Stable Isotopic Tracer Phospholipidomics Reveals Contributions of Key Phospholipid Biosynthetic Pathways to Low Hepatocyte Phosphatidylcholine to Phosphatidylethanolamine Ratio Induced by Free Fatty Acids
Abstract
1. Introduction
2. Results
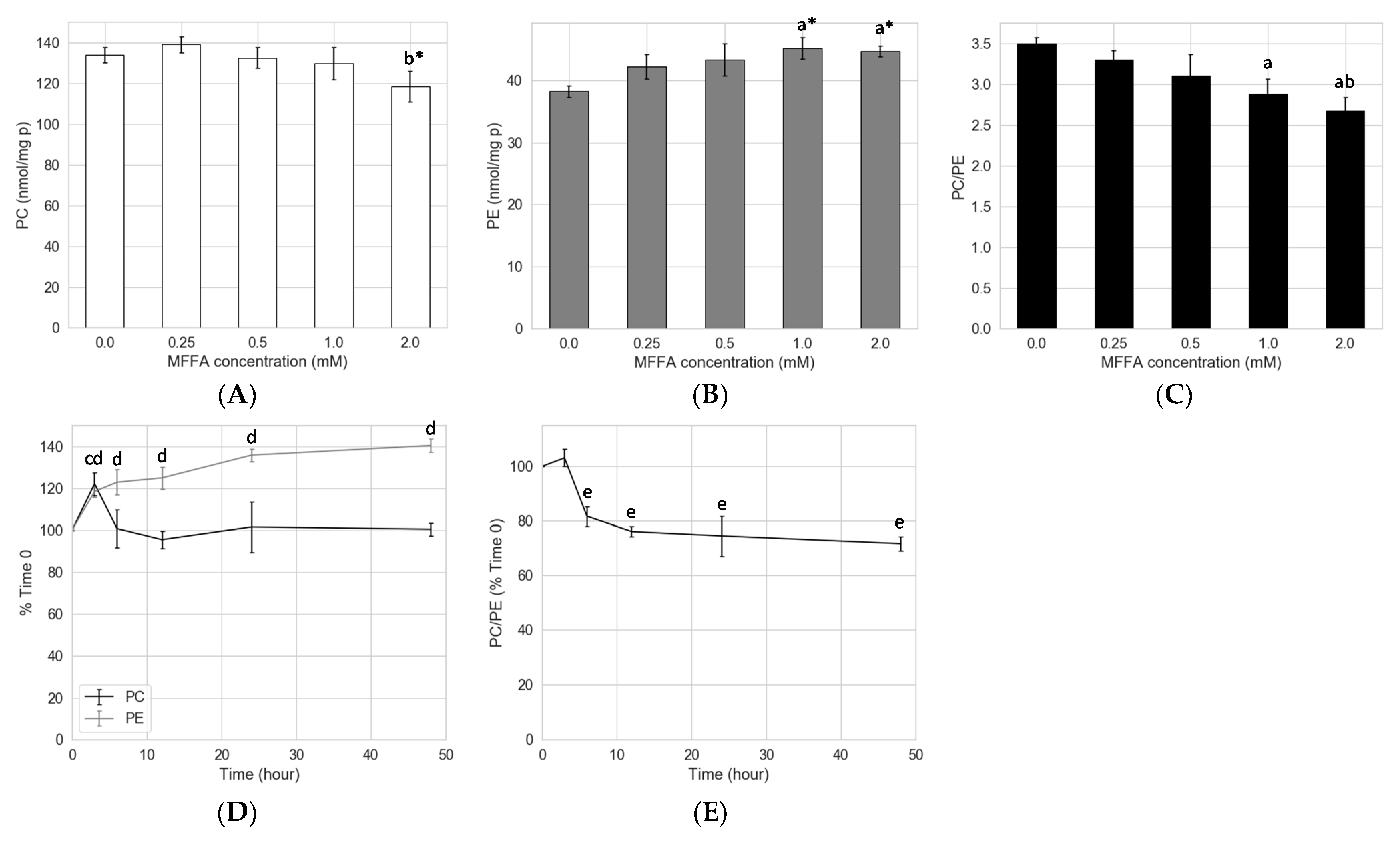
2.1. Effects of FFAs on Hepatocyte PC/PE
2.2. Concentration and Kinetic Effects of MFFA on Hepatocyte PC/PE
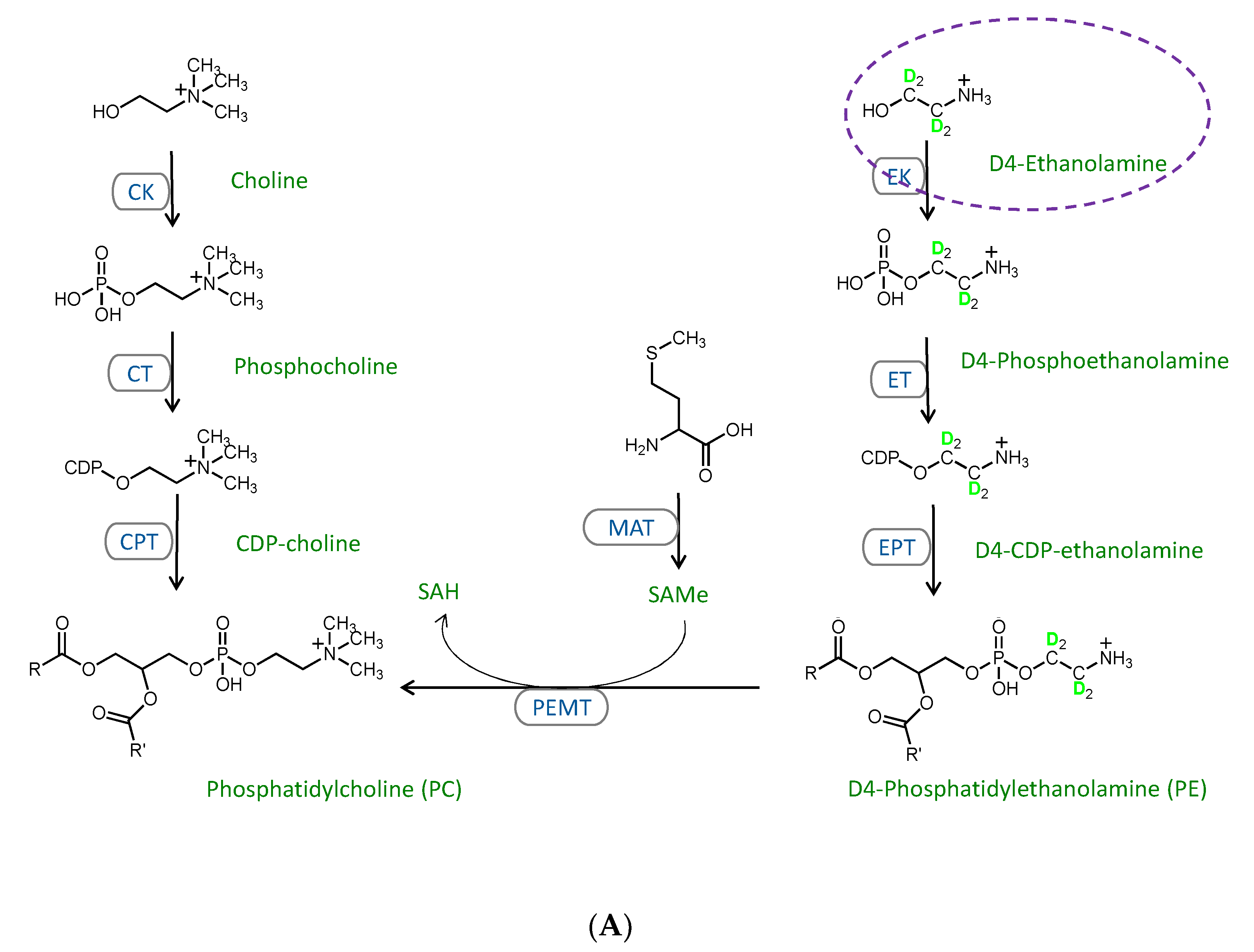
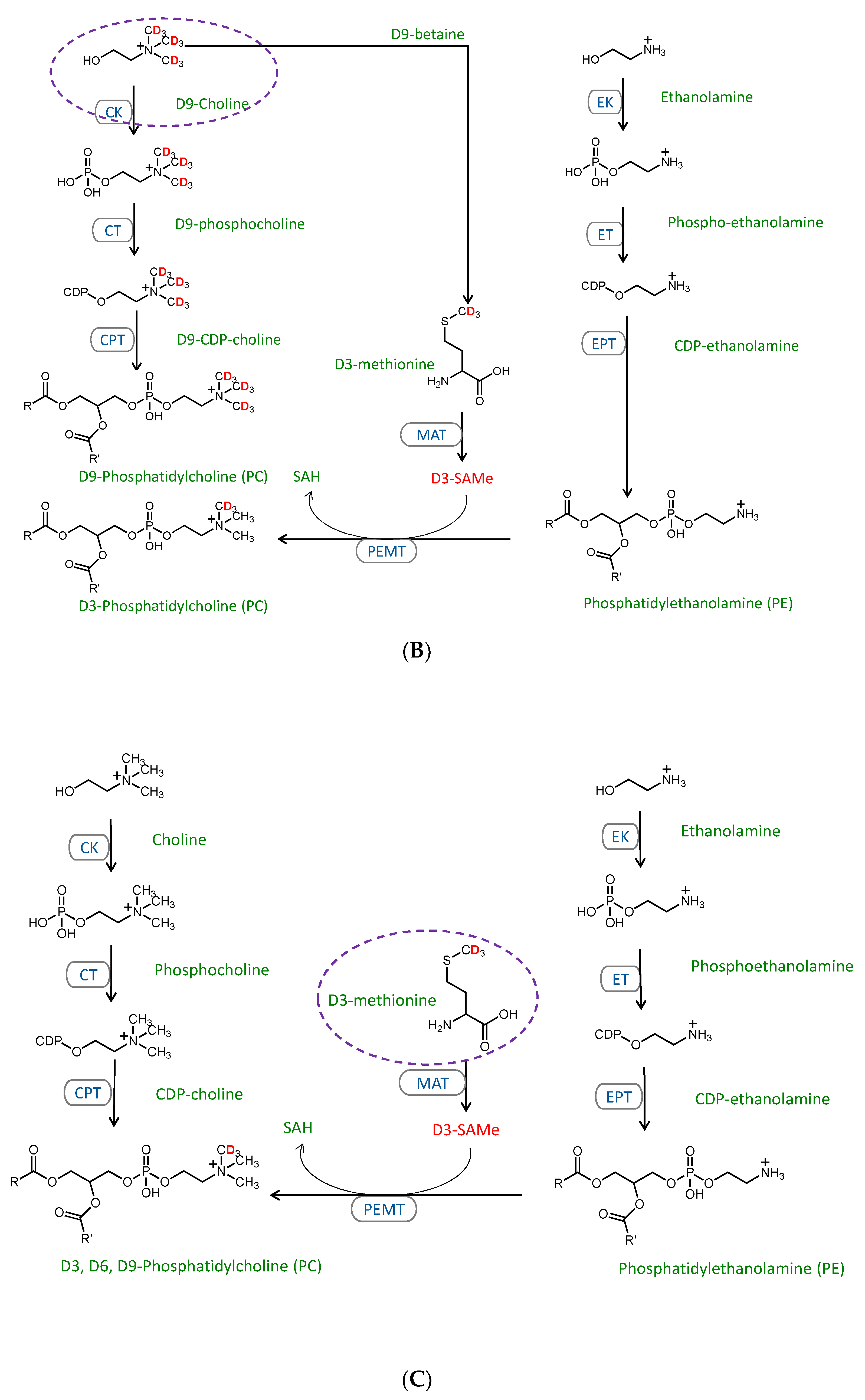
2.3. Stable Isotope Tracer Analysis of Hepatocyte Phospholipid Turnover
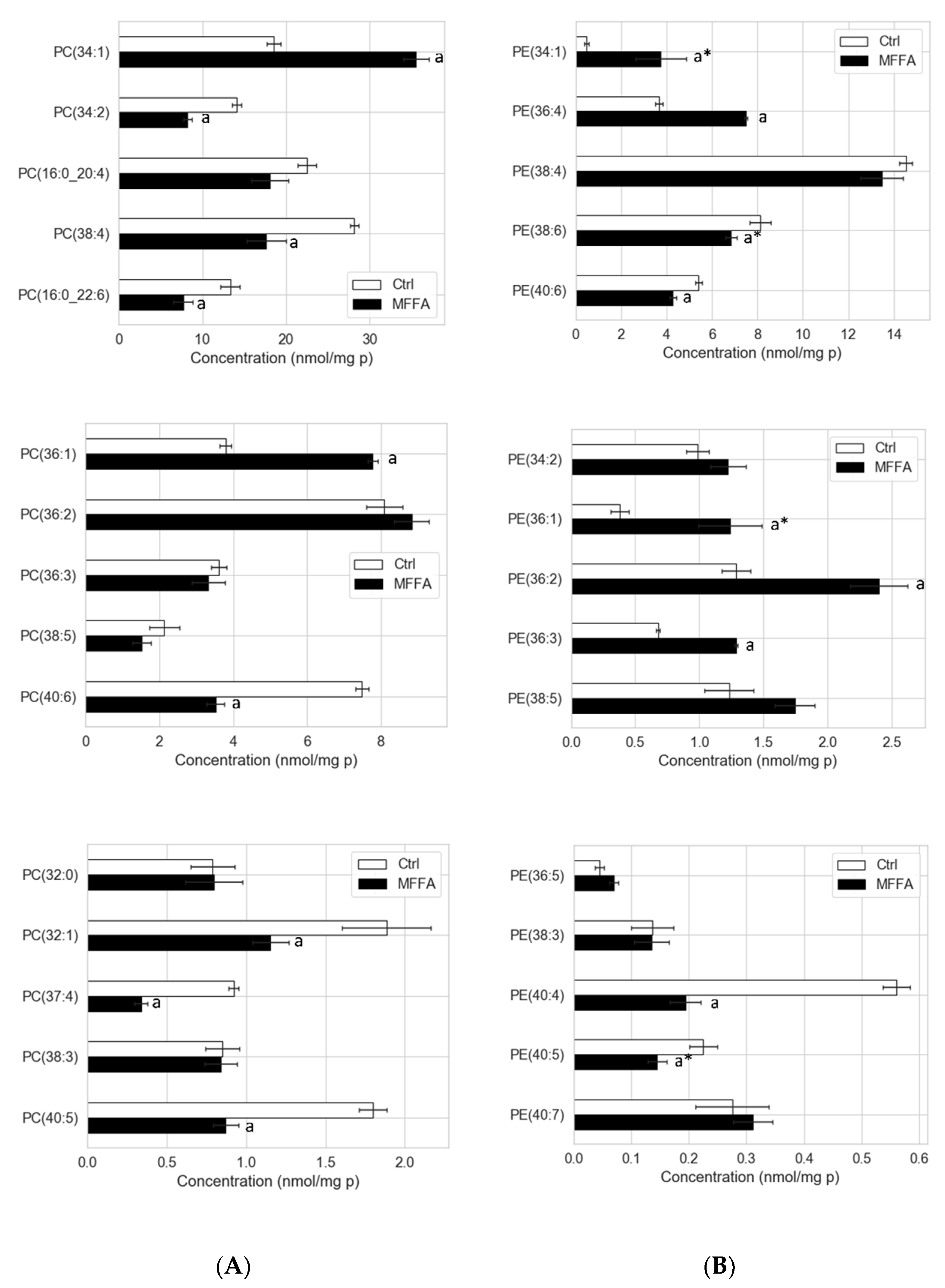
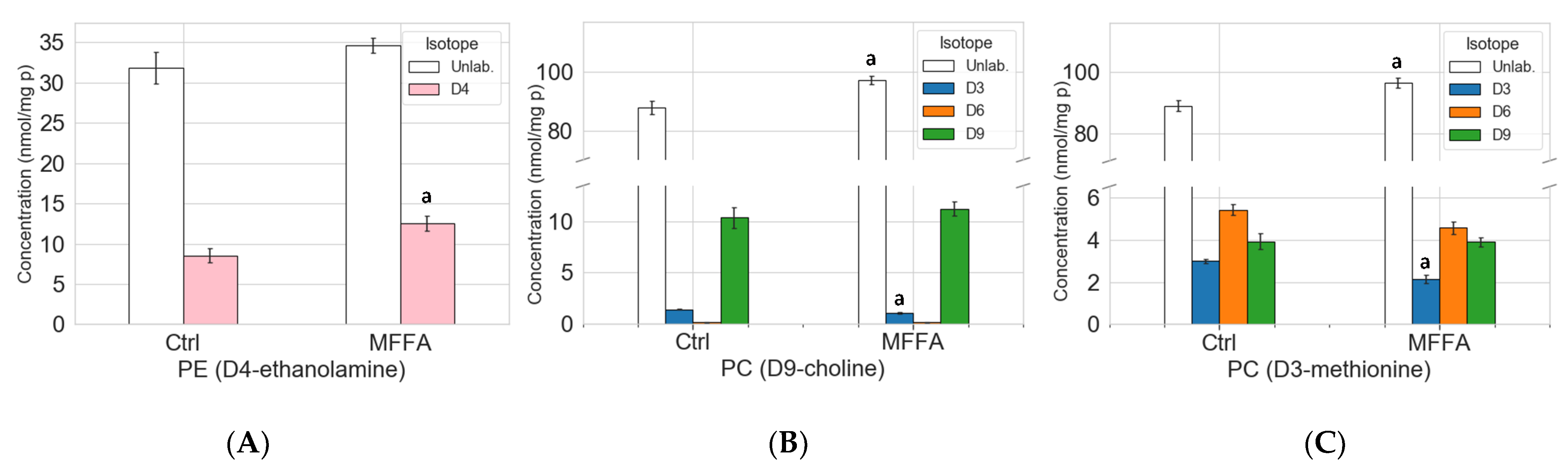
2.4. Phospholipid Isotopic Enrichment Profiles
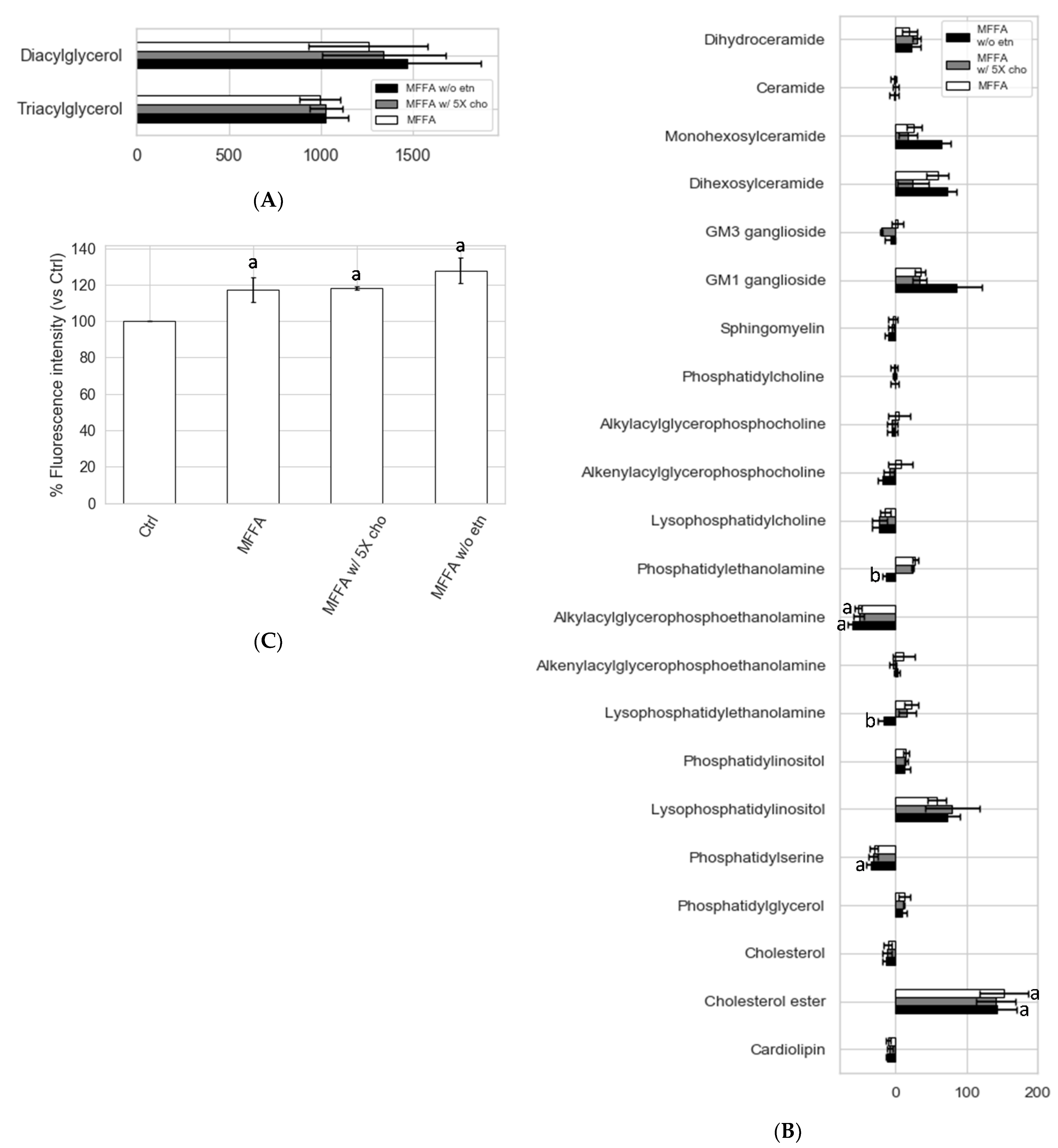
2.5. Effects of Altered Media Phospholipid Precursors on the Hepatocyte Lipidome and Cell Death
3. Discussion
3.1. Prolonged Treatment with Excessive MFFA Lowers Hepatocyte PC/PE
3.2. CDP-Ethanolamine and PEMT Pathways Contribute to the Altered PC/PE
3.3. Mechanisms Contributing to the MFFA-Induced Disruptions to CDP-Ethanolamine and PEMT Pathways
3.4. Hepatocytes Show Tolerance to Aberrant PC/PE Induced by MFFA
4. Materials and Methods
4.1. Materials
4.2. Rat Primary Hepatocytes Isolation, Culture, and Fatty Acid Treatment
4.3. Sample Preparation and Lipid Extraction
4.4. Lipidomic Analysis
4.5. Determination of Cell Death
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CDP | cytidine diphosphate |
| FFA | free fatty acid |
| LPC | lysophosphatidylcholine |
| LPE | lysophosphatidylethanolamine |
| MFFA | mixed free fatty acid |
| NAFLD | non-alcoholic fatty liver disease |
| PC | phosphatidylcholine |
| PC/PE | phosphatidylcholine to phosphatidylethanolamine ratio |
| PE | phosphatidylethanolamine |
| PEMT | phosphatidylethanolamine N-methyl transferase |
