Isolation and Proteomics of the Insulin Secretory Granule
Abstract
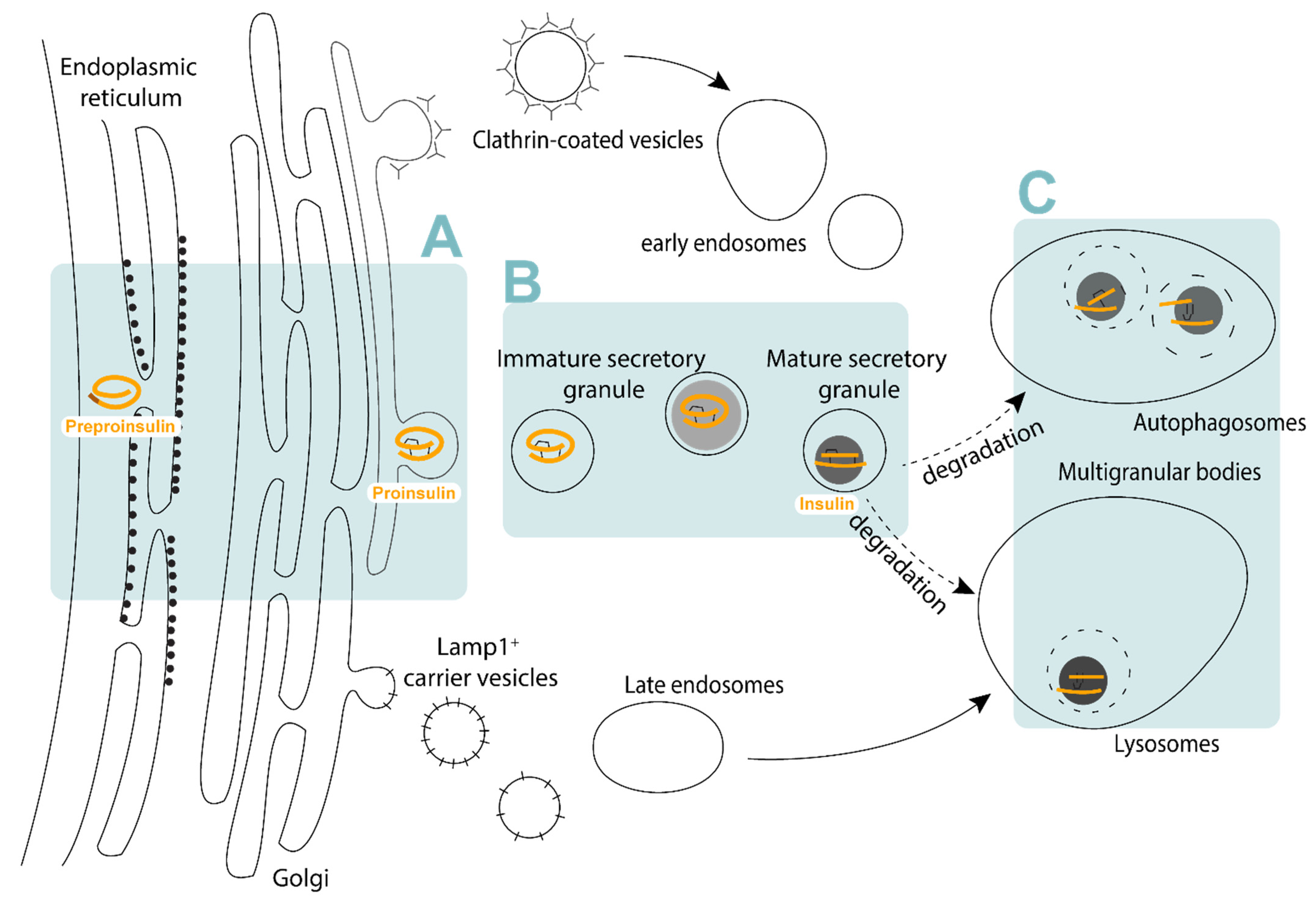
1. Insulin Granule Biogenesis and Function
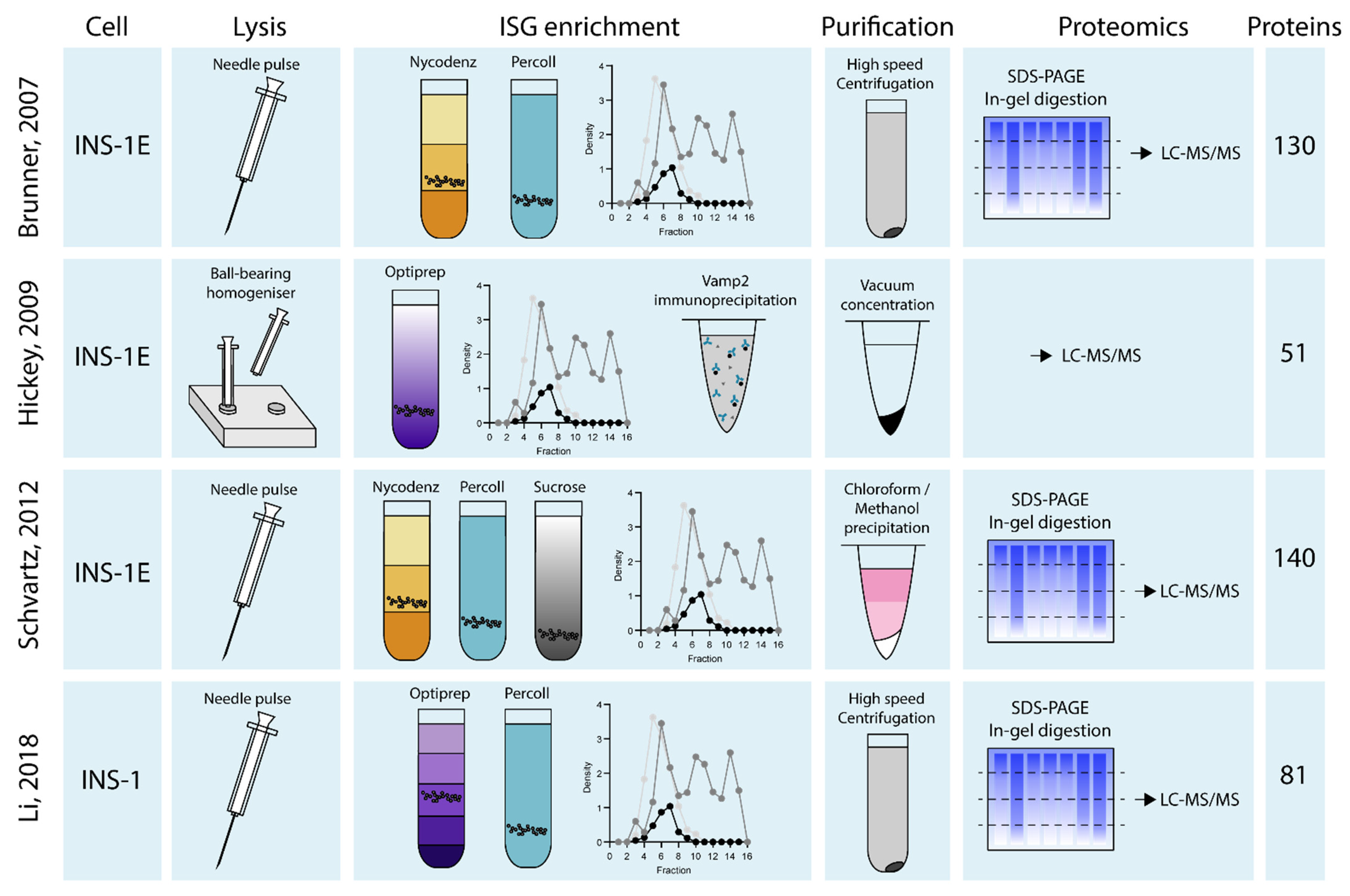
2. Isolating the Insulin Granule
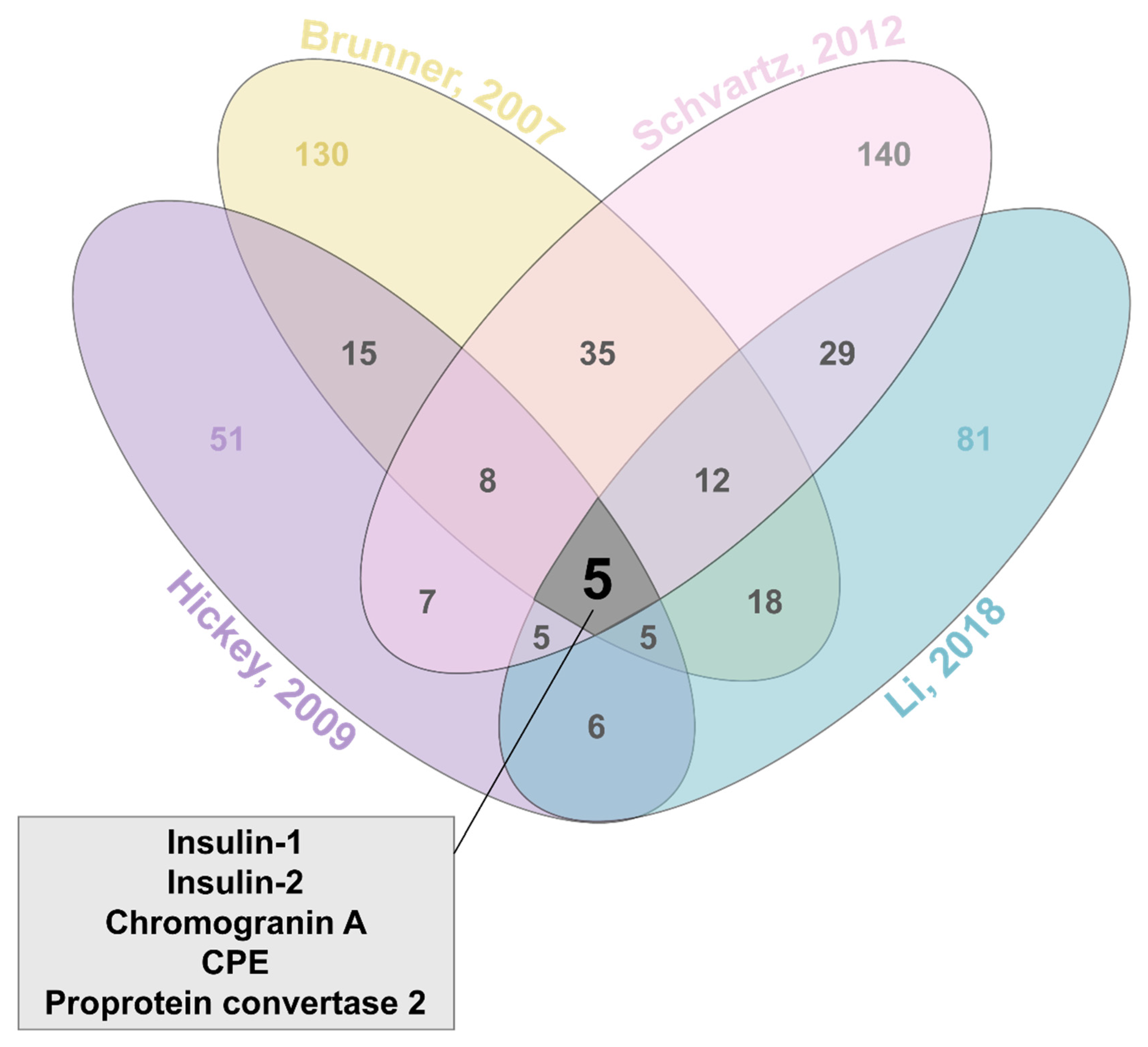
3. Identifying Insulin Granule Proteins
3.1. Intravesicular Proteins
3.2. Membrane Proteins
3.3. Other Proteins
4. Understanding ISG Function through the Proteome
5. Moving Forward
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rorsman, P.; Ashcroft, F.M. Pancreatic β-cell electrical activity and insulin secretion: Of mice and men. Physiol. Rev. 2018, 98, 117–214. [Google Scholar] [CrossRef] [PubMed]
- Weiss, M.; Steiner, D.F.; Philipson, L.H. Insulin Biosynthesis, Secretion, Structure, and Structure-Activity Relationships. In Endotext; Feingold, K.R., Anawalt, B., Boyce, A., Chrousos, G., de Herder, W.W., Dungan, K., Grossman, A., Hershman, J.M., Hofland, H.J., Kaltsas, G., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Gehart, H.; Ricci, R. Saving the neck from scission. Commun. Integr. Biol. 2013, 6, e23098. [Google Scholar] [CrossRef]
- Arvan, P.; Halban, P.A. Sorting ourselves out: Seeking consensus on trafficking in the beta-cell. Traffic 2004, 5, 53–61. [Google Scholar] [CrossRef]
- Davidson, H.W.; Rhodes, C.J.; Hutton, J.C. Intraorganellar calcium and pH control proinsulin cleavage in the pancreatic beta cell via two distinct site-specific endopeptidases. Nature 1988, 333, 93–96. [Google Scholar] [CrossRef]
- Sun, J.; Cui, J.; He, Q.; Chen, Z.; Arvan, P.; Liu, M. Proinsulin misfolding and endoplasmic reticulum stress during the development and progression of diabetes. Mol. Aspects Med. 2015, 42, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.-G.; Choi, K.-D.; Jang, S.-H.; Shin, H.-C. Role of disulfide bonds in the structure and activity of human insulin. Mol. Cells 2003, 16, 323–330. [Google Scholar] [PubMed]
- Orci, L.; Ravazzola, M.; Amherdt, M.; Madsen, O.; Perrelet, A.; Vassalli, J.D.; Anderson, R.G. Conversion of proinsulin to insulin occurs coordinately with acidification of maturing secretory vesicles. J. Cell Biol. 1986, 103, 2273–2281. [Google Scholar] [CrossRef] [PubMed]
- Orci, L.; Ravazzola, M.; Amherdt, M.; Madsen, O.; Vassalli, J.-D.; Perrelet, A. Direct identification of prohormone conversion site in insulin-secreting cells. Cell 1985, 42, 671–681. [Google Scholar] [CrossRef]
- Dunn, M.F. Zinc-ligand interactions modulate assembly and stability of the insulin hexamer—A review. Biometals 2005, 18, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Marsh, B.J.; Soden, C.; Alarcón, C.; Wicksteed, B.L.; Yaekura, K.; Costin, A.J.; Morgan, G.P.; Rhodes, C.J. Regulated autophagy controls hormone content in secretory-deficient pancreatic endocrine β-cells. Mol. Endocrinol. 2007, 21, 2255–2269. [Google Scholar] [CrossRef] [PubMed]
- Rorsman, P.; Renstrom, E. Insulin granule dynamics in pancreatic beta cells. Diabetologia 2003, 46, 1029–1045. [Google Scholar] [CrossRef] [PubMed]
- Kasai, H.; Hatakeyama, H.; Ohno, M.; Takahashi, N. Exocytosis in islet beta-cells. Adv. Exp. Med. Biol. 2010, 654, 305–338. [Google Scholar]
- Daniel, S.; Noda, M.; Straub, S.G.; Sharp, G. Identification of the docked granule pool responsible for the first phase of glucose-stimulated insulin secretion. Diabetes 1999, 48, 1686–1690. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Gandasi, N.R.; Arora, S.; Omar-Hmeadi, M.; Saras, J.; Barg, S. Syntaxin clusters at secretory granules in a munc18-bound conformation. Mol. Biol. Cell 2018, 29, 2700–2708. [Google Scholar] [CrossRef] [PubMed]
- Hoboth, P.; Muller, A.; Ivanova, A.; Mziaut, H.; Dehghany, J.; Sonmez, A.; Lachnit, M.; Meyer-Hermann, M.; Kalaidzidis, Y.; Solimena, M. Aged insulin granules display reduced microtubule-dependent mobility and are disposed within actin-positive multigranular bodies. Proc. Natl. Acad. Sci. USA 2015, 112, E667–E676. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, A.; Kalaidzidis, Y.; Dirkx, R.; Sarov, M.; Gerlach, M.; Schroth-Diez, B.; Muller, A.; Liu, Y.; Andree, C.; Mulligan, B.; et al. Age-dependent labeling and imaging of insulin secretory granules. Diabetes 2013, 62, 3687–3696. [Google Scholar] [CrossRef]
