Effect of the Andean Geography and Climate on the Specialized Metabolism of Its Vegetation: The Subtribe Espeletiinae (Asteraceae) as a Case Example
Abstract
1. Introduction
2. Results
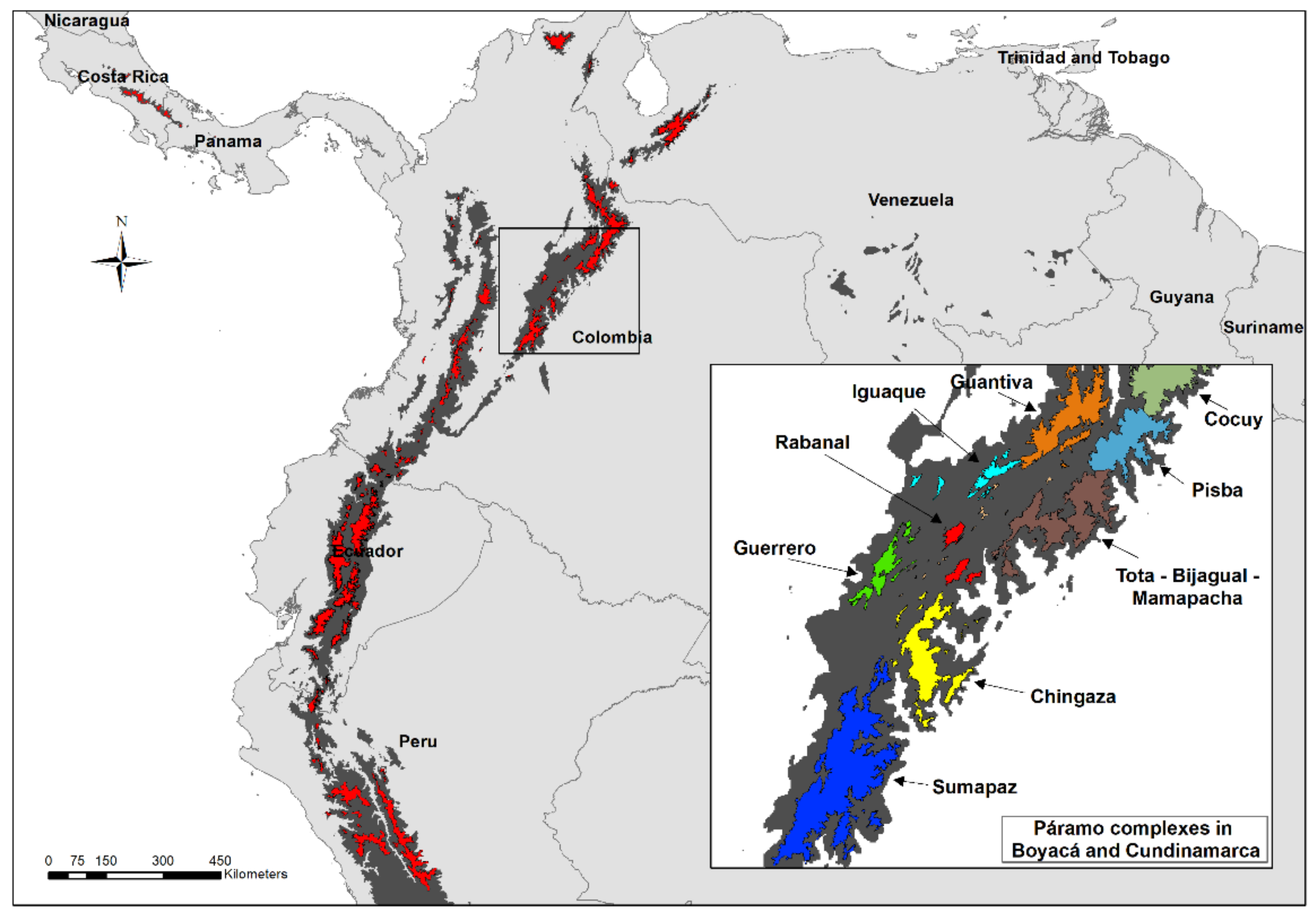
2.1. Metabolic Fingerprinting and Correlation with Biogeographic Data
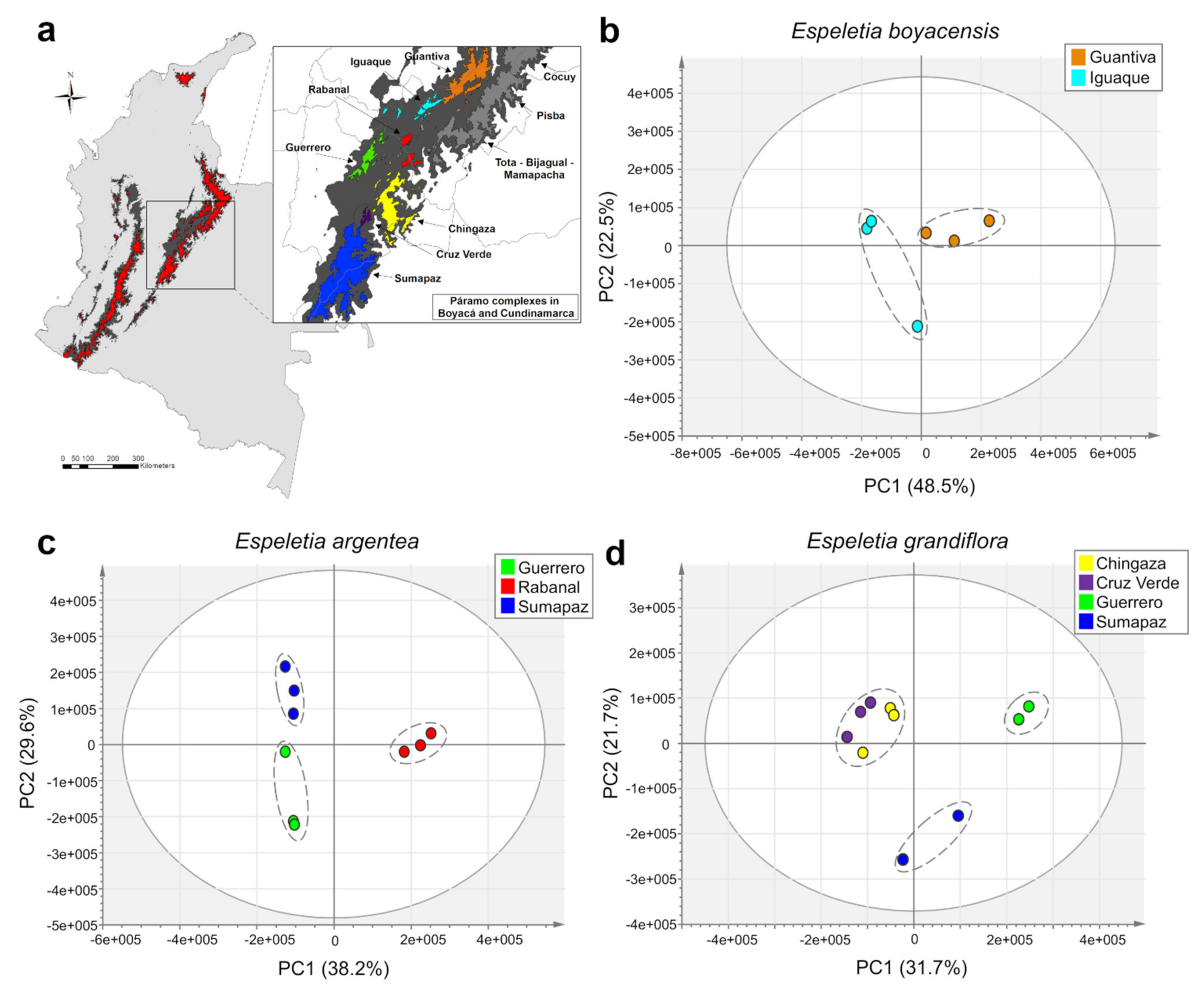
2.2. Accumulation Patterns of Specialized Metabolites in Different Geographic Locations
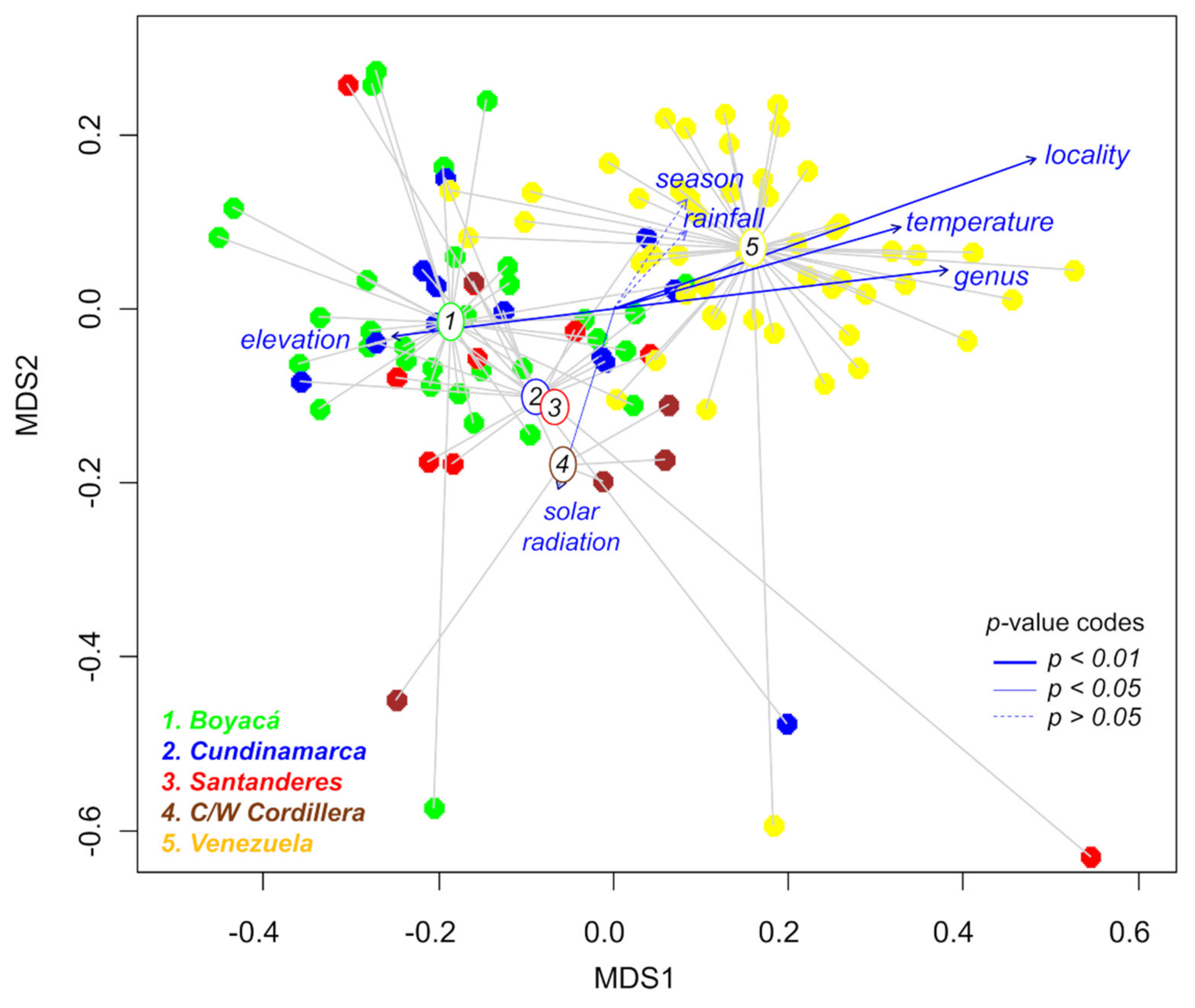
2.3. Effects of Geography, Taxonomy, and Climate on Metabolic Variation
3. Discussion
3.1. Metabolomic-Biogeographic Patterns Resemble Phylogenetic Hypotheses in Espeletiinae
3.2. Combined Effects of Geography, Taxonomy, Phylogeny, and Climate on Plant Metabolic Variation
3.3. Perspectives of Metabolic Variation in the Context of Plant Diversification
4. Materials and Methods
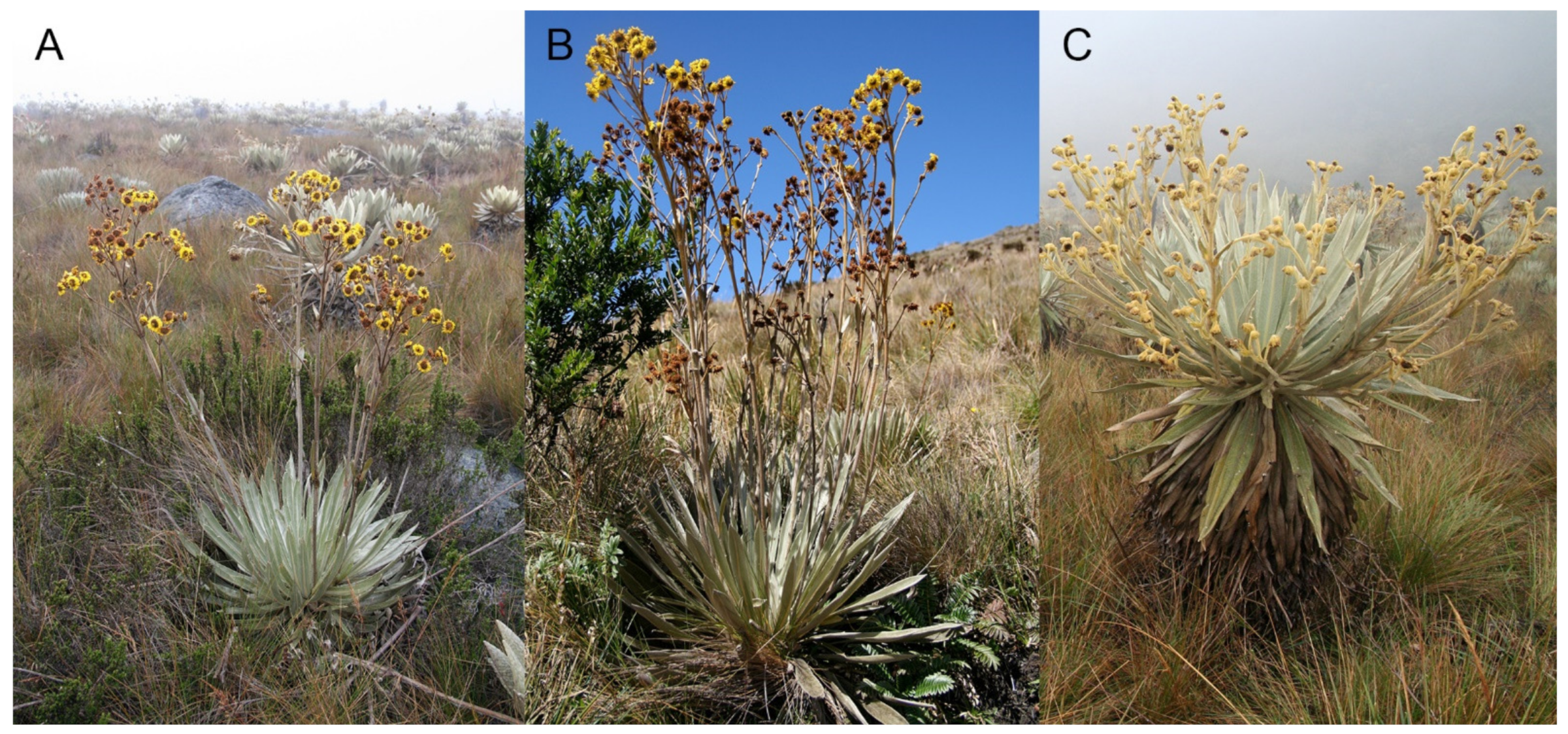
4.1. Plant Material
4.2. Environmental Variables
4.3. Sample Preparation
4.4. UHPLC-UV-HRMS Analysis
4.5. Data Preprocessing
4.6. Multivariate Analyses and Correlation with Biogeographic and Environmental Variables
4.7. Selection and Annotation of Discriminant Metabolites
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barthlott, W.; Mutke, J.; Rafiqpoor, M.D.; Kier, G.; Kreft, H. Global centres of vascular plant diversity. Nov. Acta. Leopold. 2005, 92, 61–83. [Google Scholar]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; da Fonseca, G.A.B.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef]
- Diazgranados, M.; Barber, J.C. Geography shapes the phylogeny of frailejones (Espeletiinae Cuatrec., Asteraceae): A remarkable example of recent rapid radiation in sky islands. PeerJ 2017, 5, e2968. [Google Scholar] [CrossRef]
- Gentry, A.H. Neotropical floristic diversity: Phytogeographical connections between Central and South America, pleistocene climatic fluctuations, or an accident of the Andean orogeny? Ann. Mo. Bot. Gard. 1982, 69, 557–593. [Google Scholar] [CrossRef]
- Gómez-Gutiérrez, M.C.; Pennington, R.T.; Neaves, L.E.; Milne, R.I.; Madriñán, S.; Richardson, J.E. Genetic diversity in the Andes: Variation within and between the South American species of Oreobolus R. Br. (Cyperaceae). Alp. Bot. 2017, 127, 155–170. [Google Scholar] [CrossRef]
- Hughes, C.; Eastwood, R. Island radiation on a continental scale: Exceptional rates of plant diversification after uplift of the Andes. Proc. Natl. Acad. Sci. USA 2006, 103, 10334–10339. [Google Scholar] [CrossRef]
