial review. Metabolomics 2010, 6, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Smilde, A.K.; Jansen, J.J.; Hoefsloot, H.C.J.; Lamers, R.A.N.; van der Greef, J.; Timmerman, M.E. ANO-VA—Simultaneous component analysis (ASCA): A new tool for analyzing designed metabolomics data. Bioinformatics 2005, 21, 3043–3048. [Google Scholar] [CrossRef] [PubMed]
- Bertinetto, C.; Engel, J.; Jansen, J. ANOVA simultaneous component analysis: A tutorial review. Anal. Chim. Acta X 2020, 6, 100061. [Google Scholar] [CrossRef] [PubMed]
- Nyamundanda, G.; Gormley, I.C.; Brennan, L. A dynamic probabilistic principal components model for the analysis of longitudinal metabolomics data. J. R. Stat. Soc. C Appl. 2014, 63, 763–782. [Google Scholar] [CrossRef]
- Yamamoto, H.; Yamaji, H.; Abe, Y.; Harada, K.; Waluyo, D.; Fukusaki, E.; Kondo, A.; Ohno, H.; Fukuda, H. Dimensionality reduction for metabolome data using PCA, PLS, OPLS, and RFDA with differential penalties to latent variables. Chemom. Intell. Lab. Syst. 2009, 98, 136–142. [Google Scholar] [CrossRef]
- Yamamoto, H.; Fujimori, T.; Sato, H.; Ishikawa, G.; Kami, K.; Ohashi, Y. Statistical hypothesis testing of factor loading in principal component analysis and its application to metabolite set enrichment analysis. BMC Bioinform. 2014, 15, 51. [Google Scholar] [CrossRef] [PubMed]
- Vinaixa, M.; Samino, S.; Saez, I.; Duran, J.; Guinovart, J.J.; Yanes, O. A Guideline to Univariate Statistical Analysis for LC/MS-Based Untargeted Metabolomics-Derived Data univariate analysis. Metabolites 2012, 2, 775–795. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.; Mei, Z.; Zeng, C.; Liu, S. metaX: A flexible and comprehensive software for processing metabolomics data. BMC Bioinform. 2017, 18, 183. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H. PLS-ROG: Partial least squares with rank order of groups. J. Chemom. 2017, 31, e2883. [Google Scholar] [CrossRef]
- Nakayama, Y.; Tamada, Y.; Tsugawa, H.; Bamba, T.; Fukusaki, E. Novel Strategy for Non-Targeted Isotope-Assisted Metabolomics by Means of Metabolic Turnover and Multivariate Analysis. Metabolites 2014, 4, 722–739. [Google Scholar] [CrossRef]
- Pongsuwan, W.; Fukusaki, E.; Bamba, T.; Yonetani, T.; Yamahara, A.T.; Kobayashi, A. Prediction of Japanese Green Tea Ranking by Gas Chromatography/Mass Spectrometry-Based Hydrophilic Metabolite Fingerprinting. J. Agric. Food Chem. 2007, 55, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Tsujimoto, Y.; Arita, M.; Bamba, T.; Fukusaki, E. GC/MS based metabolomics: Development of a data mining system for metabolite identification by using soft independent modeling of class analogy (SIMCA). BMC Bioinform. 2011, 12, 131. [Google Scholar] [CrossRef] [PubMed]
- Eilers, P.H.C. A Perfect Smoother. Anal. Chem. 2003, 75, 3631–3636. [Google Scholar] [CrossRef]


 ) 46, (⊕) 51.
) 46, (⊕) 51.
 ) 46, (⊕) 51.
) 46, (⊕) 51.
 ) 46, (⊕) 51.
) 46, (⊕) 51.
 ) 46, (⊕) 51.
) 46, (⊕) 51.
| Method | Equation | Eigenvector | Hypothesis Testing |
|---|---|---|---|
| PCA | (1/n)X′Xwx = λwx | wx∝corr(t,xp) | Applicable |
| Smoothed PCA | (1/n)X′Xwx = λ{(1 − κ)I + κX′D′DX}wx | Not Available | Not Applicable |
| OS-PCA | (1/n2)X′XP−1X′Xwx = λwx(1/n2)X′XX′Xwy = λPwy | wx∝corr(s,xp) | Applicable |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamamoto, H.; Nakayama, Y.; Tsugawa, H. OS-PCA: Orthogonal Smoothed Principal Component Analysis Applied to Metabolome Data. Metabolites 2021, 11, 149. https://doi.org/10.3390/metabo11030149
Yamamoto H, Nakayama Y, Tsugawa H. OS-PCA: Orthogonal Smoothed Principal Component Analysis Applied to Metabolome Data. Metabolites. 2021; 11(3):149. https://doi.org/10.3390/metabo11030149
Chicago/Turabian StyleYamamoto, Hiroyuki, Yasumune Nakayama, and Hiroshi Tsugawa. 2021. "OS-PCA: Orthogonal Smoothed Principal Component Analysis Applied to Metabolome Data" Metabolites 11, no. 3: 149. https://doi.org/10.3390/metabo11030149
APA StyleYamamoto, H., Nakayama, Y., & Tsugawa, H. (2021). OS-PCA: Orthogonal Smoothed Principal Component Analysis Applied to Metabolome Data. Metabolites, 11(3), 149. https://doi.org/10.3390/metabo11030149