| SM | sphingomyelin |
References
- Vance, J.E.; Vance, D.E. Phospholipid Biosynthesis in Mammalian Cells. Biochem. Cell Biol. 2004, 82, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Vance, J.E.; Tasseva, G. Formation and Function of Phosphatidylserine and Phosphatidylethanolamine in Mammalian Cells. Biochim. Biophys. Acta 2013, 1831, 543–554. [Google Scholar] [CrossRef]
- Cano, A.; Alonso, C. Deciphering Non-Alcoholic Fatty Liver Disease through Metabolomics: Figure 1. Biochem. Soc. Trans. 2014, 42, 1447–1452. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Agellon, L.B.; Allen, T.M.; Umeda, M.; Jewell, L.; Mason, A.; Vance, D.E. The Ratio of Phosphatidylcholine to Phosphatidylethanolamine Influences Membrane Integrity and Steatohepatitis. Cell Metab. 2006, 3, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Mas, E.; Danjoux, M.; Garcia, V.; Carpentier, S.; Ségui, B.; Levade, T. The Pro-Inflammatory Action of Tumour Necrosis Factor-α in Non-Alcoholic Steatohepatitis Is Independent of the NSMAF Gene Product. Dig. Liver Dis. 2013, 45, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Caballero, F.; Fernández, A.; Matías, N.; Martínez, L.; Fucho, R.; Elena, M.; Caballeria, J.; Morales, A.; Fernández-Checa, J.C.; García-Ruiz, C. Specific Contribution of Methionine and Choline in Nutritional Nonalcoholic Steatohepatitis: Impact on Mitochondrial S-Adenosyl-l-Methionine and Glutathione. J. Biol. Chem. 2010, 285, 18528–18536. [Google Scholar] [CrossRef]
- Chiappini, F.; Coilly, A.; Kadar, H.; Gual, P.; Tran, A.; Desterke, C.; Samuel, D.; Duclos-Vallée, J.-C.; Touboul, D.; Bertrand-Michel, J.; et al. Metabolism Dysregulation Induces a Specific Lipid Signature of Nonalcoholic Steatohepatitis in Patients. Sci. Rep. 2017, 7, 46658. [Google Scholar] [CrossRef]
