Optical Microscopy-Guided Laser Ablation Electrospray Ionization Ion Mobility Mass Spectrometry: Ambient Single Cell Metabolomics with Increased Confidence in Molecular Identification
Abstract
1. Introduction
2. Results and Discussion
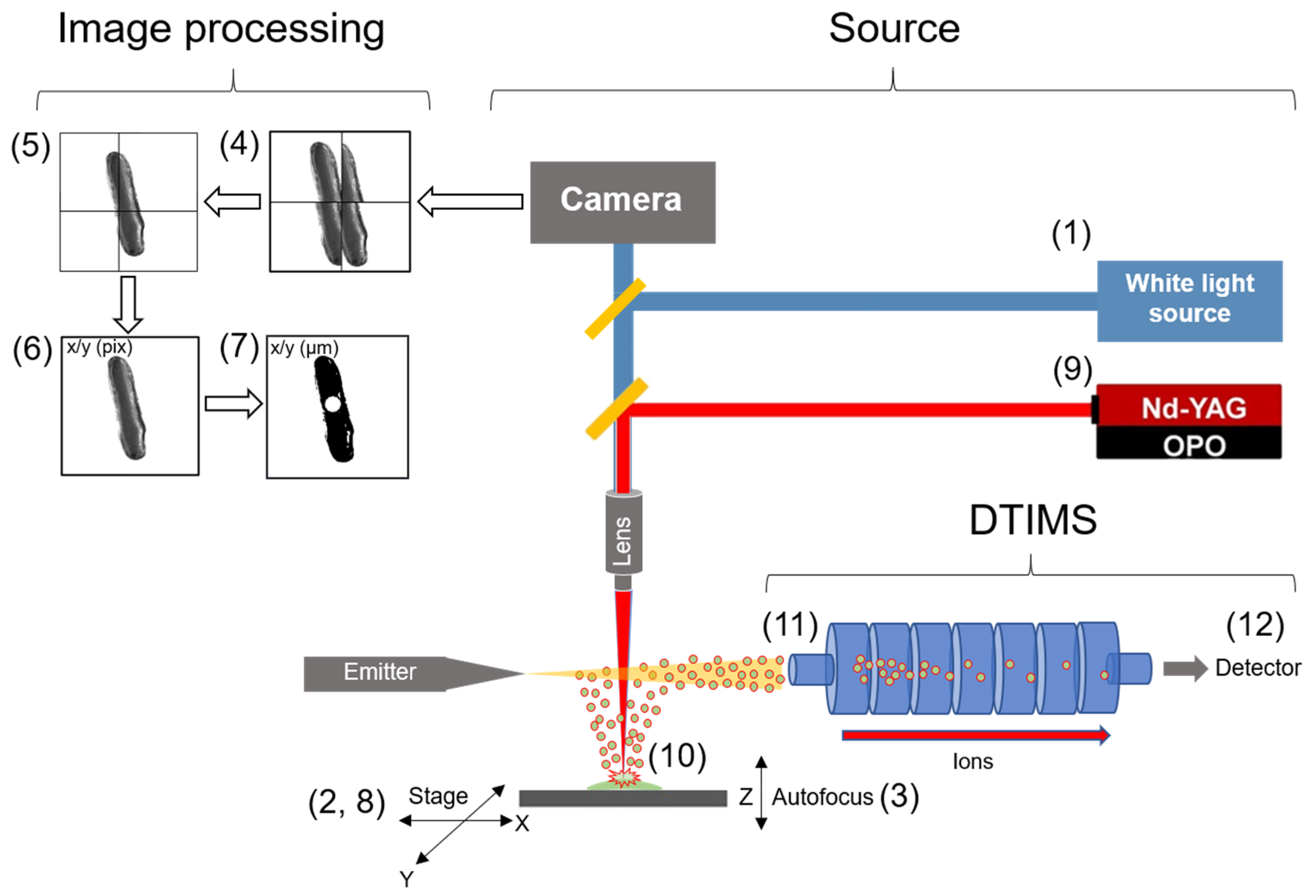
2.1. Optimization of the Molecular Microscope Sampling Modality
2.2. Configuration of the Optical Train for Single Cell Identification and Ablation
2.3. LAESI-DTIMS Analysis of Single A. Cepa Epidermal Cells
2.4. Mobility Separation of Isomeric Species in Single A. Cepa Epidermal Cells
| Species | Structure | Observed Mass (m/z) | Expected Mass (m/z) | Deviation (ppm) | DTCCS (Å2) |
|---|---|---|---|---|---|
| Tyrosine 1,3,5 | C9H11NO3 + H | 182.082 | 182.082 | 0.204 | (II) 146.42 |
| Alliin 3,4 | C6H11NO3S + Na | 200.035 | 200.035 | 1.724 | 144.15 |
| Lauric acid 2,5 | C12H24O2 + H | 201.184 | 201.184 | 1.827 | 154.01 |
| Monosaccharide 1,2,5 | C6H12O6 + K | 219.026 | 219.026 | 2.077 | 146.42 |
| Major (unknown) | n/a | 261.144 | n/a | n/a | 144.15 |
| Indigotin 6 | C16H10N2O2 + H | 263.084 | 263.082 | 7.760 | 145.43 |
| Cri/(Ca/Vi) 1,6 | C16H17NO3 + H | 272.127 | 272.128 | −4.900 | (144.22/153.61) |
| Glut/cystox 6 | C9H16N2O6S + H | 281.089 | 281.089 | −1.242 | 152.54 |
| Catechin 2,6 | C15H14O6 + H | 291.084 | 291.086 | −7.158 | (I) 165.17 |
| Quercetin 6 | C15H10O7 + H | 303.052 | 303.05 | 5.231 | 175.45 |
| 14-EET 6 | C20H32O3 + H | 321.247 | 321.247 | 1.037 | 181.24 |
| Erosone 6 | C20H16O6 + H | 353.102 | 353.102 | 0.377 | 154.89 |
| Unknown | n/a | 362.097 | n/a | n/a | 199.11 |
| Disaccharide 2,5 | C12H22O11 + Na | 365.105 | 365.105 | 1.096 | (I, II) 178.48 |
| Disaccharide 1,2,5 | C12H22O6 + K | 381.080 | 381.079 | 2.100 | (II) 180.54 |
| Thy-diphosphate 5 | C12H19N4O7P2S | 425.045 | 425.046 | −1.178 | (II) 182.21 |
| Cya-ramnoside 5 | C21H24O9 + H | 434.120 | 434.121 | −1.844 | 161.61 |
| Davidioside 6 | C21H24O9 + Na | 443.132 | 443.131 | 1.130 | 169.62 |
| Major (unknown) | n/a | 524.148 | n/a | n/a | 201.79 |
| Trisaccharide 1,2,5,6 | C18H32O16 + Na | 527.157 | 527.157 | 0.731 | (I) 215.94 |
| Trisaccharide 1,5,6 | C18H32O16 + K | 543.131 | 543.132 | −1.415 | (II) 217.26 |
| Major (unknown) | n/a | 558.115 | n/a | n/a | 213.87 |
| Gustroside 6 | C27H34O14 + Na | 605.183 | 605.184 | −2.067 | 194.57 |
| AsnGluGln 6 | C25H33N7O9 + K | 614.192 | 614.197 | −7.463 | 196.50 |
| Oligosacharide 6 | C24H42O21 + Na | 689.210 | 689.211 | −0.792 | 231.66 |
| Major (unknown) | n/a | 695.229 | n/a | n/a | 201.81 |
| Oligosacharide 1,6 | C24H42O21 + K | 705.182 | 705.185 | −4.930 | 236.69 |
| Major (unknown) | n/a | 720.192 | n/a | n/a | 240.71 |
2.5. Comparison of In Situ Single Cell and Bulk Analysis
2.6. Variance in Sugar Abundance within Cell Population
3. Materials and Methods
3.1. Materials
3.2. Instrument Design
3.3. Sensitivity Measurements
3.4. Optimizing LAESI-DTIMS Analysis
3.5. A. Cepa Cell Analysis