- Yau, B.; Hays, L.; Liang, C.; Laybutt, D.R.; Thomas, H.E.; Gunton, J.E.; Williams, L.; Hawthorne, W.J.; Thorn, P.; Rhodes, C.J.; et al. A fluorescent timer reporter enables sorting of insulin secretory granules by age. J. Biol. Chem. 2020, 295, 8901–8911. [Google Scholar] [CrossRef]
- Hanna, S.T.; Pigeau, G.M.; Galvanovskis, J.; Clark, A.; Rorsman, P.; Macdonald, P.E. Kiss-and-run exocytosis and fusion pores of secretory vesicles in human beta-cells. Pflugers Arch. 2009, 457, 1343–1350. [Google Scholar] [CrossRef]
- Pasquier, A.; Vivot, K.; Erbs, E.; Spiegelhalter, C.; Zhang, Z.; Aubert, V.; Liu, Z.; Senkara, M.; Maillard, E.; Pinget, M. Lysosomal degradation of newly formed insulin granules contributes to β cell failure in diabetes. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Schuit, F.; In’t Veld, P.; Pipeleers, D. Glucose stimulates proinsulin biosynthesis by a dose-dependent recruitment of pancreatic beta cells. Proc. Natl. Acad. Sci. USA 1988, 85, 3865–3869. [Google Scholar] [CrossRef] [PubMed]
- Riahi, Y.; Wikstrom, J.D.; Bachar-Wikstrom, E.; Polin, N.; Zucker, H.; Lee, M.-S.; Quan, W.; Haataja, L.; Liu, M.; Arvan, P. Autophagy is a major regulator of beta cell insulin homeostasis. Diabetologia 2016, 59, 1480–1491. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Mziaut, H.; Neukam, M.; Knoch, K.P.; Solimena, M. A 4D view on insulin secretory granule turnover in the β-cell. Diabetes Obes. Metab. 2017, 19, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Orci, L.; Ravazzola, M.; Amherdt, M.; Yanaihara, C.; Yanaihara, N.; Halban, P.; Renold, A.; Perrelet, A. Insulin, not C-peptide (proinsulin), is present in crinophagic bodies of the pancreatic B-cell. J. Cell Biol. 1984, 98, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Díaz, C.M.; Merino, B.; López-Acosta, J.F.; Cidad, P.; de la Fuente, M.A.; Lobatón, C.D.; Moreno, A.; Leissring, M.A.; Perdomo, G.; Cózar-Castellano, I. Pancreatic β-cell-specific deletion of insulin-degrading enzyme leads to dysregulated insulin secretion and β-cell functional immaturity. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E805–E819. [Google Scholar] [CrossRef] [PubMed]
- Steneberg, P.; Bernardo, L.; Edfalk, S.; Lundberg, L.; Backlund, F.; Östenson, C.-G.; Edlund, H. The type 2 diabetes–associated gene Ide is required for insulin secretion and suppression of α-synuclein levels in β-cells. Diabetes 2013, 62, 2004–2014. [Google Scholar] [CrossRef] [PubMed]
- Marselli, L.; Suleiman, M.; Masini, M.; Campani, D.; Bugliani, M.; Syed, F.; Martino, L.; Focosi, D.; Scatena, F.; Olimpico, F.; et al. Are we overestimating the loss of beta cells in type 2 diabetes? Diabetologia 2014, 57, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Cinti, F.; Bouchi, R.; Kim-Muller, J.Y.; Ohmura, Y.; Sandoval, P.R.; Masini, M.; Marselli, L.; Suleiman, M.; Ratner, L.E.; Marchetti, P. Evidence of β-cell dedifferentiation in human type 2 diabetes. J. Clin. Endocrinol. Metab. 2016, 101, 1044–1054. [Google Scholar] [CrossRef]
- Obermüller, S.; Calegari, F.; King, A.; Lindqvist, A.; Lundquist, I.; Salehi, A.; Francolini, M.; Rosa, P.; Rorsman, P.; Huttner, W.B. Defective secretion of islet hormones in chromogranin-B deficient mice. PLoS ONE 2010, 5, e8936. [Google Scholar] [CrossRef]
- Burns, C.H.; Yau, B.; Rodriguez, A.; Triplett, J.; Maslar, D.; An, Y.S.; van der Welle, R.E.; Kossina, R.G.; Fisher, M.R.; Strout, G.W. Pancreatic β-Cell–Specific Deletion of VPS41 Causes Diabetes Due to Defects in Insulin Secretion. Diabetes 2021, 70, 436–448. [Google Scholar] [CrossRef]
- White, M.G.; Shaw, J.A.; Taylor, R. Type 2 Diabetes: The Pathologic Basis of Reversible beta-Cell Dysfunction. Diabetes Care 2016, 39, 2080–2088. [Google Scholar] [CrossRef]
- Nichols, C.G.; Remedi, M.S. The diabetic beta-cell: Hyperstimulated vs. hyperexcited. Diabetes Obes. Metab. 2012, 14 (Suppl. 3), 129–135. [Google Scholar] [CrossRef] [PubMed]