- Fu, S.; Yang, L.; Li, P.; Hofmann, O.; Dicker, L.; Hide, W.; Lin, X.; Watkins, S.M.; Ivanov, A.R.; Hotamisligil, G.S. Aberrant Lipid Metabolism Disrupts Calcium Homeostasis Causing Liver Endoplasmic Reticulum Stress in Obesity. Nature 2011, 473, 528–531. [Google Scholar] [CrossRef]
- Palomer, X.; Pizarro-Delgado, J.; Barroso, E.; Vázquez-Carrera, M. Palmitic and Oleic Acid: The Yin and Yang of Fatty Acids in Type 2 Diabetes Mellitus. Trends Endocrinol. Metab. 2018, 29, 178–190. [Google Scholar] [CrossRef]
- Sundler, R.; Akesson, B. Regulation of Phospholipid Biosynthesis in Isolated Rat Hepatocytes. Effect of Different Substrates. J. Biol. Chem. 1975, 250, 3359–3367. [Google Scholar] [CrossRef]
- Audubert, F.; Pelech, S.L.; Vance, D.E. Fatty Acids Inhibit N-Methylation of Phosphatidylethanolamine in Rat Hepatocytes and Liver Microsomes. Biochim. Biophys. Acta 1984, 792, 348–357. [Google Scholar] [CrossRef]
- Niklas, J.; Schneider, K.; Heinzle, E. Metabolic Flux Analysis in Eukaryotes. Curr. Opin. Biotechnol. 2010, 21, 63–69. [Google Scholar] [CrossRef]
- Mueller, D.; Heinzle, E. Stable Isotope-Assisted Metabolomics to Detect Metabolic Flux Changes in Mammalian Cell Cultures. Curr. Opin. Biotechnol. 2013, 24, 54–59. [Google Scholar] [CrossRef] [PubMed]
- DeLong, C.J.; Shen, Y.-J.; Thomas, M.J.; Cui, Z. Molecular Distinction of Phosphatidylcholine Synthesis between the CDP-Choline Pathway and Phosphatidylethanolamine Methylation Pathway. J. Biol. Chem. 1999, 274, 29683–29688. [Google Scholar] [CrossRef]
- DeLong, C.J.; Hicks, A.M.; Cui, Z. Disruption of Choline Methyl Group Donation for Phosphatidylethanolamine Methylation in Hepatocarcinoma Cells. J. Biol. Chem. 2002, 277, 17217–17225. [Google Scholar] [CrossRef]