3.6. MPLEx Preparation and Analysis
3.7. Data Analysis & Software
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Macaulay, I.C.; Ponting, C.P.; Voet, T. Single-Cell Multiomics: Multiple Measurements from Single Cells. Trends Genet. 2017, 33, 155–168. [Google Scholar] [CrossRef]
- Taylor, M.J.; Lukowski, J.K.; Anderton, C.R. Spatially Resolved Mass Spectrometry at the Single Cell: Recent Innovations in Proteomics and Metabolomics. J. Am. Soc. Mass Spectrom. 2021. [Google Scholar] [CrossRef]
- Choi, H.; Yang, Z.; Weisshaar, J.C. Single-cell, real-time detection of oxidative stress induced inEscherichia coliby the antimicrobial peptide CM15. Proc. Natl. Acad. Sci. USA 2015, 112, E303–E310. [Google Scholar] [CrossRef] [PubMed]
- Brock, A.; Chang, H.; Huang, S. Non-genetic heterogeneity—A mutation-independent driving force for the somatic evolution of tumours. Nat. Rev. Genet. 2009, 10, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; Van Oudenaarden, A. Nature, Nurture, or Chance: Stochastic Gene Expression and Its Consequences. Cell 2008, 135, 216–226. [Google Scholar] [CrossRef]
- Wilson, N.K.; Kent, D.G.; Buettner, F.; Shehata, M.; Macaulay, I.C.; Calero-Nieto, F.J.; Castillo, M.S.; Oedekoven, C.A.; Diamanti, E.; Schulte, R.; et al. Combined Single-Cell Functional and Gene Expression Analysis Resolves Heterogeneity within Stem Cell Populations. Cell Stem Cell 2015, 16, 712–724. [Google Scholar] [CrossRef] [PubMed]
- Bendall, S.C.; Simonds, E.F.; Qiu, P.; Amir, E.-A.D.; Krutzik, P.O.; Finck, R.; Bruggner, R.V.; Melamed, R.; Trejo, A.; Ornatsky, O.I.; et al. Single-Cell Mass Cytometry of Differential Immune and Drug Responses Across a Human Hematopoietic Continuum. Science 2011, 332, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Neumann, E.K.; Ellis, J.F.; Triplett, A.E.; Rubakhin, S.S.; Sweedler, J.V. Lipid Analysis of 30 000 Individual Rodent Cerebellar Cells Using High-Resolution Mass Spectrometry. Anal. Chem. 2019, 91, 7871–7878. [Google Scholar] [CrossRef]
- Neumann, E.K.; Comi, T.J.; Rubakhin, S.S.; Sweedler, J.V. Lipid Heterogeneity between Astrocytes and Neurons Revealed by Single-Cell MALDI-MS Combined with Immunocytochemical Classification. Angew. Chem. 2019, 131, 5971–5975. [Google Scholar] [CrossRef]
- Amantonico, A.; Urban, P.L.; Fagerer, S.R.; Balabin, R.M.; Zenobi, R. Single-Cell MALDI-MS as an Analytical Tool for Studying Intrapopulation Metabolic Heterogeneity of Unicellular Organisms. Anal. Chem. 2010, 82, 7394–7400. [Google Scholar] [CrossRef]
- Dong, Y.; Li, B.; Malitsky, S.; Rogachev, I.; Aharoni, A.; Kaftan, F.; Svatoš, A.; Franceschi, P. Sample Preparation for Mass Spectrometry Imaging of Plant Tissues: A Review. Front. Plant Sci. 2016, 7, 60. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Lee, T.G. Biological tissue sample preparation for time-of-flight secondary ion mass spectrometry (ToF–SIMS) imaging. Nano Converg. 2018, 5, 1–13. [Google Scholar] [CrossRef]