- Kolář, F.; Dušková, E.; Sklenář, P. Niche shifts and range expansions along cordilleras drove diversification in a high-elevation endemic plant genus in the tropical Andes. Mol. Ecol. 2016, 25, 4593–4610. [Google Scholar] [CrossRef]
- Vásquez, D.L.A.; Balslev, H.; Hansen, M.M.; Sklenář, P.; Romoleroux, K. Low genetic variation and high differentiation across sky island populations of Lupinus alopecuroides (Fabaceae) in the northern Andes. Alp. Bot. 2016, 126, 135–142. [Google Scholar] [CrossRef]
- Luteyn, J.L. Páramos: A Checklist of Plant Diversity, Geographical Distribution and Botanical Literature; New York Botanical Garden Press: New York, NY, USA, 1999. [Google Scholar]
- Pérez-Escobar, O.A.; Chomicki, G.; Condamine, F.L.; Karremans, A.P.; Bogarín, D.; Matzke, N.J.; Silvestro, D.; Antonelli, A. Recent origin and rapid speciation of Neotropical orchids in the world’s richest plant biodiversity hotspot. New Phytol. 2017, 215, 891–905. [Google Scholar] [CrossRef] [PubMed]
- Madriñán, S.; Cortés, A.J.; Richardson, J.E. Páramo is the world’s fastest evolving and coolest biodiversity hotspot. Front. Genet. 2013, 4, 192. [Google Scholar] [CrossRef]
- Luebert, F.; Weigend, M. Phylogenetic insights into Andean plant diversification. Front. Ecol. Evol. 2014, 2, 27. [Google Scholar] [CrossRef]
- Flantua, S.G.A.; O’Dea, A.; Onstein, R.E.; Giraldo, C.; Hooghiemstra, H. The flickering connectivity system of the north Andean páramos. J. Biogeogr. 2019, 46, 1808–1825. [Google Scholar] [CrossRef]
- Hooghiemstra, H.; Ran, E.T.H. Late and middle pleistocene climatic change and forest development in Colombia: Pollen record Funza II (2–158 m core interval). Palaeogeogr. Palaeoclimatol. Palaeoecol. 1994, 109, 211–246. [Google Scholar] [CrossRef]
- Hooghiemstra, H.; Van der Hammen, T. Quaternary Ice-Age dynamics in the Colombian Andes: Developing an understanding of our legacy. Philos. Trans. R. Soc. Lond. 2004, 359, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Roberts, N.J.; Barendregt, R.W.; Clague, J.J. Multiple tropical Andean glaciations during a period of late Pliocene warmth. Sci. Rep. 2017, 7, 41878. [Google Scholar] [CrossRef]
- Contreras-Ortiz, N.; Atchison, G.W.; Hughes, C.E.; Madriňán, S. Convergent evolution of high elevation plant growth forms and geographically structured variation in Andean Lupinus (Fabaceae). Bot. J. Linn. Soc. 2018, 187, 118–136. [Google Scholar] [CrossRef]