- Kebede, M.A.; Oler, A.T.; Gregg, T.; Balloon, A.J.; Johnson, A.; Mitok, K.; Rabaglia, M.; Schueler, K.; Stapleton, D.; Thorstenson, C.; et al. SORCS1 is necessary for normal insulin secretory granule biogenesis in metabolically stressed beta cells. J. Clin. Investig. 2014, 124, 4240–4256. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Huang, Y.; Xu, X.; Li, X.; Alam, M.; Arunagiri, A.; Haataja, L.; Ding, L.; Wang, S.; Itkin-Ansari, P. Normal and defective pathways in biogenesis and maintenance of the insulin storage pool. J. Clin. Investig. 2021, 131, e142240. [Google Scholar] [CrossRef]
- Docherty, K.; Hutton, J.; Steiner, D. Cathepsin B-related proteases in the insulin secretory granule. J. Biol. Chem. 1984, 259, 6041–6044. [Google Scholar] [CrossRef]
- Kuliawat, R.; Klumperman, J.; Ludwig, T.; Arvan, P. Differential sorting of lysosomal enzymes out of the regulated secretory pathway in pancreatic beta-cells. J. Cell Biol. 1997, 137, 595–608. [Google Scholar] [CrossRef]
- Naik, A.R.; Formosa, B.J.; Pulvender, R.G.; Liyanaarachchi, A.G.; Jena, B.P. vH+-ATPase-induced intracellular acidification is critical to glucose-stimulated insulin secretion in beta cells. Histochem. Cell Biol. 2020, 153, 279–285. [Google Scholar] [CrossRef]
- Chimienti, F.; Devergnas, S.; Favier, A.; Seve, M. Identification and cloning of a β-cell–specific zinc transporter, ZnT-8, localized into insulin secretory granules. Diabetes 2004, 53, 2330–2337. [Google Scholar] [CrossRef]
- Wang, Y.-S.; Tzeng, H.-T.; Tsai, C.-H.; Cheng, H.-C.; Lai, W.-W.; Liu, H.-S.; Wang, Y.-C. VAMP8, a vesicle-SNARE required for RAB37-mediated exocytosis, possesses a tumor metastasis suppressor function. Cancer Lett. 2018, 437, 79–88. [Google Scholar] [CrossRef]
- Rindler, M.J. Carboxypeptidase E, a peripheral membrane protein implicated in the targeting of hormones to secretory granules, co-aggregates with granule content proteins at acidic pH. J. Biol. Chem. 1998, 273, 31180–31185. [Google Scholar] [CrossRef][Green Version]
- Stephens, S.B.; Edwards, R.J.; Sadahiro, M.; Lin, W.-J.; Jiang, C.; Salton, S.R.; Newgard, C.B. The prohormone VGF regulates β cell function via insulin secretory granule biogenesis. Cell Rep. 2017, 20, 2480–2489. [Google Scholar] [CrossRef]
- Hendy, G.N.; Li, T.; Girard, M.; Feldstein, R.C.; Mulay, S.; Desjardins, R.; Day, R.; Karaplis, A.C.; Tremblay, M.L.; Canaff, L. Targeted ablation of the chromogranin a (Chga) gene: Normal neuroendocrine dense-core secretory granules and increased expression of other granins. Mol. Endocrinol. 2006, 20, 1935–1947. [Google Scholar] [CrossRef]
- Yoshie, S.; Hagn, C.; Ehrhart, M.; Fischer-Colbrie, R.; Grube, D.; Winkler, H.; Gratzl, M. Immunological characterization of chromogranins A and B and secretogranin II in the bovine pancreatic islet. Histochemistry 1987, 87, 99–106. [Google Scholar] [CrossRef]
- Bearrows, S.C.; Bauchle, C.J.; Becker, M.; Haldeman, J.M.; Swaminathan, S.; Stephens, S.B. Chromogranin B regulates early-stage insulin granule trafficking from the Golgi in pancreatic islet beta-cells. J. Cell Sci. 2019, 132, jcs231373. [Google Scholar] [CrossRef]
- Chen, Y.; Xia, Z.; Wang, L.; Yu, Y.; Liu, P.; Song, E.; Xu, T. An efficient two-step subcellular fractionation method for the enrichment of insulin granules from INS-1 cells. Biophys. Rep. 2015, 1, 34–40. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hutton, J.; Penn, E.; Peshavaria, M. Isolation and characterisation of insulin secretory granules from a rat islet cell tumour. Diabetologia 1982, 23, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.M.; Saermark, T.; Howell, S.L. A small-scale method for the isolation of insulin-containing secretory granules from islets of Langerhans. Anal. Biochem. 1987, 166, 142–149. [Google Scholar] [CrossRef]
- Hickey, A.J.; Bradley, J.W.; Skea, G.L.; Middleditch, M.J.; Buchanan, C.M.; Phillips, A.R.; Cooper, G.J. Proteins associated with immunopurified granules from a model pancreatic islet beta-cell system: Proteomic snapshot of an endocrine secretory granule. J. Proteome Res. 2009, 8, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Neukam, M.; Gans, K.; Vasiljević, J.; Broichhagen, J.; Johnsson, K.; Kurth, T.; Solimena, M. Purification of age-distinct insulin secretory granules through antigen restriction using the CLIP-tag. bioRxiv 2020. [Google Scholar] [CrossRef]
- Guest, P.C. Multiplex Sequential Immunoprecipitation of Insulin Secretory Granule Proteins from Radiolabeled Pancreatic Islets. Methods Mol. Biol. 2017, 1546, 177–185. [Google Scholar] [PubMed]