- Van Nuffel, S.; Elie, N.; Yang, E.; Nouet, J.; Touboul, D.; Chaurand, P.; Brunelle, A. Insights into the MALDI Process after Matrix Deposition by Sublimation Using 3D ToF-SIMS Imaging. Anal. Chem. 2018, 90, 1907–1914. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhang, Y.; Ge, J.; Liu, K.; Li, P. Sample preparation for mass spectrometry imaging of leaf tissues: A case study on analyte delocalization. Anal. Bioanal. Chem. 2018, 410, 7449–7456. [Google Scholar] [CrossRef]
- Anderson, D.M.G.; Floyd, K.A.; Barnes, S.; Clark, J.M.; Clark, J.I.; Mchaourab, H.; Schey, K.L. A method to prevent protein delocalization in imaging mass spectrometry of non-adherent tissues: Application to small vertebrate lens imaging. Anal. Bioanal. Chem. 2015, 407, 2311–2320. [Google Scholar] [CrossRef] [PubMed]
- Bokhart, M.T.; Manni, J.; Garrard, K.P.; Ekelöf, M.; Nazari, M.; Muddiman, D.C. IR-MALDESI Mass Spectrometry Imaging at 50 Micron Spatial Resolution. J. Am. Soc. Mass Spectrom. 2017, 28, 2099–2107. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, B.; Sripadi, P.; Reschke, B.R.; Henderson, H.D.; Powell, M.J.; Moody, S.A.; Vertes, A. Subcellular Metabolite and Lipid Analysis of Xenopus laevis Eggs by LAESI Mass Spectrometry. PLoS ONE 2014, 9, e115173. [Google Scholar] [CrossRef] [PubMed]
- Stopka, S.A.; Agtuca, B.J.; Koppenaal, D.W.; Paša-Tolić, L.; Stacey, G.; Vertes, A.; Anderton, C.R. Laser-ablation electrospray ionization mass spectrometry with ion mobility separation reveals metabolites in the symbiotic interactions of soybean roots and rhizobia. Plant J. 2017, 91, 340–354. [Google Scholar] [CrossRef] [PubMed]
- Agtuca, B.J.; Stopka, S.A.; Evans, S.; Samarah, L.; Liu, Y.; Xu, D.; Stacey, M.G.; Koppenaal, D.W.; Paša-Tolić, L.; Anderton, C.R.; et al. Metabolomic profiling of wild-type and mutant soybean root nodules using laser-ablation electrospray ionization mass spectrometry reveals altered metabolism. Plant J. 2020, 103, 1937–1958. [Google Scholar] [CrossRef] [PubMed]
- Samarah, L.Z.; Khattar, R.; Tran, T.H.; Stopka, S.A.; Brantner, C.A.; Parlanti, P.; Veličković, D.; Shaw, J.B.; Agtuca, B.J.; Stacey, G.; et al. Single-Cell Metabolic Profiling: Metabolite Formulas from Isotopic Fine Structures in Heterogeneous Plant Cell Populations. Anal. Chem. 2020, 92, 7289–7298. [Google Scholar] [CrossRef]
- Shrestha, B.; Vertes, A. In Situ Metabolic Profiling of Single Cells by Laser Ablation Electrospray Ionization Mass Spectrometry. Anal. Chem. 2009, 81, 8265–8271. [Google Scholar] [CrossRef] [PubMed]
- Stolee, J.A.; Shrestha, B.; Mengistu, G.; Vertes, A. Observation of Subcellular Metabolite Gradients in Single Cells by Laser Ablation Electrospray Ionization Mass Spectrometry. Angew. Chem. Int. Ed. 2012, 51, 10386–10389. [Google Scholar] [CrossRef]
