Analyses of Lipid A Diversity in Gram-Negative Intestinal Bacteria Using Liquid Chromatography–Quadrupole Time-of-Flight Mass Spectrometry
Abstract
:1. Introduction
2. Results
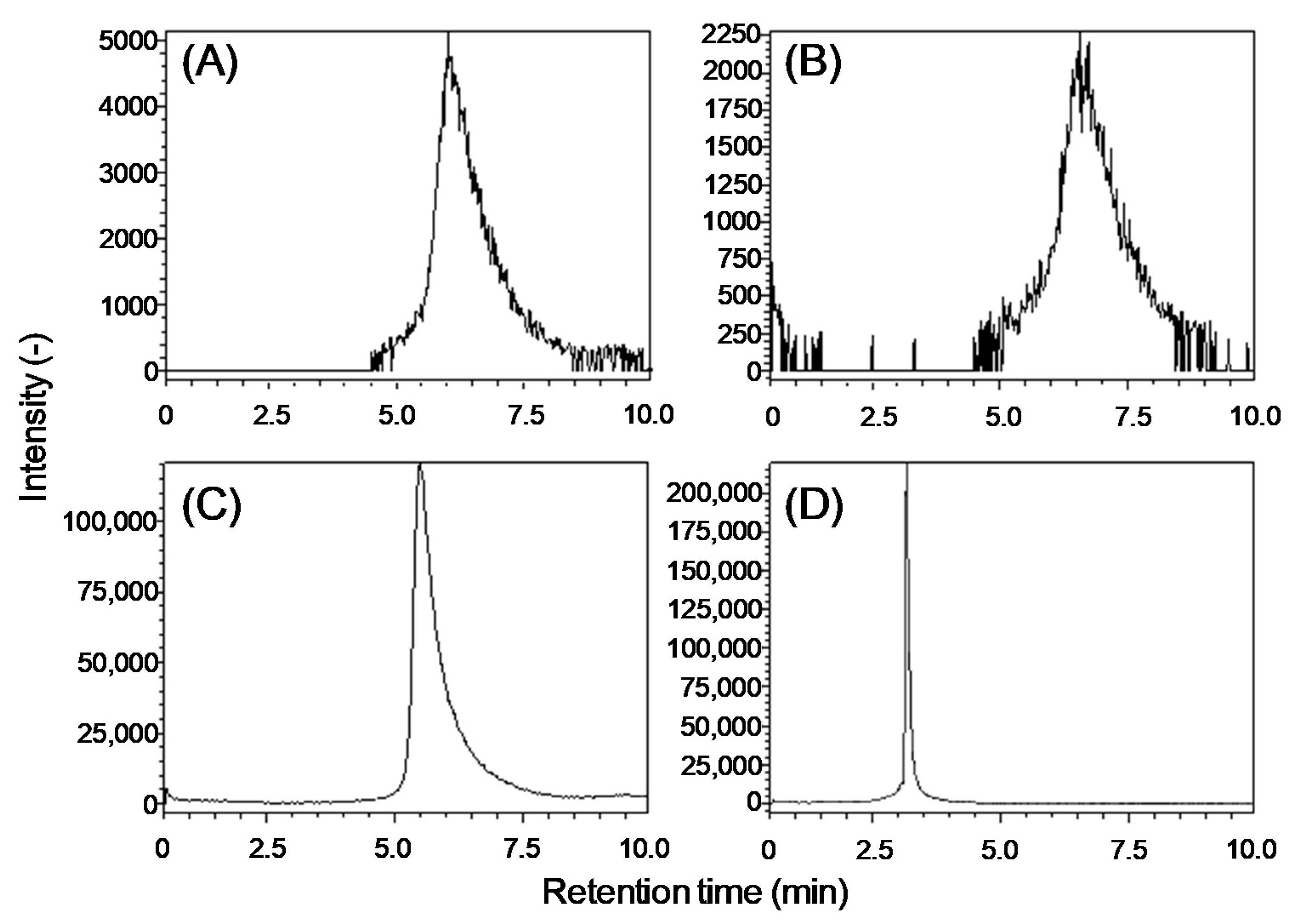
2.1. Development of an Analytical Method for Lipid A
2.2. Analysis of Lipid A in E. coli Wild Type and Gene Knockout Strains
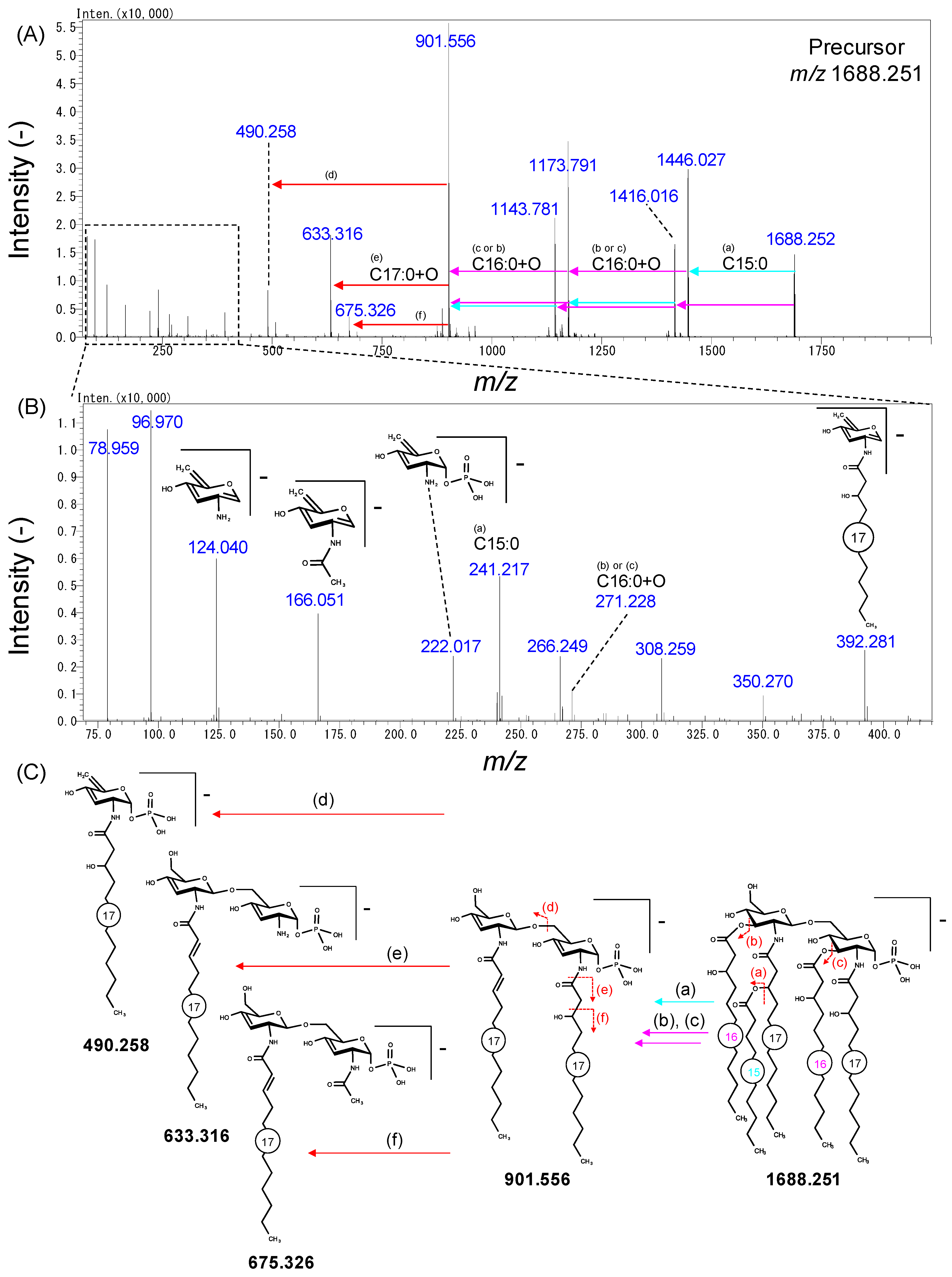
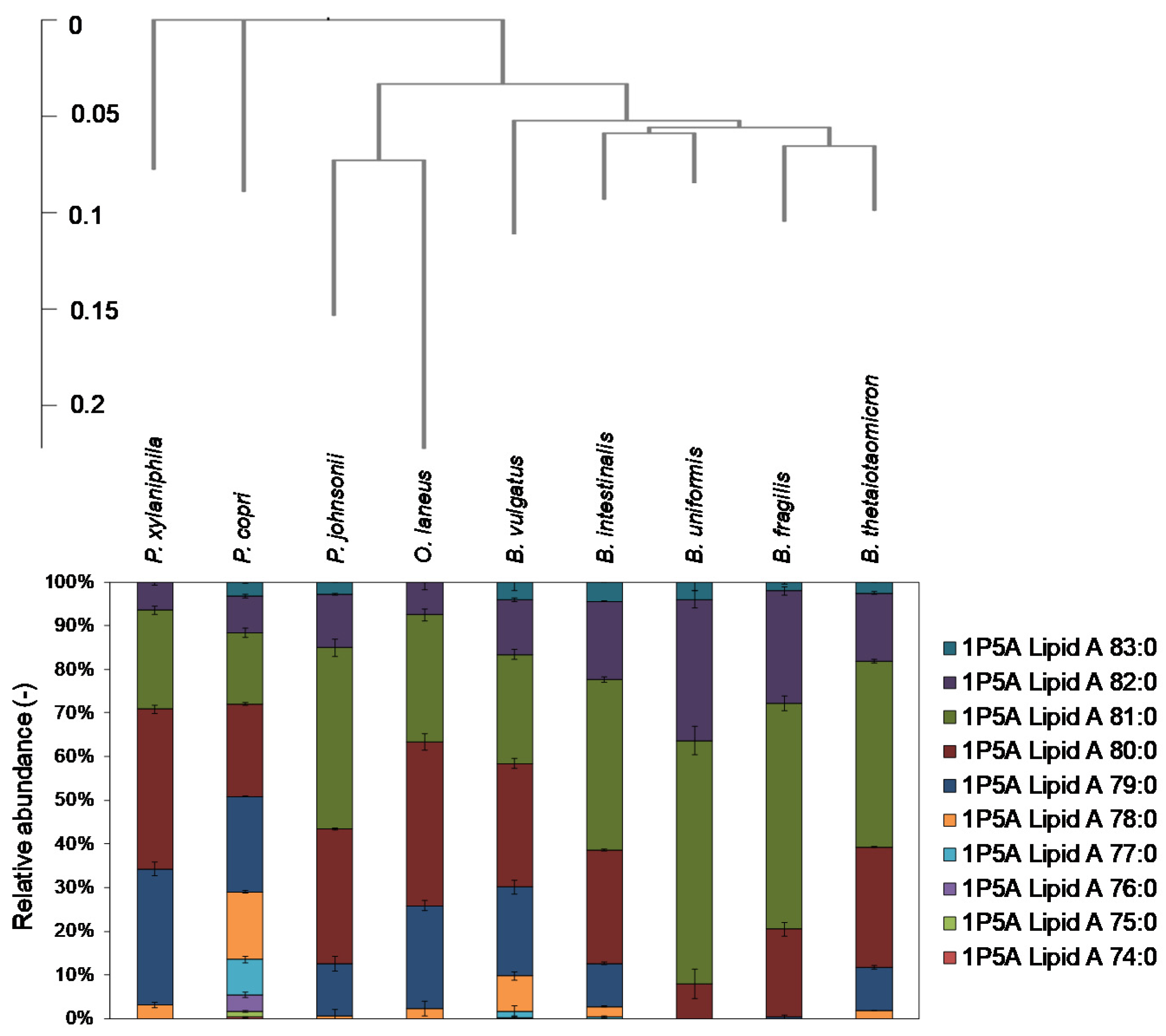
2.3. Analysis of Lipid A in Bacteroidetes Bacteria
3. Discussion
4. Materials and Methods
4.1. Strains and Cultures
4.2. Lipid A Extraction and Analysis
4.3. Creation of Phylogenic Tree
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miller, S.I.; Ernst, R.K.; Bader, M.W. LPS, TLR4 and infectious disease diversity. Nat. Rev. Microbiol. 2005, 3, 36–46. [Google Scholar] [CrossRef]
- Steimle, A.; Autenrieth, I.B.; Frick, J.S. Structure and function: Lipid A modifications in commensals and pathogens. Int. J. Med. Microbiol. 2016, 306, 290–301. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Quinn, P.J.; Yan, A. Kdo2-lipid A: Structural diversity and impact on immunopharmacology. Biol. Rev. 2015, 90, 408–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, I.; Blaser, M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef] [Green Version]
- Schirmer, M.; Garner, A.; Vlamakis, H.; Xavier, R.J. Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 2019, 17, 497–511. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D. Toll-like Receptors and Inflammatory Bowel Disease. Front. Immunol. 2018, 9, 72. [Google Scholar] [CrossRef] [Green Version]