- Nevado, B.; Contreras-Ortiz, N.; Hughes, C.; Filatov, D.A. Pleistocene glacial cycles drive isolation, gene flow and speciation in the high-elevation Andes. New Phytol. 2018, 219, 779–793. [Google Scholar] [CrossRef]
- Chacón, J.; Madriñán, S.; Chase, M.W.; Bruhl, J.J. Molecular phylogenetics of Oreobolus (Cyperaceae) and the origin and diversification of the American species. Taxon 2006, 55, 359–366. [Google Scholar] [CrossRef]
- Scherson, R.A.; Vidal, R.; Sanderson, M.J. Phylogeny, biogeography, and rates of diversification of new world Astragalus (Leguminosae) with an emphasis on South American radiations. Am. J. Bot. 2008, 95, 1030–1039. [Google Scholar] [CrossRef]
- Vargas, O.M.; Ortiz, E.M.; Simpson, B.B. Conflicting phylogenomic signals reveal a pattern of reticulate evolution in a recent high-Andean diversification (Asteraceae: Astereae: Diplostephium). New Phytol. 2017, 214, 1736–1750. [Google Scholar] [CrossRef]
- Bell, C.D.; Donoghue, M.J. Phylogeny and biogeography of Valerianaceae (Dipsacales) with special reference to the South American valerians. Org. Divers. Evol. 2005, 5, 147–159. [Google Scholar] [CrossRef]
- Nürk, N.M.; Scheriau, C.; Madriñán, S. Explosive radiation in high Andean Hypericum—rates of diversification among New World lineages. Front. Genet. 2013, 4, 175. [Google Scholar] [CrossRef]
- Jabaily, R.S.; Sytsma, K.J. Historical biogeography and life-history evolution of Andean Puya (Bromeliaceae). Bot. J. Linn. Soc. 2013, 171, 201–224. [Google Scholar] [CrossRef]
- Pouchon, C.; Fernández, A.; Nassar, J.M.; Boyer, F.; Aubert, S.; Lavergne, S.; Mavárez, J. Phylogenomic analysis of the explosive adaptive radiation of the Espeletia complex (Asteraceae) in the tropical Andes. Syst. Biol. 2018, 67, 1041–1060. [Google Scholar] [CrossRef]
- Diazgranados, M. A nomenclator for the frailejones (Espeletiinae Cuatrec., Asteraceae). PhytoKeys 2012, 16, 1–52. [Google Scholar] [CrossRef]
- Mavárez, J. A Taxonomic Revision of Espeletia (Asteraceae). The Venezuelan Radiation. Harv. Pap. Bot. 2019, 24, 131–244. [Google Scholar] [CrossRef]
- Cuatrecasas, J. A Systematic Study of the Subtribe Espeletiinae: Heliantheae, Asteraceae; The New York Botanical Garden Press: New York, NY, USA, 2013; ISBN 089327514X. [Google Scholar]
- Diazgranados, M. Una mirada biológica a los páramos circundantes a la Sabana de Bogotá. In Los Páramos Circundantes a la Sabana de Bogotá; Guhl, E., Ed.; Jardín Botánico de Bogotá: Bogotá, Colombia, 2015; pp. 175–205. [Google Scholar]