- Stopka, S.A.; Khattar, R.; Agtuca, B.J.; Anderton, C.R.; Paša-Tolić, L.; Stacey, G.; Vertes, A. Metabolic Noise and Distinct Subpopulations Observed by Single Cell LAESI Mass Spectrometry of Plant Cells in situ. Front. Plant Sci. 2018, 9, 9. [Google Scholar] [CrossRef]
- Kulkarni, P.; Wilschut, R.A.; Verhoeven, K.J.F.; Van Der Putten, W.H.; Garbeva, P. LAESI mass spectrometry imaging as a tool to differentiate the root metabolome of native and range-expanding plant species. Planta 2018, 248, 1515–1523. [Google Scholar] [CrossRef] [PubMed]
- Stopka, S.A.; Samarah, L.Z.; Shaw, J.B.; Liyu, A.V.; Veličković, D.; Agtuca, B.J.; Kukolj, C.; Koppenaal, D.W.; Stacey, G.; Paša-Tolić, L.; et al. Ambient Metabolic Profiling and Imaging of Biological Samples with Ultrahigh Molecular Resolution Using Laser Ablation Electrospray Ionization 21 Tesla FTICR Mass Spectrometry. Anal. Chem. 2019, 91, 5028–5035. [Google Scholar] [CrossRef]
- Qi, Y.; Fu, P.; Volmer, D.A. Analysis of natural organic matter via fourier transform ion cyclotron resonance mass spectrometry: An overview of recent non-petroleum applications. Mass Spectrom. Rev. 2020, 21634. [Google Scholar] [CrossRef] [PubMed]
- Ekelöf, M.; Dodds, J.; Khodjaniyazova, S.; Garrard, K.P.; Baker, E.S.; Muddiman, D.C. Coupling IR-MALDESI with Drift Tube Ion Mobility-Mass Spectrometry for High-Throughput Screening and Imaging Applications. J. Am. Soc. Mass Spectrom. 2020, 31, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Rivera, E.S.; Djambazova, K.V.; Neumann, E.K.; Caprioli, R.M.; Spraggins, J.M. Integrating Ion Mobility and Imaging Mass Spectrometry for Comprehensive Analysis of Biological Tissues: A brief review and perspective. J. Mass Spectrom. 2020, 55, e4614. [Google Scholar] [CrossRef]
- Shrestha, B.; Vertes, A. High-Throughput Cell and Tissue Analysis with Enhanced Molecular Coverage by Laser Ablation Electrospray Ionization Mass Spectrometry Using Ion Mobility Separation. Anal. Chem. 2014, 86, 4308–4315. [Google Scholar] [CrossRef]
- Stopka, S.A.; Shrestha, B.; Maréchal, É.; Falconet, D.; Vertes, A. Metabolic transformation of microalgae due to light acclimation and genetic modifications followed by laser ablation electrospray ionization mass spectrometry with ion mobility separation. Analyst 2014, 139, 5945–5953. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Smith, B.K.; Márk, L.; Nemes, P.; Nazarian, J.; Vertes, A. Ambient molecular imaging by laser ablation electrospray ionization mass spectrometry with ion mobility separation. Int. J. Mass Spectrom. 2015, 377, 681–689. [Google Scholar] [CrossRef]
- Stopka, S.A.; Mansour, T.R.; Shrestha, B.; Maréchal, É.; Falconet, D.; Vertes, A. Turnover rates in microorganisms by laser ablation electrospray ionization mass spectrometry and pulse-chase analysis. Anal. Chim. Acta 2016, 902, 1–7. [Google Scholar] [CrossRef]
- Comi, T.J.; Neumann, E.K.; Do, T.D.; Sweedler, J.V. microMS: A Python Platform for Image-Guided Mass Spectrometry Profiling. J. Am. Soc. Mass Spectrom. 2017, 28, 1919–1928. [Google Scholar] [CrossRef]