- Fukata, M.; Chen, A.; Vamadevan, A.S.; Cohen, J.; Breglio, K.; Krishnareddy, S.; Hsu, D.; Xu, R.; Harpaz, N.; Dannenberg, A.J.; et al. Toll-Like Receptor-4 Promotes the Development of Colitis-Associated Colorectal Tumors. Gastroenterology 2007, 133, 1869–1881. [Google Scholar] [CrossRef] [Green Version]
- Huang, N.Q.; Jin, H.; Zhou, S.Y.; Shi, J.S.; Jin, F. TLR4 is a link between diabetes and Alzheimer’s disease. Behav. Brain Res. 2017, 316, 234–244. [Google Scholar] [CrossRef]
- Perez-Pardo, P.; Dodiya, H.B.; Engen, P.A.; Forsyth, C.B.; Huschens, A.M.; Shaikh, M.; Voigt, R.M.; Naqib, A.; Green, S.J.; Kordower, J.H.; et al. Role of TLR4 in the gut-brain axis in Parkinson’s disease: A translational study from men to mice. Gut 2018, 829–843. [Google Scholar] [CrossRef]
- Hennezel, E.; Abubucker, S.; Murphy, L.O.; Cullen, T.W. Total Lipopolysaccharide from the Human Gut Microbiome Silences Toll-Like Receptor Signaling. mSystems 2017, 2, e00046-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vatanen, T.; Kostic, A.D.; D’Hennezel, E.; Siljander, H.; Franzosa, E.A.; Yassour, M.; Kolde, R.; Vlamakis, H.; Arthur, T.D.; Hämäläinen, A.M.; et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 2016, 165, 842–853. [Google Scholar] [CrossRef] [Green Version]
- Weintraub, A.; Zäharinger, U.; Wollenweber, H.-W.; Seydel, U.; Rietschel, E.T. Structural characterization of the lipid A component of Bacteroides fragilis strain NCTC 9343 lipopolysaccharide. Eur. J. Biochem. 1989, 183, 425–431. [Google Scholar] [CrossRef]