- Diazgranados, M.; Castellanos, C. Evaluación del Riesgo de Extinción de Frailejones en Colombia; Official Report; Instituto de Investigaciones en Recursos Biológicos Alexander von Humboldt: Bogotá, Colombia, 2018. [Google Scholar]
- Sampaio, B.L.; Edrada-Ebel, R.; Da Costa, F.B. Effect of the environment on the secondary metabolic profile of Tithonia diversifolia: A model for environmental metabolomics of plants. Sci. Rep. 2016, 6, 29265. [Google Scholar] [CrossRef] [PubMed]
- Sardans, J.; Gargallo-Garriga, A.; Urban, O.; Klem, K.; Walker, T.W.N.; Holub, P.; Janssens, I.A.; Peñuelas, J. Ecometabolomics for a better understanding of plant responses and acclimation to abiotic factors linked to global change. Metabolites 2020, 10, 239. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef]
- Ernst, M.; Nothias, L.-F.; van der Hooft, J.J.J.; Silva, R.R.; Saslis-Lagoudakis, C.H.; Grace, O.M.; Martinez-Swatson, K.; Hassemer, G.; Funez, L.A.; Simonsen, H.T.; et al. Assessing specialized metabolite diversity in the cosmopolitan plant genus Euphorbia L. Front. Plant. Sci. 2019, 10, 846. [Google Scholar] [CrossRef]
- Sedio, B.E. Recent breakthroughs in metabolomics promise to reveal the cryptic chemical traits that mediate plant community composition, character evolution and lineage diversification. New Phytol. 2017, 214, 952–958. [Google Scholar] [CrossRef]
- Padilla-González, G.F.; Frey, M.; Gómez-Zeledón, J.; Da Costa, F.B.; Spring, O. Metabolomic and gene expression approaches reveal the developmental and environmental regulation of the secondary metabolism of yacón (Smallanthus sonchifolius, Asteraceae). Sci. Rep. 2019, 9, 13178. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.B.; Ernst, M.; Hooft, J.J.J.; Silva, R.R.; Park, J.; Medema, M.H.; Sung, S.H.; Dorrestein, P.C. Comprehensive mass spectrometry-guided phenotyping of plant specialized metabolites reveals metabolic diversity in the cosmopolitan plant family Rhamnaceae. Plant. J. 2019, 98, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Padilla-González, G.F.; Diazgranados, M.; Da Costa, F.B. Biogeography shaped the metabolome of the genus Espeletia: A phytochemical perspective on an Andean adaptive radiation. Sci. Rep. 2017, 7, 8835. [Google Scholar] [CrossRef]
- Witten, I.H.; Eibe, F.; Hall, M.A. Data Mining: Practical Machine Learning Tools and Techniques, 3rd ed.; Morgan Kaufmann: San Francisco, CA, USA, 2005. [Google Scholar]
