Abstract
AKR1B10 is a human nicotinamide adenine dinucleotide phosphate (NADPH)-dependent reductase belonging to the aldo-keto reductase (AKR) 1B subfamily. It catalyzes the reduction of aldehydes, some ketones and quinones, and interacts with acetyl-CoA carboxylase and heat shock protein 90α. The enzyme is highly expressed in epithelial cells of the stomach and intestine, but down-regulated in gastrointestinal cancers and inflammatory bowel diseases. In contrast, AKR1B10 expression is low in other tissues, where the enzyme is upregulated in cancers, as well as in non-alcoholic fatty liver disease and several skin diseases. In addition, the enzyme’s expression is elevated in cancer cells resistant to clinical anti-cancer drugs. Thus, growing evidence supports AKR1B10 as a potential target for diagnosing and treating these diseases. Herein, we reviewed the literature on the roles of AKR1B10 in a healthy gastrointestinal tract, the development and progression of cancers and acquired chemoresistance, in addition to its gene regulation, functions, and inhibitors.
1. Introduction
Aldo-keto reductases (AKRs) are a group of NAD(P)(H)-dependent enzymes catalyzing interconversions between the carbonyl and alcohol groups of endogenous and xenobiotic compounds [1]. The AKR superfamily is systematized into 16 families: AKR1 (aldehyde reductases, aldose reductases, hydroxysteroid dehydrogenases, and steroid 5b-reductases); AKR2 (mannose and xylose reductases); AKR3 (yeast AKRs); AKR4 (chalcone and codeinone reductases); AKR5 (gluconic acid reductases); AKR6 (β-subunits of the potassiumgated voltage channels); AKR7 (aflatoxin dialdehyde and succinic semialdehyde reductases); AKR8 (pyridoxal reductases); AKR9 (aryl alcohol dehydrogenases); AKR10 (Streptomyces AKRs); AKR11 (Bacillus AKRs); AKR12 (Streptomyces sugar aldehyde reductases); AKR13 (hyperthermophilic bacteria reductases); AKR14 (Escherichia coli reductases), AKR15 (Mycobacterium reductases), and AKR16 (Vibrio cholerae reductases). Each family is further divided into several subfamilies based on a >60% amino acid sequence identity. To date, fifteen AKR members have been identified in humans and belong to the AKR1A, AKR1B, AKR1C, AKR1E, AKR6A, and AKR7A subfamilies. There are three members of the human AKR1B subfamily: AKR1B1 (aldose reductase), AKR1B10 (aldose reductase-like protein-1), and AKR1B15, whose genes are clustered at chromosome 7q33 [1]. AKR1B1, AKR1B10, and an enzymatically active isoform of AKR1B15 are 36-kDa soluble monomeric proteins consisting of 316 amino acids and sharing >68% amino acid sequence identity, of which 91.5% are shared between AKR1B10 and AKR1B15 [2,3]. The three AKRs are NADPH-dependent reductases and display overlapping substrate specificities for aromatic and aliphatic aldehydes but differ in their catalytic efficiencies [2,3,4,5,6], which is notably higher for retinal (all-trans-retinaldehyde) in AKR1B10 [5]. In addition, the glucose reductase activity characteristics of AKR1B1 are very low for AKR1B10 and AKR1B15 [2,4,5], and prostaglandin F synthase activity is observed with AKR1B1, but not with AKR1B10 [7]. In contrast to AKR1B1, AKR1B10 and AKR1B15 exhibit low 17β-hydroxysteroid dehydrogenase activity for estrone and 4-androstene-3,17-dione [3,6]. For subcellular localization, AKR1B1 and AKR1B10 are cytosolic, whereas AKR1B15 is in the mitochondria [3]. The three AKR1Bs also have different tissue distributions. While AKR1B1 is ubiquitous, AKR1B10 protein is predominantly expressed in the human stomach and intestine [2,8], although its mRNA is detected in many other tissues [3,8,9]. The mRNA for AKR1B15 is predominantly expressed in the placenta, testis, skeletal muscle, and adipose tissue, where its level is lower than that of mRNA for AKR1B10 [3].
While the most studied enzyme of the AKR1B subfamily is AKR1B1, which has been implicated in the pathogenesis of diabetes complications and inflammatory disease [10,11], many studies of AKR1B10 have focused on its association with cancers and other diseases since its overexpression in hepatocellular carcinomas (HCC) was found in 1998 [12]. In addition to its role in gastrointestinal homeostasis, increasing evidence suggests that the aberrant expression of AKR1B10 promotes its diagnostic and prognostic utility as a potential tumor biomarker and elucidates its role in carcinogenesis, tumor progression, and the development of chemoresistance. In this review, we summarize recent progress towards understanding the gene regulation of AKR1B10 and its functions in gastrointestinal physiology, the pathogenesis of several cancers and skin diseases, and acquired drug resistance. Based on the available evidence, we propose that AKR1B10 is thus a potential target for drug discovery. We also provide a brief overview of AKR1B10 inhibitors.
2. Gene Regulation of AKR1B10
2.1. Factors Regulating AKR1B10 Expression
AKR1B10 is induced in various types of cancer tissues and down-regulated in gastrointestinal cancers. In addition to tumor progression, changes in AKR1B10 expression levels are associated with several noncanceraous diseases and chemoresistance (as described later). Thus, AKR1B10 expression is an important factor in the pathogenesis of these diseases. However, the molecular mechanisms of AKR1B10 gene regulation have not been fully elucidated.