- Siegel, T.P.; Hamm, G.; Bunch, J.; Cappell, J.; Fletcher, J.S.; Schwamborn, K. Mass Spectrometry Imaging and Integration with Other Imaging Modalities for Greater Molecular Understanding of Biological Tissues. Mol. Imaging Biol. 2018, 20, 888–901. [Google Scholar] [CrossRef]
- Zheng, X.; Aly, N.A.; Zhou, Y.; Dupuis, K.T.; Bilbao, A.; Paurus, V.L.; Orton, D.J.; Wilson, R.; Payne, S.H.; Smith, R.D.; et al. A structural examination and collision cross section database for over 500 metabolites and xenobiotics using drift tube ion mobility spectrometry. Chem. Sci. 2017, 8, 7724–7736. [Google Scholar] [CrossRef] [PubMed]
- Compton, L.R.; Reschke, B.; Friend, J.; Powell, M.; Vertes, A. Remote laser ablation electrospray ionization mass spectrometry for non-proximate analysis of biological tissues. Rapid Commun. Mass Spectrom. 2014, 29, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Hines, K.M.; Ross, D.H.; Davidson, K.L.; Bush, M.F.; Xu, L. Large-Scale Structural Characterization of Drug and Drug-Like Compounds by High-Throughput Ion Mobility-Mass Spectrometry. Anal. Chem. 2017, 89, 9023–9030. [Google Scholar] [CrossRef]
- Rosen, E.P.; Bokhart, M.T.; Nazari, M.; Muddiman, D.C. Influence of C-Trap Ion Accumulation Time on the Detectability of Analytes in IR-MALDESI MSI. Anal. Chem. 2015, 87, 10483–10490. [Google Scholar] [CrossRef]
- Nemes, P.; Vertes, A. Laser Ablation Electrospray Ionization for Atmospheric Pressure, in Vivo, and Imaging Mass Spectrometry. Anal. Chem. 2007, 79, 8098–8106. [Google Scholar] [CrossRef]
- Tang, K.; Shvartsburg, A.A.; Lee, H.-N.; Prior, D.C.; Buschbach, M.A.; Li, F.; Tolmachev, A.V.; Anderson, G.A.; Smith, R.D. High-Sensitivity Ion Mobility Spectrometry/Mass Spectrometry Using Electrodynamic Ion Funnel Interfaces. Anal. Chem. 2005, 77, 3330–3339. [Google Scholar] [CrossRef] [PubMed]
- Kiss, A.; Heeren, R.M.A. Size, weight and position: Ion mobility spectrometry and imaging MS combined. Anal. Bioanal. Chem. 2011, 399, 2623–2634. [Google Scholar] [CrossRef] [PubMed]
- Ewing, M.A.; Glover, M.S.; Clemmer, D.E. Hybrid ion mobility and mass spectrometry as a separation tool. J. Chromatogr. A 2016, 1439, 3–25. [Google Scholar] [CrossRef]
- Hoaglund, C.S.; Valentine, S.J.; Sporleder, C.R.; Reilly, J.P.; Clemmer, D.E. Three-Dimensional Ion Mobility/TOFMS Analysis of Electrosprayed Biomolecules. Anal. Chem. 1998, 70, 2236–2242. [Google Scholar] [CrossRef]
- Doutre, C.; Nasiopoulos, P. Fast vignetting correction and color matching for panoramic image stitching. In Proceedings of the 2009 16th IEEE International Conference on Image Processing (ICIP), Cairo, Egypt, 7–10 November 2009; pp. 709–712. [Google Scholar]
- Reddy, J.P.; Rhim, J.-W. Extraction and Characterization of Cellulose Microfibers from Agricultural Wastes of Onion and Garlic. J. Nat. Fibers 2018, 15, 465–473. [Google Scholar] [CrossRef]