- Padilla-Gonzalez, G.F.; Diazgranados, M.; Ccana-Ccapatinta, G.; Casoti, R.; Da Costa, F.B. Caffeic acid derivatives and further compounds from Espeletia barclayana Cuatrec. (Asteraceae, Espeletiinae). Biochem. Syst. Ecol. 2017, 70, 291–293. [Google Scholar] [CrossRef]
- Clifford, M.N.; Knight, S.; Kuhnert, N. Discriminating between the six isomers of dicaffeoylquinic acid by LC-MSn. J. Agric. Food Chem. 2005, 53, 3821–3832. [Google Scholar] [CrossRef] [PubMed]
- Žiarovská, J.; Padilla-González, G.F.F.; Viehmannová, I.; Fernández, E. Genetic and chemical diversity among yacon [Smallanthus sonchifolius (Poepp. et Endl.) H. Robinson] accessions based on iPBS markers and metabolomic fingerprinting. Plant. Physiol. Biochem. 2019, 141, 183–192. [Google Scholar] [CrossRef]
- Martucci, M.E.P.; De Vos, R.C.H.; Carollo, C.A.; Gobbo-Neto, L. Metabolomics as a potential chemotaxonomical tool: Application in the genus Vernonia schreb. PLoS ONE 2014, 9, e93149. [Google Scholar] [CrossRef] [PubMed]
- Bohlmann, F.; Suding, H.; Cuatrecasas, J.; King, R.M.; Robinson, H. New diterpenes from subtribe Espeletiinae. Phytochemistry 1980, 19, 267–271. [Google Scholar] [CrossRef]
- Usubillaga, A.; de Hernandez, J.; Perez, N.; Kiriakidis, M. Kauranoid diterpenes in Espletia species. Phytochemistry 1973, 12, 2999. [Google Scholar] [CrossRef]
- Kim, H.Y.; Park, H.M.; Lee, C.H. Mass spectrometry-based chemotaxonomic classification of Penicillium species (P. echinulatum, P. expansum, P. solitum, and P. oxalicum) and its correlation with antioxidant activity. J. Microbiol. Methods 2012, 90, 327–335. [Google Scholar] [CrossRef]
- Tellez, A.N.; Torrenegra, R.D.; Pedrozo, J.; Gray, A. Cycloartan-2 beta-2-methyl butanoate isolated from genus Espeletia (Asteraceae). Molecules 1998, 3, M49-U4. [Google Scholar]
- Tellez, A.N.; Torrenegra, R.D.; Pedrozo, J.; Martínez, A. Cycloeucalen-3 beta-(2-methyl butanoate). New cycloeucalen isolated from the Espeletia barclayana Cuatr. (Asteraceae). Molecules 2000, 5, M163-U6. [Google Scholar]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Flantua, S.G.A.; Hooghiemstra, H. Historical Connectivity and Mountain Biodiversity. In Mountains, Climate and Biodiversity; Hoorn, C., Perrigo, A., Antonelli, A., Eds.; Wiley Blackwell: Hoboken, NJ, USA, 2018; pp. 171–185. ISBN 9781119159872. [Google Scholar]
- Defossez, E.; Pitteloud, C.; Descombes, P.; Glauser, G.; Allard, P.-M.; Walker, T.W.N.; Fernandez-Conradi, P.; Wolfender, J.-L.; Pellissier, L.; Rasmann, S. Spatial and evolutionary predictability of phytochemical diversity. Proc. Natl. Acad. Sci. USA 2021, 118, e2013344118. [Google Scholar] [CrossRef]