- Pynn, C.J.; Henderson, N.G.; Clark, H.; Koster, G.; Bernhard, W.; Postle, A.D. Specificity and Rate of Human and Mouse Liver and Plasma Phosphatidylcholine Synthesis Analyzed in Vivo. J. Lipid Res. 2011, 52, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Bleijerveld, O.B.; Brouwers, J.F.H.M.; Vaandrager, A.B.; Helms, J.B.; Houweling, M. The CDP-Ethanolamine Pathway and Phosphatidylserine Decarboxylation Generate Different Phosphatidylethanolamine Molecular Species. J. Biol. Chem. 2007, 282, 28362–28372. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Jiang, X.; West, A.A.; Perry, C.A.; Malysheva, O.V.; Brenna, J.T.; Stabler, S.P.; Allen, R.H.; Gregory, J.F.; Caudill, M.A. Pregnancy Alters Choline Dynamics: Results of a Randomized Trial Using Stable Isotope Methodology in Pregnant and Nonpregnant Women. Am. J. Clin. Nutr. 2013, 98, 1459–1467. [Google Scholar] [CrossRef]
- Feldstein, A.E.; Werneburg, N.W.; Canbay, A.; Guicciardi, M.E.; Bronk, S.F.; Rydzewski, R.; Burgart, L.J.; Gores, G.J. Free Fatty Acids Promote Hepatic Lipotoxicity by Stimulating TNF-Alpha Expression via a Lysosomal Pathway. Hepatology 2004, 40, 185–194. [Google Scholar] [CrossRef]
- Gómez-Lechón, M.J.; Donato, M.T.; Martínez-Romero, A.; Jiménez, N.; Castell, J.V.; O’Connor, J.-E. A Human Hepatocellular in Vitro Model to Investigate Steatosis. Chem. Biol. Interact. 2007, 165, 106–116. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, L.; Gurley, E.; Studer, E.; Shang, J.; Wang, T.; Wang, C.; Yan, M.; Jiang, Z.; Hylemon, P.B.; et al. Prevention of Free Fatty Acid-Induced Hepatic Lipotoxicity by 18beta-Glycyrrhetinic Acid through Lysosomal and Mitochondrial Pathways. Hepatology 2008, 47, 1905–1915. [Google Scholar] [CrossRef]