- Lee, Y.H.; Tan, H.T.; Chung, M.C. Subcellular fractionation methods and strategies for proteomics. Proteomics 2010, 10, 3935–3956. [Google Scholar] [CrossRef] [PubMed]
- Ferri, G.; Digiacomo, L.; Lavagnino, Z.; Occhipinti, M.; Bugliani, M.; Cappello, V.; Caracciolo, G.; Marchetti, P.; Piston, D.W.; Cardarelli, F. Insulin secretory granules labelled with phogrin-fluorescent proteins show alterations in size, mobility and responsiveness to glucose stimulation in living beta-cells. Sci. Rep. 2019, 9, 2890. [Google Scholar] [CrossRef]
- Giordano, T.; Brigatti, C.; Podini, P.; Bonifacio, E.; Meldolesi, J.; Malosio, M. Beta cell chromogranin B is partially segregated in distinct granules and can be released separately from insulin in response to stimulation. Diabetologia 2008, 51, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Ramm, G.; Slot, J.W.; James, D.E.; Stoorvogel, W. Insulin recruits GLUT4 from specialized VAMP2-carrying vesicles as well as from the dynamic endosomal/trans-Golgi network in rat adipocytes. Mol. Biol. Cell 2000, 11, 4079–4091. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, T.; Shibasaki, T.; Minami, K.; Takahashi, H.; Mizoguchi, A.; Uriu, Y.; Numata, T.; Mori, Y.; Miyazaki, J.-I.; Miki, T. Rim2α determines docking and priming states in insulin granule exocytosis. Cell Metab. 2010, 12, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Matsunaga, K.; Wang, H.; Ishizaki, R.; Kobayashi, E.; Kiyonari, H.; Mukumoto, Y.; Okunishi, K.; Izumi, T. Exophilin-8 assembles secretory granules for exocytosis in the actin cortex via interaction with RIM-BP2 and myosin-VIIa. eLife 2017, 6, e26174. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, T.; Liu, X.; Yuan, Q.; Xiao, T.; Yuan, X.; Zhang, Y.; Yuan, L.; Wang, Y. CASK, APBA1, and STXBP1 collaborate during insulin secretion. Mol. Cell. Endocrinol. 2021, 520, 111076. [Google Scholar] [CrossRef]
- Tooze, S.A.; Flatmark, T.; Tooze, J.; Huttner, W.B. Characterization of the immature secretory granule, an intermediate in granule biogenesis. J. Cell Biol. 1991, 115, 1491–1503. [Google Scholar] [CrossRef]
- Li, M.; Du, W.; Zhou, M.; Zheng, L.; Song, E.; Hou, J. Proteomic analysis of insulin secretory granules in INS-1 cells by protein correlation profiling. Biophys. Rep. 2018, 4, 329–338. [Google Scholar] [CrossRef]
- MacDonald, M.J.; Ade, L.; Ntambi, J.M.; Ansari, I.-U.H.; Stoker, S.W. Characterization of phospholipids in insulin secretory granules and mitochondria in pancreatic beta cells and their changes with glucose stimulation. J. Biol. Chem. 2015, 290, 11075–11092. [Google Scholar] [CrossRef]
- Albrethsen, J.; Goetze, J.P.; Johnsen, A.H. Mining the granule proteome: A potential source of endocrine biomarkers. Biomark. Med. 2015, 9, 259–265. [Google Scholar] [CrossRef]
- Brunner, Y.; Coute, Y.; Iezzi, M.; Foti, M.; Fukuda, M.; Hochstrasser, D.F.; Wollheim, C.B.; Sanchez, J.C. Proteomics analysis of insulin secretory granules. Mol. Cell. Proteom. 2007, 6, 1007–1017. [Google Scholar] [CrossRef]
- Chandramouli, K.; Qian, P.Y. Proteomics: Challenges, techniques and possibilities to overcome biological sample complexity. Hum. Genom. Proteom. 2009, 2009, 239204. [Google Scholar] [CrossRef]
- Rindler, M.J.; Xu, C.F.; Gumper, I.; Smith, N.N.; Neubert, T.A. Proteomic analysis of pancreatic zymogen granules: Identification of new granule proteins. J. Proteome Res. 2007, 6, 2978–2992. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schvartz, D.; Brunner, Y.; Coute, Y.; Foti, M.; Wollheim, C.B.; Sanchez, J.C. Improved characterization of the insulin secretory granule proteomes. J. Proteom. 2012, 75, 4620–4631. [Google Scholar] [CrossRef] [PubMed]
- Lomedico, P.; Rosenthal, N.; Efstratiadis, A.; Gilbert, W.; Kolodner, R.; Tizard, R. The structure and evolution of the two nonallelic rat preproinsulin genes. Cell 1979, 18, 545–558. [Google Scholar] [CrossRef]
- Ullrich, A.; Shine, J.; Chirgwin, J.; Pictet, R.; Tischer, E.; Rutter, W.J.; Goodman, H.M. Rat insulin genes: Construction of plasmids containing the coding sequences. Science 1977, 196, 1313–1319. [Google Scholar] [CrossRef]