- Maldonado, C.; Barnes, C.J.; Cornett, C.; Holmfred, E.; Hansen, S.H.; Persson, C.; Antonelli, A.; Rønsted, N. Phylogeny predicts the quantity of antimalarial alkaloids within the iconic yellow Cinchona bark (Rubiaceae: Cinchona calisaya). Front. Plant Sci. 2017, 8, 391. [Google Scholar] [CrossRef] [PubMed]
- Cortés, A.J.; Garzón, L.N.; Valencia, J.B.; Madriñán, S. On the causes of rapid diversification in the Páramos: Isolation by ecology and genomic divergence in Espeletia. Front. Plant Sci. 2018, 9, 1700. [Google Scholar] [CrossRef] [PubMed]
- Nevado, B.; Atchison, G.W.; Hughes, C.E.; Filatov, D.A. Widespread adaptive evolution during repeated evolutionary radiations in New World lupins. Nat. Commun. 2016, 7, 12384. [Google Scholar] [CrossRef] [PubMed]
- Rangel-Ch, O. La Región de Vida Paramuna; Instituto de Ciencias Naturales-Instituto Alexander von Humboldt: Bogotá, Colombia, 2000. [Google Scholar]
- De Vos, R.C.; Moco, S.; Lommen, A.; Keurentjes, J.J.B.; Bino, R.J.; Hall, R.D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007, 2, 778–791. [Google Scholar] [CrossRef] [PubMed]
- Padilla-González, G.F.G.F.; Aldana, J.A.J.A.; Da Costa, F.B.; Costa, F.B. Da Chemical characterization of two morphologically related Espeletia (Asteraceae) species and chemometric analysis based on essential oil components. Rev. Bras. Farmacogn. 2016, 26, 694–700. [Google Scholar] [CrossRef]
- Padilla-González, G.F.; Diazgranados, M.; Oliveira, T.B.; Chagas-Paula, D.A.; Da Costa, F.B. Chemistry of the subtribe Espeletiinae (Asteraceae) and its correlation with phylogenetic data: An in silico chemosystematic approach. Bot. J. Linn. Soc. 2018, 186, 18–46. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- van den Berg, R.A.; Hoefsloot, H.C.J.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metab. 2012, 1, 92–107. [Google Scholar]
- Gromski, P.S.; Xu, Y.; Hollywood, K.A.; Turner, M.L.; Goodacre, R. The influence of scaling metabolomics data on model classification accuracy. Metabolomics 2015, 11, 684–695. [Google Scholar] [CrossRef]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A tutorial review: Metabolomics and partial least squares-discriminant analysis—A marriage of convenience or a shotgun wedding. Anal. Chim. Acta 2015, 879, 10–23. [Google Scholar] [CrossRef]
- Triba, M.N.; Le Moyec, L.; Amathieu, R.; Goossens, C.; Bouchemal, N.; Nahon, P.; Rutledge, D.N.; Savarin, P. PLS/OPLS models in metabolomics: The impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. Biosyst. 2015, 11, 13–19. [Google Scholar] [CrossRef]