- Sundler, R.; Akesson, B.; Nilsson, A. Effect of Different Fatty Acids on Glycerolipid Synthesis in Isolated Rat Hepatocytes. J. Biol. Chem. 1974, 249, 5102–5107. [Google Scholar] [CrossRef]
- Homan, R.; Grossman, J.E.; Pownall, H.J. Differential Effects of Eicosapentaenoic Acid and Oleic Acid on Lipid Synthesis and Secretion by HepG2 Cells. J. Lipid Res. 1991, 32, 231–241. [Google Scholar] [CrossRef]
- Mato, J.M.; Martínez-Chantar, M.L.; Lu, S.C. S-Adenosylmethionine Metabolism and Liver Disease. Ann. Hepatol. 2013, 12, 183–189. [Google Scholar] [CrossRef]
- Karpe, F.; Dickmann, J.R.; Frayn, K.N. Fatty Acids, Obesity, and Insulin Resistance: Time for a Reevaluation. Diabetes 2011, 60, 2441–2449. [Google Scholar] [CrossRef] [PubMed]
- Bergman, R.; Ader, M. Free Fatty Acids and Pathogenesis of Type 2 Diabetes Mellitus. Trends Endocrinol. Metab. 2000, 11, 351–356. [Google Scholar] [CrossRef]
- Svedberg, J.; Strömblad, G.; Wirth, A.; Smith, U.; Björntorp, P. Fatty Acids in the Portal Vein of the Rat Regulate Hepatic Insulin Clearance. J. Clin. Investig. 1991, 88, 2054–2058. [Google Scholar] [CrossRef]
- Barak, A.J.; Beckenhauer, H.C.; Badakhsh, S.; Tuma, D.J. The Effect of Betaine in Reversing Alcoholic Steatosis. Alcohol. Clin. Exp. Res. 1997, 21, 1100–1102. [Google Scholar] [CrossRef]
- Lu, S.C.; Huang, Z.-Z.; Yang, H.; Mato, J.M.; Avila, M.A.; Tsukamoto, H. Changes in Methionine Adenosyltransferase and S-Adenosylmethionine Homeostasis in Alcoholic Rat Liver. Am. J. Physiol. Gastrointest. Liver Physiol. 2000, 279, G178–G185. [Google Scholar] [CrossRef]
- Cano, A.; Buqué, X.; Martínez-Uña, M.; Aurrekoetxea, I.; Menor, A.; García-Rodríguez, J.L.; Lu, S.C.; Martínez-Chantar, M.L.; Mato, J.M.; Ochoa, B.; et al. Methionine Adenosyltransferase 1A Gene Deletion Disrupts Hepatic Very Low-Density Lipoprotein Assembly in Mice. Hepatology 2011, 54, 1975–1986. [Google Scholar] [CrossRef]