- Lindberg, A.A.; Weintraub, A.; Zähringer, U.; Rietschel, E.T. Structure-activity relationships in Lipopolysaccharides of bacteroides fragilis. Rev. Infect. Dis. 1990, 12, S133–S141. [Google Scholar] [CrossRef]
- Guo, L.; Lim, K.B.; Poduje, C.M.; Daniel, M.; Gunn, J.S.; Hackett, M.; Miller, S.I. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 1998, 95, 189–198. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.W.; Shaffer, S.A.; Ernst, R.K.; Goodlett, D.R.; Tureček, F. Determination of pyrophosphorylated forms of lipid A in Gram-negative bacteria using a multivaried mass spectrometric approach. Proc. Natl. Acad. Sci. USA 2008, 105, 12742–12747. [Google Scholar] [CrossRef] [Green Version]
- Zhou, P.; Altman, E.; Perry, M.B.; Li, J. Study of matrix additives for sensitive analysis of lipid a by matrix-assisted laser desorption ionization mass spectrometry. Appl. Environ. Microbiol. 2010, 76, 3437–3443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larrouy-Maumus, G.; Clements, A.; Filloux, A.; McCarthy, R.R.; Mostowy, S. Direct detection of lipid A on intact Gram-negative bacteria by MALDI-TOF mass spectrometry. J. Microbiol. Methods 2016, 120, 68–71. [Google Scholar] [CrossRef] [Green Version]
- Kussak, A.; Weintraub, A. Quadrupole ion-trap mass spectrometry to locate fatty acids on lipid A from Gram-negative bacteria. Anal. Biochem. 2002, 307, 131–137. [Google Scholar] [CrossRef]