- Wasmeier, C.; Hutton, J.C. Molecular cloning of phogrin, a protein-tyrosine phosphatase homologue localized to insulin secretory granule membranes. J. Biol. Chem. 1996, 271, 18161–18170. [Google Scholar] [CrossRef]
- Hoflehner, J.; Eder, U.; Laslop, A.; Seidah, N.G.; Fischer-Colbrie, R.; Winkler, H. Processing of secretogranin II by prohormone convertases: Importance ofPC1 in generation of secretoneurin. FEBS Lett. 1995, 360, 294–298. [Google Scholar] [PubMed]
- Laslop, A.; Weiss, C.; Savaria, D.; Eiter, C.; Tooze, S.A.; Seidah, N.G.; Winkler, H. Proteolytic processing of chromogranin B and secretogranin II by prohormone convertases. J. Neurochem. 1998, 70, 374–383. [Google Scholar] [CrossRef]
- Davidson, H.W.; Peshavaria, M.; Hutton, J.C. Proteolytic conversion of proinsulin into insulin. Identification of a Ca2+-dependent acidic endopeptidase in isolated insulin-secretory granules. Biochem. J. 1987, 246, 279–286. [Google Scholar] [CrossRef]
- Hutton, J.C.; Davidson, H.W.; Peshavaria, M. Proteolytic processing of chromogranin A in purified insulin granules. Formation of a 20 kDa N-terminal fragment (betagranin) by the concerted action of a Ca2+-dependent endopeptidase and carboxypeptidase H (EC 3.4. 17.10). Biochem. J. 1987, 244, 457–464. [Google Scholar] [CrossRef]
- Muller, L.; Lindberg, I. The cell biology of the prohormone convertases PCI and PC2. Prog. Nucleic Acid Res. Mol. Biol. 1999, 63, 69–108. [Google Scholar] [PubMed]
- Bennett, D.L.; Bailyes, E.; Nielsen, E.; Guest, P.; Rutherford, N.; Arden, S.; Hutton, J. Identification of the type 2 proinsulin processing endopeptidase as PC2, a member of the eukaryote subtilisin family. J. Biol. Chem. 1992, 267, 15229–15236. [Google Scholar] [CrossRef]
- Guest, P.C.; Ravazzola, M.; Davidson, H.W.; Orci, L.; Hutton, J.C. Molecular heterogeneity and cellular localization of carboxypeptidase H in the islets of Langerhans. Endocrinology 1991, 129, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Tao-Cheng, J.-H.; Eiden, L.E.; Loh, Y.P. Chromogranin A, an “on/off” switch controlling dense-core secretory granule biogenesis. Cell 2001, 106, 499–509. [Google Scholar] [CrossRef]
- Portela-Gomes, G.; Gayen, J.; Grimelius, L.; Stridsberg, M.; Mahata, S. The importance of chromogranin A in the development and function of endocrine pancreas. Regul. Pept. 2008, 151, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Udupi, V.; Lee, H.-M.; Kurosky, A.; Greeley, G.H., Jr. Prohormone convertase-1 is essential for conversion of chromogranin A to pancreastatin. Regul. Pept. 1999, 83, 123–127. [Google Scholar] [CrossRef]
- Schmid, G.M.; Meda, P.; Caille, D.; Wargent, E.; O’Dowd, J.; Hochstrasser, D.F.; Cawthorne, M.A.; Sanchez, J.-C. Inhibition of insulin secretion by betagranin, an N-terminal chromogranin A fragment. J. Biol. Chem. 2007, 282, 12717–12724. [Google Scholar] [CrossRef]
- Hutton, J.C. Insulin secretory granule biogenesis and the proinsulin-processing endopeptidases. Diabetologia 1994, 37, S48–S56. [Google Scholar] [CrossRef]
- Davidson, H.W.; Hutton, J.C. The insulin-secretory-granule carboxypeptidase H. Purification and demonstration of involvement in proinsulin processing. Biochem. J. 1987, 245, 575–582. [Google Scholar] [CrossRef]
- Sandberg, M.; Borg, L.H. Intracellular degradation of insulin and crinophagy are maintained by nitric oxide and cyclo-oxygenase 2 activity in isolated pancreatic islets. Biol. Cell 2006, 98, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Klumperman, J.; Kuliawat, R.; Griffith, J.M.; Geuze, H.J.; Arvan, P. Mannose 6–phosphate receptors are sorted from immature secretory granules via adaptor protein AP-1, clathrin, and syntaxin 6–positive vesicles. J. Cell Biol. 1998, 141, 359–371. [Google Scholar] [CrossRef]
- Arvan, P.; Castle, D. Sorting and storage during secretory granule biogenesis: Looking backward and looking forward. Biochem. J. 1998, 332, 593–610. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.C.; Scheller, R.H. Mechanisms of synaptic vesicle exocytosis. Annu. Rev. Cell Dev. Biol. 2000, 16, 19–49. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.P.; Ohno, M.; Dolai, S.; He, Y.; Qin, T.; Liang, T.; Zhu, D.; Kang, Y.; Liu, Y.; Kauppi, M. Munc18b is a major mediator of insulin exocytosis in rat pancreatic β-cells. Diabetes 2013, 62, 2416–2428. [Google Scholar] [CrossRef] [PubMed]