- Dahlhoff, C.; Desmarchelier, C.; Sailer, M.; Fürst, R.W.; Haag, A.; Ulbrich, S.E.; Hummel, B.; Obeid, R.; Geisel, J.; Bader, B.L.; et al. Hepatic Methionine Homeostasis Is Conserved in C57BL/6N Mice on High-Fat Diet Despite Major Changes in Hepatic One-Carbon Metabolism. PLoS ONE 2013, 8, e57387. [Google Scholar] [CrossRef]
- Dahlhoff, C.; Worsch, S.; Sailer, M.; Hummel, B.A.; Fiamoncini, J.; Uebel, K.; Obeid, R.; Scherling, C.; Geisel, J.; Bader, B.L.; et al. Methyl-Donor Supplementation in Obese Mice Prevents the Progression of NAFLD, Activates AMPK and Decreases Acyl-Carnitine Levels. Mol. Metab. 2014, 3, 565–580. [Google Scholar] [CrossRef]
- Day, C.P.; James, O.F. Steatohepatitis: A Tale of Two “Hits”? Gastroenterology 1998, 114, 842–845. [Google Scholar] [CrossRef]
- Kalhan, S.C.; Edmison, J.; Marczewski, S.; Dasarathy, S.; Gruca, L.L.; Bennett, C.; Duenas, C.; Lopez, R. Methionine and Protein Metabolism in Non-Alcoholic Steatohepatitis: Evidence for Lower Rate of Transmethylation of Methionine. Clin. Sci. 2011, 121, 179–189. [Google Scholar] [CrossRef]
- Ahrens, M.; Ammerpohl, O.; von Schönfels, W.; Kolarova, J.; Bens, S.; Itzel, T.; Teufel, A.; Herrmann, A.; Brosch, M.; Hinrichsen, H.; et al. DNA Methylation Analysis in Nonalcoholic Fatty Liver Disease Suggests Distinct Disease-Specific and Remodeling Signatures after Bariatric Surgery. Cell Metab. 2013, 18, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Mato, J.M.; Martínez-Chantar, M.L.; Lu, S.C. Methionine Metabolism and Liver Disease. Annu. Rev. Nutr. 2008, 28, 273–293. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.C.; Alvarez, L.; Huang, Z.-Z.; Chen, L.; An, W.; Corrales, F.J.; Avila, M.A.; Kanel, G.; Mato, J.M. Methionine Adenosyltransferase 1A Knockout Mice Are Predisposed to Liver Injury and Exhibit Increased Expression of Genes Involved in Proliferation. Proc. Natl. Acad. Sci. USA 2001, 98, 5560–5565. [Google Scholar] [CrossRef] [PubMed]