- Kaszycki, J.L.; La Rotta, A.; Colsch, B.; Fenaille, F.; Dauly, C.; Kamleh, A.; Wu, C. Separation of biologically relevant isomers on an Orbitrap mass spectrometer using high-resolution drift tube ion mobility and varied drift gas mixtures. Rapid Commun. Mass Spectrom. 2019, 33, 3–10. [Google Scholar] [CrossRef]
- Picache, J.A.; Rose, B.S.; Balinski, A.; Leaptrot, K.L.; Sherrod, S.D.; May, J.C.; McLean, J.A. Collision cross section compendium to annotate and predict multi-omic compound identities. Chem. Sci. 2018, 10, 983–993. [Google Scholar] [CrossRef]
- Pang, X.; Jia, C.; Chen, Z.; Li, L. Structural Characterization of Monomers and Oligomers of D-Amino Acid-Containing Peptides Using T-Wave Ion Mobility Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2016, 28, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.; Lietz, C.B.; Yu, Q.; Li, L. Site-Specific Characterization of d-Amino Acid Containing Peptide Epimers by Ion Mobility Spectrometry. Anal. Chem. 2014, 86, 2972–2981. [Google Scholar] [CrossRef] [PubMed]
- Fouque, K.J.D.; Garabedian, A.; Porter, J.; Baird, M.; Pang, X.; Williams, T.D.; Li, L.; Shvartsburg, A.; Fernandez-Lima, F. Fast and Effective Ion Mobility–Mass Spectrometry Separation ofd-Amino-Acid-Containing Peptides. Anal. Chem. 2017, 89, 11787–11794. [Google Scholar] [CrossRef]
- Zhou, Z.; Shen, X.; Tu, J.; Zhu, Z.-J. Large-Scale Prediction of Collision Cross-Section Values for Metabolites in Ion Mobility-Mass Spectrometry. Anal. Chem. 2016, 88, 11084–11091. [Google Scholar] [CrossRef]
- Böttcher, C.; Krähmer, A.; Stürtz, M.; Widder, S.; Schulz, H. Comprehensive metabolite profiling of onion bulbs (Allium cepa) using liquid chromatography coupled with electrospray ionization quadrupole time-of-flight mass spectrometry. Metabolomics 2017, 13, 35. [Google Scholar] [CrossRef]
- Nakayasu, E.S.; Nicora, C.D.; Sims, A.C.; Burnum-Johnson, K.E.; Kim, Y.-M.; Kyle, J.E.; Matzke, M.M.; Shukla, A.K.; Chu, R.K.; Schepmoes, A.A.; et al. MPLEx: A Robust and Universal Protocol for Single-Sample Integrative Proteomic, Metabolomic, and Lipidomic Analyses. mSystems 2016, 1, e00043-16. [Google Scholar] [CrossRef]
- Zhang, L.; Khattar, N.; Kemenes, I.; Kemenes, G.; Zrinyi, Z.; Pirger, Z.; Vertes, A. Subcellular Peptide Localization in Single Identified Neurons by Capillary Microsampling Mass Spectrometry. Sci. Rep. 2018, 8, 12227. [Google Scholar] [CrossRef] [PubMed]
- Evers, T.M.J.; Hochane, M.; Tans, S.J.; Heeren, R.M.A.; Semrau, S.; Nemes, P.; Mashaghi, A. Deciphering Metabolic Heterogeneity by Single-Cell Analysis. Anal. Chem. 2019, 91, 13314–13323. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Page, J.S.; Smith, R.D. Charge competition and the linear dynamic range of detection in electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2004, 15, 1416–1423. [Google Scholar] [CrossRef] [PubMed]