- Iorio, V.; Festa, M.; Rosati, A.; Hahne, M.; Tiberti, C.; Capunzo, M.; De Laurenzi, V.; Turco, M. BAG3 regulates formation of the SNARE complex and insulin secretion. Cell Death Dis. 2015, 6, e1684. [Google Scholar] [CrossRef]
- Gaisano, H.Y. Recent new insights into the role of SNARE and associated proteins in insulin granule exocytosis. Diabetes Obes. Metab. 2017, 19 (Suppl. 1), 115–123. [Google Scholar] [CrossRef] [PubMed]
- Qin, T.; Liang, T.; Zhu, D.; Kang, Y.; Xie, L.; Dolai, S.; Sugita, S.; Takahashi, N.; Ostenson, C.-G.; Banks, K. Munc18b increases insulin granule fusion, restoring deficient insulin secretion in type-2 diabetes human and Goto-Kakizaki rat islets with improvement in glucose homeostasis. EBioMedicine 2017, 16, 262–274. [Google Scholar] [CrossRef] [PubMed]
- Regazzi, R.; Wollheim, C.; Lang, J.; Theler, J.; Rossetto, O.; Montecucco, C.; Sadoul, K.; Weller, U.; Palmer, M.; Thorens, B. VAMP-2 and cellubrevin are expressed in pancreatic beta-cells and are essential for Ca(2+)-but not for GTP gamma S-induced insulin secretion. EMBO J. 1995, 14, 2723–2730. [Google Scholar] [CrossRef]
- Martinez, O.; Goud, B. Rab proteins. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 1998, 1404, 101–112. [Google Scholar] [CrossRef]
- Cheviet, S.V.; Coppola, T.; Haynes, L.P.; Burgoyne, R.D.; Regazzi, R. The Rab-binding protein Noc2 is associated with insulin-containing secretory granules and is essential for pancreatic β-cell exocytosis. Mol. Endocrinol. 2004, 18, 117–126. [Google Scholar] [CrossRef]
- Wang, Z.; Thurmond, D.C. Mechanisms of biphasic insulin-granule exocytosis–roles of the cytoskeleton, small GTPases and SNARE proteins. J. Cell Sci. 2009, 122, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, K.; Fujita, T.; Gomi, H.; Izumi, T. Granuphilin exclusively mediates functional granule docking to the plasma membrane. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hatanaka, M.; Tanabe, K.; Yanai, A.; Ohta, Y.; Kondo, M.; Akiyama, M.; Shinoda, K.; Oka, Y.; Tanizawa, Y. Wolfram syndrome 1 gene (WFS1) product localizes to secretory granules and determines granule acidification in pancreatic β-cells. Hum. Mol. Genet. 2011, 20, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Barg, S.; Huang, P.; Eliasson, L.; Nelson, D.J.; Obermüller, S.; Rorsman, P.; Thévenod, F.; Renström, E. Priming of insulin granules for exocytosis by granular Cl− uptake and acidification. J. Cell Sci. 2001, 114, 2145–2154. [Google Scholar] [CrossRef]
- Gharanei, S.; Zatyka, M.; Astuti, D.; Fenton, J.; Sik, A.; Nagy, Z.; Barrett, T.G. Vacuolar-type H+-ATPase V1A subunit is a molecular partner of Wolfram syndrome 1 (WFS1) protein, which regulates its expression and stability. Hum. Mol. Genet. 2013, 22, 203–217. [Google Scholar] [CrossRef]
- Orci, L.; Ravazzola, M.; Storch, M.-J.; Anderson, R.; Vassalli, J.-D.; Perrelet, A. Proteolytic maturation of insulin is a post-Golgi event which occurs in acidifying clathrin-coated secretory vesicles. Cell 1987, 49, 865–868. [Google Scholar] [CrossRef]
- Heaslip, A.T.; Nelson, S.R.; Lombardo, A.T.; Previs, S.B.; Armstrong, J.; Warshaw, D.M. Cytoskeletal dependence of insulin granule movement dynamics in INS-1 beta-cells in response to glucose. PLoS ONE 2014, 9, e109082. [Google Scholar] [CrossRef]
- Wang, B.; Lin, H.; Li, X.; Lu, W.; Kim, J.B.; Xu, A.; Cheng, K.K. The adaptor protein APPL2 controls glucose-stimulated insulin secretion via F-actin remodeling in pancreatic β-cells. Proc. Natl. Acad. Sci. USA 2020, 117, 28307–28315. [Google Scholar] [CrossRef]
- Ghiasi, S.M.; Dahlby, T.; Andersen, C.H.; Haataja, L.; Petersen, S.; Omar-Hmeadi, M.; Yang, M.; Pihl, C.; Bresson, S.E.; Khilji, M.S. Endoplasmic reticulum chaperone glucose-regulated protein 94 is essential for proinsulin handling. Diabetes 2019, 68, 747–760. [Google Scholar] [CrossRef]
- Varghese, R.; Wong, C.K.; Deol, H.; Wagner, G.F.; DiMattia, G.E. Comparative analysis of mammalian stanniocalcin genes. Endocrinology 1998, 139, 4714–4725. [Google Scholar] [CrossRef]