- Foretz, M.; Guichard, C.; Ferré, P.; Foufelle, F. Sterol Regulatory Element Binding Protein-1c Is a Major Mediator of Insulin Action on the Hepatic Expression of Glucokinase and Lipogenesis-Related Genes. Proc. Natl. Acad. Sci. USA 1999, 96, 12737–12742. [Google Scholar] [CrossRef] [PubMed]
- Karaskov, E.; Scott, C.; Zhang, L.; Teodoro, T.; Ravazzola, M.; Volchuk, A. Chronic Palmitate but Not Oleate Exposure Induces Endoplasmic Reticulum Stress, Which May Contribute to INS-1 Pancreatic Beta-Cell Apoptosis. Endocrinology 2006, 147, 3398–3407. [Google Scholar] [CrossRef]
- Meikle, P.J.; Wong, G.; Tsorotes, D.; Barlow, C.K.; Weir, J.M.; Christopher, M.J.; MacIntosh, G.L.; Goudey, B.; Stern, L.; Kowalczyk, A.; et al. Plasma Lipidomic Analysis of Stable and Unstable Coronary Artery Disease. Arter. Thromb. Vasc. Biol. 2011, 31, 2723–2732. [Google Scholar] [CrossRef]
- Weir, J.M.; Wong, G.; Barlow, C.K.; Greeve, M.A.; Kowalczyk, A.; Almasy, L.; Comuzzie, A.G.; Mahaney, M.C.; Jowett, J.B.M.; Shaw, J.; et al. Plasma Lipid Profiling in a Large Population-Based Cohort. J. Lipid Res. 2013, 54, 2898–2908. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.-Y.; Watt, M.J.; Rensen, S.; Greve, J.W.; Huynh, K.; Jayawardana, K.S.; Meikle, P.J.; Meex, R.C.R. Mitochondrial Dysfunction-Related Lipid Changes Occur in Nonalcoholic Fatty Liver Disease Progression. J. Lipid Res. 2018, 59, 1977–1986. [Google Scholar] [CrossRef] [PubMed]