- Nagy, G.; Veličković, D.; Chu, R.K.; Carrell, A.A.; Weston, D.J.; Ibrahim, Y.M.; Anderton, C.R.; Smith, R.D. Towards resolving the spatial metabolome with unambiguous molecular annotations in complex biological systems by coupling mass spectrometry imaging with structures for lossless ion manipulations. Chem. Commun. 2019, 55, 306–309. [Google Scholar] [CrossRef]
- Edelstein, A.D.; Amodaj, N.; Hoover, K.H.; Vale, R.D.; Stuurman, N. Computer Control of Microscopes Using µManager. Curr. Protoc. Mol. Biol. 2010, 92, 14–20. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Sternberg, S.R. Biomedical Image Processing. IEEE Comput. 1983, 16, 22–34. [Google Scholar] [CrossRef]
- Bitter, R.; Mohiuddin, T.; Nawrocki, M. LabVIEW™ Advanced Programming Techniques; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar]
- Ibrahim, Y.M.; Baker, E.S.; Danielson, W.F.; Norheim, R.V.; Prior, D.C.; Anderson, G.A.; Belov, M.E.; Smith, R.D. Development of a new ion mobility time-of-flight mass spectrometer. Int. J. Mass Spectrom. 2015, 377, 655–662. [Google Scholar] [CrossRef]
- Shah, A.R.; Davidson, J.; Monroe, M.E.; Mayampurath, A.M.; Danielson, W.F.; Shi, Y.; Robinson, A.C.; Clowers, B.H.; Belov, M.E.; Anderson, G.A.; et al. An efficient data format for mass spectrometry-based proteomics. J. Am. Soc. Mass Spectrom. 2010, 21, 1784–1788. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Niedermeyer, T.H.J.; Strohalm, M. mMass as a Software Tool for the Annotation of Cyclic Peptide Tandem Mass Spectra. PLoS ONE 2012, 7, e44913. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taylor, M.J.; Mattson, S.; Liyu, A.; Stopka, S.A.; Ibrahim, Y.M.; Vertes, A.; Anderton, C.R. Optical Microscopy-Guided Laser Ablation Electrospray Ionization Ion Mobility Mass Spectrometry: Ambient Single Cell Metabolomics with Increased Confidence in Molecular Identification. Metabolites 2021, 11, 200. https://doi.org/10.3390/metabo11040200
Taylor MJ, Mattson S, Liyu A, Stopka SA, Ibrahim YM, Vertes A, Anderton CR. Optical Microscopy-Guided Laser Ablation Electrospray Ionization Ion Mobility Mass Spectrometry: Ambient Single Cell Metabolomics with Increased Confidence in Molecular Identification. Metabolites. 2021; 11(4):200. https://doi.org/10.3390/metabo11040200
Chicago/Turabian StyleTaylor, Michael J., Sara Mattson, Andrey Liyu, Sylwia A. Stopka, Yehia M. Ibrahim, Akos Vertes, and Christopher R. Anderton. 2021. "Optical Microscopy-Guided Laser Ablation Electrospray Ionization Ion Mobility Mass Spectrometry: Ambient Single Cell Metabolomics with Increased Confidence in Molecular Identification" Metabolites 11, no. 4: 200. https://doi.org/10.3390/metabo11040200
APA StyleTaylor, M. J., Mattson, S., Liyu, A., Stopka, S. A., Ibrahim, Y. M., Vertes, A., & Anderton, C. R. (2021). Optical Microscopy-Guided Laser Ablation Electrospray Ionization Ion Mobility Mass Spectrometry: Ambient Single Cell Metabolomics with Increased Confidence in Molecular Identification. Metabolites, 11(4), 200. https://doi.org/10.3390/metabo11040200