- Ellard, J.P.; McCudden, C.R.; Tanega, C.; James, K.A.; Ratkovic, S.; Staples, J.F.; Wagner, G.F. The respiratory effects of stanniocalcin-1 (STC-1) on intact mitochondria and cells: STC-1 uncouples oxidative phosphorylation and its actions are modulated by nucleotide triphosphates. Mol. Cell. Endocrinol. 2007, 264, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Iversen, P.; Sorensen, D.; Benestad, H. Inhibitors of angiogenesis selectively reduce the malignant cell load in rodent models of human myeloid leukemias. Leukemia 2002, 16, 376–381. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zaidi, D.; Turner, J.K.; Durst, M.A.; Wagner, G.F. Stanniocalcin-1 co-localizes with insulin in the pancreatic islets. Int. Sch. Res. Not. 2012, 2012, 834359. [Google Scholar] [CrossRef] [PubMed]
- Vakilian, M.; Tahamtani, Y.; Ghaedi, K. A review on insulin trafficking and exocytosis. Gene 2019, 706, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Olofsson, C.S.; Gopel, S.O.; Barg, S.; Galvanovskis, J.; Ma, X.; Salehi, A.; Rorsman, P.; Eliasson, L. Fast insulin secretion reflects exocytosis of docked granules in mouse pancreatic B-cells. Pflugers Arch. 2002, 444, 43–51. [Google Scholar] [CrossRef]
- Olofsson, C.S.; Salehi, A.; Holm, C.; Rorsman, P. Palmitate increases L-type Ca2+ currents and the size of the readily releasable granule pool in mouse pancreatic β-cells. J. Physiol. 2004, 557, 935–948. [Google Scholar] [CrossRef]
- Pottekat, A.; Becker, S.; Spencer, K.R.; Yates III, J.R.; Manning, G.; Itkin-Ansari, P.; Balch, W.E. Insulin biosynthetic interaction network component, TMEM24, facilitates insulin reserve pool release. Cell Rep. 2013, 4, 921–930. [Google Scholar] [CrossRef]
- Gomi, H.; Mizutani, S.; Kasai, K.; Itohara, S.; Izumi, T. Granuphilin molecularly docks insulin granules to the fusion machinery. J. Cell Biol. 2005, 171, 99–109. [Google Scholar] [CrossRef]
- Gandasi, N.R.; Yin, P.; Omar-Hmeadi, M.; Laakso, E.O.; Vikman, P.; Barg, S. Glucose-dependent granule docking limits insulin secretion and is decreased in human type 2 diabetes. Cell Metab. 2018, 27, 470–478.e4. [Google Scholar] [CrossRef]
- Hao, M.; Li, X.; Rizzo, M.A.; Rocheleau, J.V.; Dawant, B.M.; Piston, D.W. Regulation of two insulin granule populations within the reserve pool by distinct calcium sources. J. Cell Sci. 2005, 118, 5873–5884. [Google Scholar] [CrossRef]
- Gaisano, H.Y. Here come the newcomer granules, better late than never. Trends Endocrinol. Metab. 2014, 25, 381–388. [Google Scholar] [CrossRef]
- Zhu, D.; Koo, E.; Kwan, E.; Kang, Y.; Park, S.; Xie, H.; Sugita, S.; Gaisano, H. Syntaxin-3 regulates newcomer insulin granule exocytosis and compound fusion in pancreatic beta cells. Diabetologia 2013, 56, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, M.G.; Sherman, A. Newcomer insulin secretory granules as a highly calcium-sensitive pool. Proc. Natl. Acad. Sci. USA 2009, 106, 7432–7436. [Google Scholar] [CrossRef]
- Kreutzberger, A.J.; Kiessling, V.; Doyle, C.A.; Schenk, N.; Upchurch, C.M.; Elmer-Dixon, M.; Ward, A.E.; Preobraschenski, J.; Hussein, S.S.; Tomaka, W. Distinct insulin granule subpopulations implicated in the secretory pathology of diabetes types 1 and 2. eLife 2020, 9, e62506. [Google Scholar] [CrossRef]
- Hou, N.; Mogami, H.; Kubota-Murata, C.; Sun, M.; Takeuchi, T.; Torii, S. Preferential release of newly synthesized insulin assessed by a multi-label reporter system using pancreatic beta-cell line MIN6. PLoS ONE 2012, 7, e47921. [Google Scholar] [CrossRef] [PubMed]
- Tsuboi, T.; Kitaguchi, T.; Karasawa, S.; Fukuda, M.; Miyawaki, A. Age-dependent preferential dense-core vesicle exocytosis in neuroendocrine cells revealed by newly developed monomeric fluorescent timer protein. Mol. Biol. Cell 2010, 21, 87–94. [Google Scholar] [CrossRef]
- Hays, L.B.; Wicksteed, B.; Wang, Y.; McCuaig, J.F.; Philipson, L.H.; Edwardson, J.M.; Rhodes, C.J. Intragranular targeting of syncollin, but not a syncollinGFP chimera, inhibits regulated insulin exocytosis in pancreatic beta-cells. J. Endocrinol. 2005, 185, 57–67. [Google Scholar] [CrossRef]
- Gauthier, D.J.; Sobota, J.A.; Ferraro, F.; Mains, R.E.; Lazure, C. Flow cytometry-assisted purification and proteomic analysis of the corticotropes dense-core secretory granules. Proteomics 2008, 8, 3848–3861. [Google Scholar] [CrossRef] [PubMed]