- Sforza, S.; Silipo, A.; Molinaro, A.; Marchelli, R.; Parrilli, M.; Lanzetta, R. Determination of fatty acid positions in native lipid A by positive and negative electrospray ionization mass spectrometry. J. Mass Spectrom. 2004, 39, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.H.; Garrett, T.A.; Reynolds, C.M.; Shaw, W.A.; Moore, J.D.; Smith, D.C.; Ribeiro, A.A.; Murphy, R.C.; Ulevitch, R.J.; Fearns, C.; et al. Kdo2-Lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J. Lipid Res. 2006, 47, 1097–1111. [Google Scholar] [CrossRef] [Green Version]
- Sándor, V.; Dörnyei, Á.; Makszin, L.; Kilár, F.; Péterfi, Z.; Kocsis, B.; Kilár, A. Characterization of complex, heterogeneous lipid A samples using HPLC–MS/MS technique I. Overall analysis with respect to acylation, phosphorylation and isobaric distribution. J. Mass Spectrom. 2016, 51, 1043–1063. [Google Scholar] [CrossRef]
- Wakamatsu, A.; Morimoto, K.; Shimizu, M.; Kudoh, S. A severe peak tailing of phosphate compounds caused by interaction with stainless steel used for liquid chromatography and electrospray mass spectrometry. J. Sep. Sci. 2005, 28, 1823–1830. [Google Scholar] [CrossRef]
- Ito, M.; Fujii, N.; Wittwer, C.; Sasaki, A.; Tanaka, M.; Bittner, T.; Jessen, H.J.; Saiardi, A.; Takizawa, S.; Nagata, E. Hydrophilic interaction liquid chromatography–tandem mass spectrometry for the quantitative analysis of mammalian-derived inositol poly/pyrophosphates. J. Chromatogr. A 2018, 1573, 87–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, E.C.; Hackett, M. Rapid isolation method for lipopolysaccharide and lipid A from Gram-negative bacteria. Analyst 2000, 125, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Ikeda, K.; Takahashi, M.; Satoh, A.; Mori, Y.; Uchino, H.; Okahashi, N.; Yamada, Y.; Tada, I.; Bonini, P.; et al. A lipidome atlas in MS-DIAL 4. Nat. Biotechnol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Bishop, R.E.; Gibbons, H.S.; Guina, T.; Trent, M.S.; Miller, S.I.; Raetz, C.R.H. Transfer of palmitate from phospholipids to lipid A in outer membranes of Gram-negative bacteria. EMBO J. 2000, 19, 5071–5080. [Google Scholar] [CrossRef]
- Vorachek-Warren, M.K.; Ramirez, S.; Cotter, R.J.; Raetz, C.R.H. A triple mutant of Escherichia coli lacking secondary acyl chains on lipid A. J. Biol. Chem. 2002, 277, 14194–14205. [Google Scholar] [CrossRef] [Green Version]
- Carty, S.M.; Sreekumar, K.R.; Raetz, C.R.H. Effect of cold shock on lipid a biosynthesis in Escherichia coli: Induction at 12 °C of an acyltransferase specific for palmitoleoyl-acyl carrier protein. J. Biol. Chem. 1999, 274, 9677–9685. [Google Scholar] [CrossRef] [Green Version]
- Berezow, A.B.; Ernst, R.K.; Coats, S.R.; Braham, P.H.; Karimi-Naser, L.M.; Darveau, R.P. The structurally similar, penta-acylated lipopolysaccharides of Porphyromonas gingivalis and Bacteroides elicit strikingly different innate immune responses. Microb. Pathog. 2009, 47, 68–77. [Google Scholar] [CrossRef] [Green Version]
- Jacobson, A.N.; Choudhury, B.P.; Fischbach, M.A. The biosynthesis of lipooligosaccharide from Bacteroides thetaiotaomicron. mBio 2018, 9, e02289-17. [Google Scholar] [CrossRef] [Green Version]