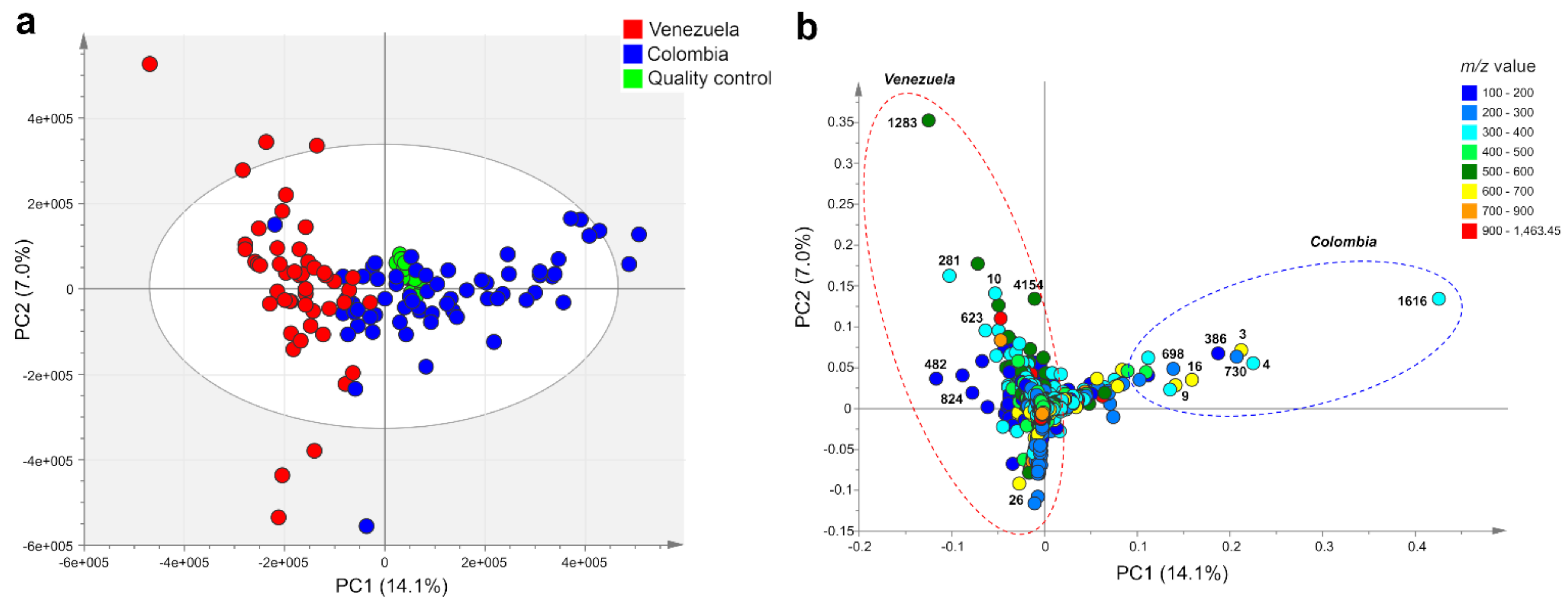


| ID | m/z | Rt min | Identity or Molecular Formula | Confidence Level * |
|---|---|---|---|---|
| Colombia | ||||
| 1616 | 315.051 | 11.1 | 3-O-methylquercetin | 1 |
| 4 | 301.035 | 10.7 | quercetin | 1 |
| 3 | 631.109 | 11.1 | dimer of 3-O-methylquercetin | 1 |
| 730 | 299.020 | 15.0 | biflavonoid (C31H20O14) fragment | 3 |
| 386 | 153.018 | 2.8 | protocatechuic acid | 1 |
| 16 | 615.079 | 15.0 | biflavonoid (C31H20O14) | 3 |
| 9 | 695.124 | 9.7 | 2,3,5 or 2,4,5-tricaffeoylaltraric acid | 1 |
| 698 | 299.020 | 11.1 | 3-O-methylquercetin fragment | 1 |
| 2217 | 301.072 | 13.6 | C16H14O6 | 4 |
| 484 | 209.029 | 0.9 | altraric acid | 2 |
| Venezuela | ||||
| 1283 | 515.119 | 8.4 | 3,5-di-O-(E)-dicaffeoylquinic acid | 1 |
| 281 | 353.088 | 4.5 | 5-O-(E)-caffeoylquinic acid | 1 |
| 10 | 329.067 | 14.5 | di-O-methylquercetin | 2 |
| 4154 | 515.119 | 8.1 | 1,5-di-O-(E)-dicaffeoylquinic acid | 1 |
| 623 | 317.212 | 22.0 | hydroxy-ent-kauren-18-oic acid | 3 |
| 482 | 191.055 | 1.0 | quinic acid | 1 |
| 824 | 171.066 | 7.6 | C8H12O4 | 4 |
| 26 | 609.125 | 9.9 | quercetin-3-O-cinnamoyl-hexoside | 2 |
| ID | m/z | Rt | Identity or Molecular Formula | Confidence Level * |
|---|---|---|---|---|
| 1208 | 511.292 | 12.9 | C27H44O9 | 4 |
| 3047 | 311.020 | 12.6 | 5-hydroxyanthraquinone-1,3-dicarboxylic acid | 3 |
| 3560 | 657.292 | 10.6 | C30H58O15 | 4 |
| 109 | 267.072 | 1.0 | glycosylated metabolite (C9H16O9) | 3 |
| 4724 | 527.235 | 4.2 | C22H40O14 | 4 |
| 2591 | 543.151 | 8.0 | methyl 3-O-(E)-caffeoyl-4-O-(E)-feruloylquinate | 2 |
| ID | m/z | Rt | Identity or Molecular Formula | VIP Score | Confidence Level * |
|---|---|---|---|---|---|
| Sumapaz | |||||
| 298 | 485.283 | 35.4 | C36H38O | 3.26 | 4 |
| 437 | 485.283 | 34.2 | C36H38O | 1.92 | 4 |
| 373 | 485.283 | 28.1 | C36H38O | 1.58 | 4 |
| 312 | 485.283 | 32.5 | C36H38O | 1.57 | 4 |
| 297 | 485.283 | 31.5 | C36H38O | 1.28 | 4 |
| Guantiva, Iguaque and Guerrero | |||||
| 9 | 515.119 | 8.1 | 1,5-di-O-(E)-dicaffeoylquinic acid | 3.05 | 1 |
| 22 | 353.088 | 4.5 | 5-O-(E)-caffeoylquinic acid | 2.73 | 1 |
| 23 | 329.234 | 12.6 | C22H34O2 | 2.10 | 4 |
| 27 | 695.124 | 9.7 | 2,3,5 or 2,4,5-tricaffeoylaltraric acid | 2.04 | 1 |
| 38 | 515.119 | 5.8 | 1,3-di-O-(E)-dicaffeoylquinic acid | 1.66 | 1 |
| 45 | 515.119 | 8.6 | 3,4-di-O-(E)-dicaffeoylquinic acid | 1.49 | 1 |
| Chingaza and Cruz Verde | |||||