- Macklin, A.; Khan, S.; Kislinger, T. Recent advances in mass spectrometry based clinical proteomics: Applications to cancer research. Clin. Proteom. 2020, 17, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Yates, J.R.; Ruse, C.I.; Nakorchevsky, A. Proteomics by mass spectrometry: Approaches, advances, and applications. Annu. Rev. Biomed. Eng. 2009, 11, 49–79. [Google Scholar] [CrossRef] [PubMed]
- Timp, W.; Timp, G. Beyond mass spectrometry, the next step in proteomics. Sci. Adv. 2020, 6, eaax8978. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, J.; Boulgakov, A.A.; Hernandez, E.T.; Bardo, A.M.; Bachman, J.L.; Marotta, J.; Johnson, A.M.; Anslyn, E.V.; Marcotte, E.M. Highly parallel single-molecule identification of proteins in zeptomole-scale mixtures. Nat. Biotechnol. 2018, 36, 1076–1082. [Google Scholar] [CrossRef]
- Cao, M.; Mao, Z.; Kam, C.; Xiao, N.; Cao, X.; Shen, C.; Cheng, K.K.; Xu, A.; Lee, K.-M.; Jiang, L. PICK1 and ICA69 control insulin granule trafficking and their deficiencies lead to impaired glucose tolerance. PLoS Biol. 2013, 11, e1001541. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wei, S.; Cheng, K.; Gounko, N.V.; Ericksen, R.E.; Xu, A.; Hong, W.; Han, W. BIG3 inhibits insulin granule biogenesis and insulin secretion. EMBO Rep. 2014, 15, 714–722. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, A.D.; Manson, J.E.; Meigs, J.B.; Rifai, N.; Buring, J.E.; Liu, S.; Ridker, P.M. Insulin, proinsulin, proinsulin:insulin ratio, and the risk of developing type 2 diabetes mellitus in women. Am. J. Med. 2003, 114, 438–444. [Google Scholar] [CrossRef]
- Brackeva, B.; De Punt, V.; Kramer, G.; Costa, O.; Verhaeghen, K.; Stangé, G.; Sadones, J.; Xavier, C.; Aerts, J.; Gorus, F. Potential of UCHL1 as biomarker for destruction of pancreatic beta cells. J. Proteom. 2015, 117, 156–167. [Google Scholar] [CrossRef]
- Martens, G. Species-related differences in the proteome of rat and human pancreatic beta cells. J. Diabetes Res. 2015, 2015, 549818. [Google Scholar] [CrossRef]
- Brackeva, B.; Kramer, G.; Vissers, J.; Martens, G. Quantitative proteomics of rat and human pancreatic beta cells. Data Brief 2015, 3, 234–239. [Google Scholar] [CrossRef]
- Clardy, S.M.; Mohan, J.F.; Vinegoni, C.; Keliher, E.J.; Iwamoto, Y.; Benoist, C.; Mathis, D.; Weissleder, R. Rapid, high efficiency isolation of pancreatic ss-cells. Sci. Rep. 2015, 5, 13681. [Google Scholar] [CrossRef]
- Jayaraman, S. A novel method for the detection of viable human pancreatic beta cells by flow cytometry using fluorophores that selectively detect labile zinc, mitochondrial membrane potential and protein thiols. Cytom. A 2008, 73, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Lukowiak, B.; Vandewalle, B.; Riachy, R.; Kerr-Conte, J.; Gmyr, V.; Belaich, S.; Lefebvre, J.; Pattou, F. Identification and purification of functional human beta-cells by a new specific zinc-fluorescent probe. J. Histochem. Cytochem. 2001, 49, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Smelt, M.J.; Faas, M.M.; de Haan, B.J.; de Vos, P. Pancreatic beta-cell purification by altering FAD and NAD(P)H metabolism. Exp. Diabetes Res. 2008, 2008, 165360. [Google Scholar] [CrossRef] [PubMed]
- Van De Winkel, M.; Pipeleers, D. Autofluorescence-activated cell sorting of pancreatic islet cells: Purification of insulin-containing B-cells according to glucose-induced changes in cellular redox state. Biochem. Biophys. Res. Commun. 1983, 114, 835–842. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Norris, N.; Yau, B.; Kebede, M.A. Isolation and Proteomics of the Insulin Secretory Granule. Metabolites 2021, 11, 288. https://doi.org/10.3390/metabo11050288
Norris N, Yau B, Kebede MA. Isolation and Proteomics of the Insulin Secretory Granule. Metabolites. 2021; 11(5):288. https://doi.org/10.3390/metabo11050288
Chicago/Turabian StyleNorris, Nicholas, Belinda Yau, and Melkam Alamerew Kebede. 2021. "Isolation and Proteomics of the Insulin Secretory Granule" Metabolites 11, no. 5: 288. https://doi.org/10.3390/metabo11050288
APA StyleNorris, N., Yau, B., & Kebede, M. A. (2021). Isolation and Proteomics of the Insulin Secretory Granule. Metabolites, 11(5), 288. https://doi.org/10.3390/metabo11050288