- Park, H.G.; Sathiyanarayanan, G.; Hwang, C.H.; Ann, D.H.; Kim, J.H.; Bang, G.; Jang, K.S.; Ryu, H.W.; Lee, Y.K.; Yang, Y.H.; et al. Chemical Structure of the Lipid A component of Pseudomonas sp. strain PAMC 28618 from Thawing Permafrost in Relation to Pathogenicity. Sci. Rep. 2017, 7, 2168. [Google Scholar] [CrossRef]
- Akram, A.R.; Chankeshwara, S.V.; Scholefield, E.; Aslam, T.; McDonald, N.; Megia-Fernandez, A.; Marshall, A.; Mills, B.; Avlonitis, N.; Craven, T.H.; et al. In situ identification of Gram-negative bacteria in human lungs using a topical fluorescent peptide targeting lipid A. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef] [Green Version]
- Sorensen, M.; Chandler, C.E.; Gardner, F.M.; Ramadan, S.; Khot, P.D.; Leung, L.M.; Farrance, C.E.; Goodlett, D.R.; Ernst, R.K.; Nilsson, E. Rapid microbial identification and colistin resistance detection via MALDI-TOF MS using a novel on-target extraction of membrane lipids. Sci. Rep. 2020, 10, 21536. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [Green Version]
- Lemoine, F.; Correia, D.; Lefort, V.; Doppelt-Azeroual, O.; Mareuil, F.; Cohen-Boulakia, S.; Gascuel, O. NGPhylogeny.fr: New generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019, 47, W260–W265. [Google Scholar] [CrossRef] [PubMed] [Green Version]

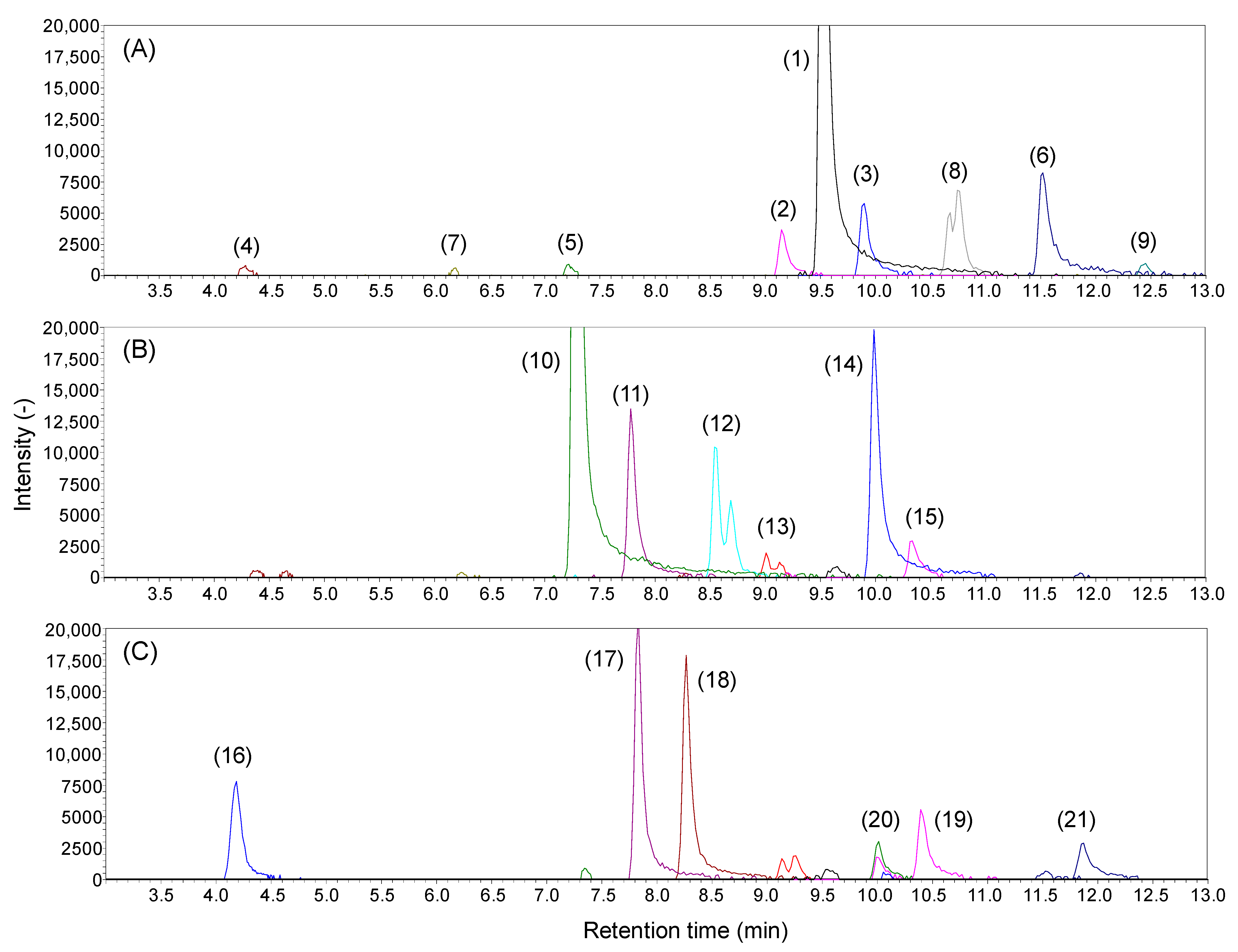


| Peak ID | Retention Time (min) | Measured m/z | Ion Types | Compounds | Class | Acyl Property | |||
|---|---|---|---|---|---|---|---|---|---|
| C3′ | C2′ | C3 | C2 | ||||||
| 7 | 6.2 | 1279.848 | [M−H]− | Lipid A 54:0 | 1P4A *1 | N.D. *2 | |||