- Huynh, K.; Barlow, C.K.; Jayawardana, K.S.; Weir, J.M.; Mellett, N.A.; Cinel, M.; Magliano, D.J.; Shaw, J.E.; Drew, B.G.; Meikle, P.J. High-Throughput Plasma Lipidomics: Detailed Mapping of the Associations with Cardiometabolic Risk Factors. Cell Chem. Biol. 2019, 26, 71–84.e4. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, K.-Y.; Barlow, C.K.; Kammoun, H.; Mellett, N.A.; Weir, J.M.; Murphy, A.J.; Febbraio, M.A.; Meikle, P.J. Stable Isotopic Tracer Phospholipidomics Reveals Contributions of Key Phospholipid Biosynthetic Pathways to Low Hepatocyte Phosphatidylcholine to Phosphatidylethanolamine Ratio Induced by Free Fatty Acids. Metabolites 2021, 11, 188. https://doi.org/10.3390/metabo11030188
Peng K-Y, Barlow CK, Kammoun H, Mellett NA, Weir JM, Murphy AJ, Febbraio MA, Meikle PJ. Stable Isotopic Tracer Phospholipidomics Reveals Contributions of Key Phospholipid Biosynthetic Pathways to Low Hepatocyte Phosphatidylcholine to Phosphatidylethanolamine Ratio Induced by Free Fatty Acids. Metabolites. 2021; 11(3):188. https://doi.org/10.3390/metabo11030188
Chicago/Turabian StylePeng, Kang-Yu, Christopher K Barlow, Helene Kammoun, Natalie A Mellett, Jacquelyn M Weir, Andrew J Murphy, Mark A Febbraio, and Peter J Meikle. 2021. "Stable Isotopic Tracer Phospholipidomics Reveals Contributions of Key Phospholipid Biosynthetic Pathways to Low Hepatocyte Phosphatidylcholine to Phosphatidylethanolamine Ratio Induced by Free Fatty Acids" Metabolites 11, no. 3: 188. https://doi.org/10.3390/metabo11030188
APA StylePeng, K.-Y., Barlow, C. K., Kammoun, H., Mellett, N. A., Weir, J. M., Murphy, A. J., Febbraio, M. A., & Meikle, P. J. (2021). Stable Isotopic Tracer Phospholipidomics Reveals Contributions of Key Phospholipid Biosynthetic Pathways to Low Hepatocyte Phosphatidylcholine to Phosphatidylethanolamine Ratio Induced by Free Fatty Acids. Metabolites, 11(3), 188. https://doi.org/10.3390/metabo11030188