| 1 | 315.051 | 11.1 | 3-O-methylquercetin | 7.11 | 1 |
| 7 | 209.029 | 0.9 | altraric acid | 4.24 | 2 |
| 2 | 631.109 | 11.1 | dimer of 3-O-methylquercetin | 3.74 | 1 |
| 26 | 153.018 | 2.8 | protocatechuic acid | 3.39 | 1 |
| 6 | 299.020 | 15.0 | fragment of biflavonoid (C31H20O14) | 3.38 | 3 |
| 10 | 615.079 | 15.0 | biflavonoid (C31H20O14) | 3.03 | 3 |
| 17 | 301.035 | 10.7 | quercetin | 1.98 | 1 |
| Rabanal | |||||
| 3 | 195.050 | 0.9 | gluconic acid or isomers | 8.90 | 3 |
| 18 | 179.055 | 0.9 | hexose | 2.76 | 3 |
| Variable | NMDS1 | NMDS2 | R2 | Pr(>r) |
|---|---|---|---|---|
| Locality | 0.9417 | 0.3364 | 0.5295 | 9.9 × 10−5 |
| Genus | 0.9932 | 0.1167 | 0.2982 | 9.9 × 10−5 |
| Temperature | 0.9249 | 0.3803 | 0.2072 | 9.9 × 10−5 |
| Elevation | −0.9964 | −0.0843 | 0.1391 | 4.0 × 10−4 |
| Solar radiation | −0.2752 | −0.9614 | 0.0814 | 0.0117 |
| Season | 0.5502 | 0.8350 | 0.0459 | 0.0853 |
| Rainfall | 0.7026 | 0.7116 | 0.0253 | 0.2505 |
| Genus | Determined to Species | Determined to Genus | Replicates | Total Samples |
|---|---|---|---|---|
| Espeletia | 64 | 28 | 86 | 178 |
| Espeletiopsis | 4 | 0 | 10 | 14 |
| Libanothamnus | ? | 8 | 2 | 10 |
| Ruilopezia | ? | 6 | 0 | 6 |
| Paramiflos | 1 | 0 | 1 | 2 |
| Total | ≥71 | 42 | 99 | 210 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padilla-González, G.F.; Diazgranados, M.; Da Costa, F.B. Effect of the Andean Geography and Climate on the Specialized Metabolism of Its Vegetation: The Subtribe Espeletiinae (Asteraceae) as a Case Example. Metabolites 2021, 11, 220. https://doi.org/10.3390/metabo11040220
Padilla-González GF, Diazgranados M, Da Costa FB. Effect of the Andean Geography and Climate on the Specialized Metabolism of Its Vegetation: The Subtribe Espeletiinae (Asteraceae) as a Case Example. Metabolites. 2021; 11(4):220. https://doi.org/10.3390/metabo11040220
Chicago/Turabian StylePadilla-González, Guillermo F., Mauricio Diazgranados, and Fernando B. Da Costa. 2021. "Effect of the Andean Geography and Climate on the Specialized Metabolism of Its Vegetation: The Subtribe Espeletiinae (Asteraceae) as a Case Example" Metabolites 11, no. 4: 220. https://doi.org/10.3390/metabo11040220
APA StylePadilla-González, G. F., Diazgranados, M., & Da Costa, F. B. (2021). Effect of the Andean Geography and Climate on the Specialized Metabolism of Its Vegetation: The Subtribe Espeletiinae (Asteraceae) as a Case Example. Metabolites, 11(4), 220. https://doi.org/10.3390/metabo11040220