| 4 | 4.3 | 1359.813 | [M−H]− | Lipid A 54:0 | 2P4A | OH | C14:0(O-C12:0) | C14:0(OH) | C14:0(OH) |
| 16 | 4.2 | 1403.841 | [M−H]− | Lipid A 56:0 | 2P4A | C14:0(OH) | C14:0(OH) | C14:0(OH) | C14:0(OH) |
| 12 | 8.6 and 8.7 | 1506.041 | [M−H]− | Lipid A 68:0 | 1P5A | C14:0(OH) | C14:0(O-C12:0) | C14:0(OH) | C14:0(OH) |
| 13 | 9.0 and 9.2 | 1534.072 | [M−H]− | Lipid A 70:0 | 1P5A | C14:0(OH) | C14:0(O-C14:0) | C14:0(OH) | C14:0(OH) |
| 5 and 10 | 7.3 | 1586.004 | [M−H]− | Lipid A 68:0 | 2P5A | C14:0(OH) | C14:0(O-C12:0) | C14:0(OH) | C14:0(OH) |
| 11 | 7.8 | 1614.038 | [M−H]− | Lipid A 70:0 | 2P5A | C14:0(OH) | C14:0(O-C14:0) | C14:0(OH) | C14:0(OH) |
| 17 | 7.8 | 1614.038 | [M−H]− | Lipid A 70:0 | 2P5A | C14:0(O-C14:0) | C14:0(OH) | C14:0(OH) | C14:0(OH) |
| 18 | 8.3 | 1642.067 | [M−H]− | Lipid A 72:0 | 2P5A | C14:0(OH) | C14:0(OH) | C14:0(OH) | C14:0(O-C16:0) |
| 8 | 10.7 and 10.8 | 1716.237 | [M−H]− | Lipid A 82:0 | 1P6A | C14:0(O-C14:0) | C14:0(O-C12:0) | C14:0(OH) | C14:0(OH) |
| 2 | 9.2 | 1768.171 | [M−H]− | Lipid A 80:0 | 2P6A | C14:0(O-C12:0) | C14:0(O-C12:0) | C14:0(OH) | C14:0(OH) |
| 1 | 9.5 | 1796.203 | [M−H]− | Lipid A 82:0 | 2P6A | C14:0(O-C14:0) | C14:0(O-C12:0) | C14:0(OH) | C14:0(OH) |
| 3 | 9.9 | 1824.231 | [M−H]− | Lipid A 84:0 | 2P6A | C14:0(O-C14:0) | C14:0(O-C14:0) | C14:0(OH) | C14:0(OH) |
| 14 | 10.1 | 1824.231 | [M−H]− | Lipid A 84:0 | 2P6A | C14:0(OH) | C14:0(O-C12:0) | C14:0(OH) | C14:0(O-C16:0) |
| 20 | 10.0 | 1850.250 | [M−H]− | Lipid A 86:1 | 2P6A | C14:0(O-C14:0) | C14:0(O-C16:1) | C14:0(OH) | C14:0(OH) |
| 15 | 10.4 | 1852.263 | [M−H]− | Lipid A 86:0 | 2P6A | C14:0(OH) | C14:0(O-C14:0) | C14:0(OH) | C14:0(O-C16:0) |
| 19 | 10.4 | 1852.263 | [M−H]− | Lipid A 86:0 | 2P6A | C14:0(O-C14:0) | C14:0(OH) | C14:0(OH) | C14:0(O-C16:0) |
| 9 | 12.4 | 1954.465 | [M−H]− | Lipid A 98:0 | 1P7A *1 | N.D. *2 | |||
| 6 | 11.5 | 1016.712 | [M−2H]2− | Lipid A 98:0 | 2P7A | N.D. *3 | |||
| 21 | 11.9 | 1043.736 | [M−2H]2− | Lipid A 102:1 | 2P7A *4 | N.D. *2 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okahashi, N.; Ueda, M.; Matsuda, F.; Arita, M. Analyses of Lipid A Diversity in Gram-Negative Intestinal Bacteria Using Liquid Chromatography–Quadrupole Time-of-Flight Mass Spectrometry. Metabolites 2021, 11, 197. https://doi.org/10.3390/metabo11040197
Okahashi N, Ueda M, Matsuda F, Arita M. Analyses of Lipid A Diversity in Gram-Negative Intestinal Bacteria Using Liquid Chromatography–Quadrupole Time-of-Flight Mass Spectrometry. Metabolites. 2021; 11(4):197. https://doi.org/10.3390/metabo11040197
Chicago/Turabian StyleOkahashi, Nobuyuki, Masahiro Ueda, Fumio Matsuda, and Makoto Arita. 2021. "Analyses of Lipid A Diversity in Gram-Negative Intestinal Bacteria Using Liquid Chromatography–Quadrupole Time-of-Flight Mass Spectrometry" Metabolites 11, no. 4: 197. https://doi.org/10.3390/metabo11040197
APA StyleOkahashi, N., Ueda, M., Matsuda, F., & Arita, M. (2021). Analyses of Lipid A Diversity in Gram-Negative Intestinal Bacteria Using Liquid Chromatography–Quadrupole Time-of-Flight Mass Spectrometry. Metabolites, 11(4), 197. https://doi.org/10.3390/metabo11040197