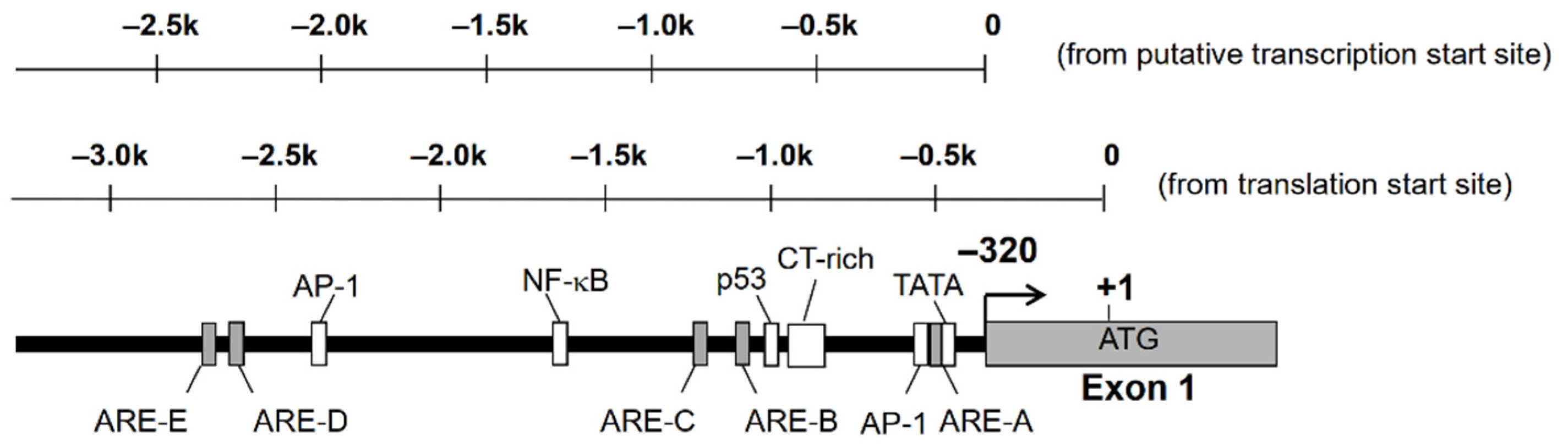
Nucleotide sequence analysis of the 5′-flanking region of AKR1B10 gene revealed the existence of putative TATA box, CAAT box, p53, AP-1, and antioxidant response elements (ARE) [13,14]. There is a complex microsatellite composed of repetitive C and T sequences, which are highly polymorphic and may affect the expression of AKR1B10 (Figure 1). We found polymorphism at this microsatellite in the human lung adenocarcinoma A549 cell line, but no significant difference in promoter activity was observed in the gene reporter assay analysis [14].
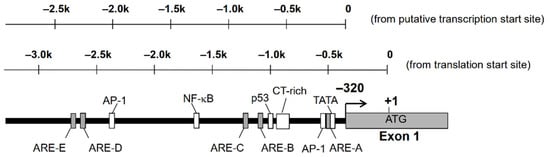
Figure 1.
Putative transcription factor binding sites on the 5′-flanking region of AKR1B10 gene.
AKR1B10 expression is affected by treatment of human cultured cells with various agents (Table 1). Most of the agents upregulate AKR1B10, whereas a phorbol ester 12-O-tetradecanoylphorbol 13-acetate (TPA) down-regulates the expression [15]. 5-Fluorouracil (5-FU) and oxaliplatin (L-OHP) exhibit opposite effects on the AKR1B10 expression depending on colorectal cancer (CRC) cell lines [16]. Regulators and signal molecules involved in the modulation of AKR1B10 expression by several agents are also listed in Table 1.

Table 1.
Agents that modulate AKR1B10 expression.
2.2. Contribution of Nrf2 to AKR1B10 Induction
An antioxidant (ethoxyquin) induces AKR1B10 expression in lung cancer cell lines A549 and H23 [14]. Ethoxyquin is known to activate the transcription factor Nrf2 (NF-E2-related factor). Furthermore, increased AKR1B10 promoter activity caused by ethoxyquin was suppressed by the introduction of dominant-negative Nrf2 protein in the gene reporter assay. In addition, the introduction of Nrf2 augmented promoter activity, indicating that AKR1B10 gene transcription is regulated by Nrf2. Nrf2 is a member of the Cap‘n’Collar family of basic leucine zipper transcription factors known to be activated by various antioxidants or reactive oxygen species (ROS). Nrf2 forms a heterodimer with small Maf proteins and binds to the ARE of target genes [24,25]. In non-stimulated cells, Nrf2 is trapped, leading to ubiquitin-dependent degradation by Kelch-like ECH-associated protein 1 (Keap1). Once the cells are stimulated, the Keap1 protein is oxidatively modified and releases Nrf2, allowing its nuclear localization [26,27]. It has been suggested that Nrf2 is involved in the up-regulation of AKR1B10 by proteasome inhibitors [17], 9,10-phenanthrenequinone [22], and doxorubicin [18]. In addition, microarray analyses consistently indicate that the Keap1/Nrf2 pathway is involved in AKR1B10 gene regulation [28,29,30]. We demonstrated in the gene reporter assay that the introduction of dominant-negative Nrf2 protein also leads to a reduction in basal transcriptional activity of AKR1B10 in A549 cells, where AKR1B10 is highly expressed. However, the treatment failed to show any effect in H23 cells where AKR1B10 expression was very low, suggesting that Nrf2 participates in the basal expression of AKR1B10 [31]. Thus, the Keap1/Nrf2 pathway is one of the major regulatory systems for AKR1B10 gene regulation.
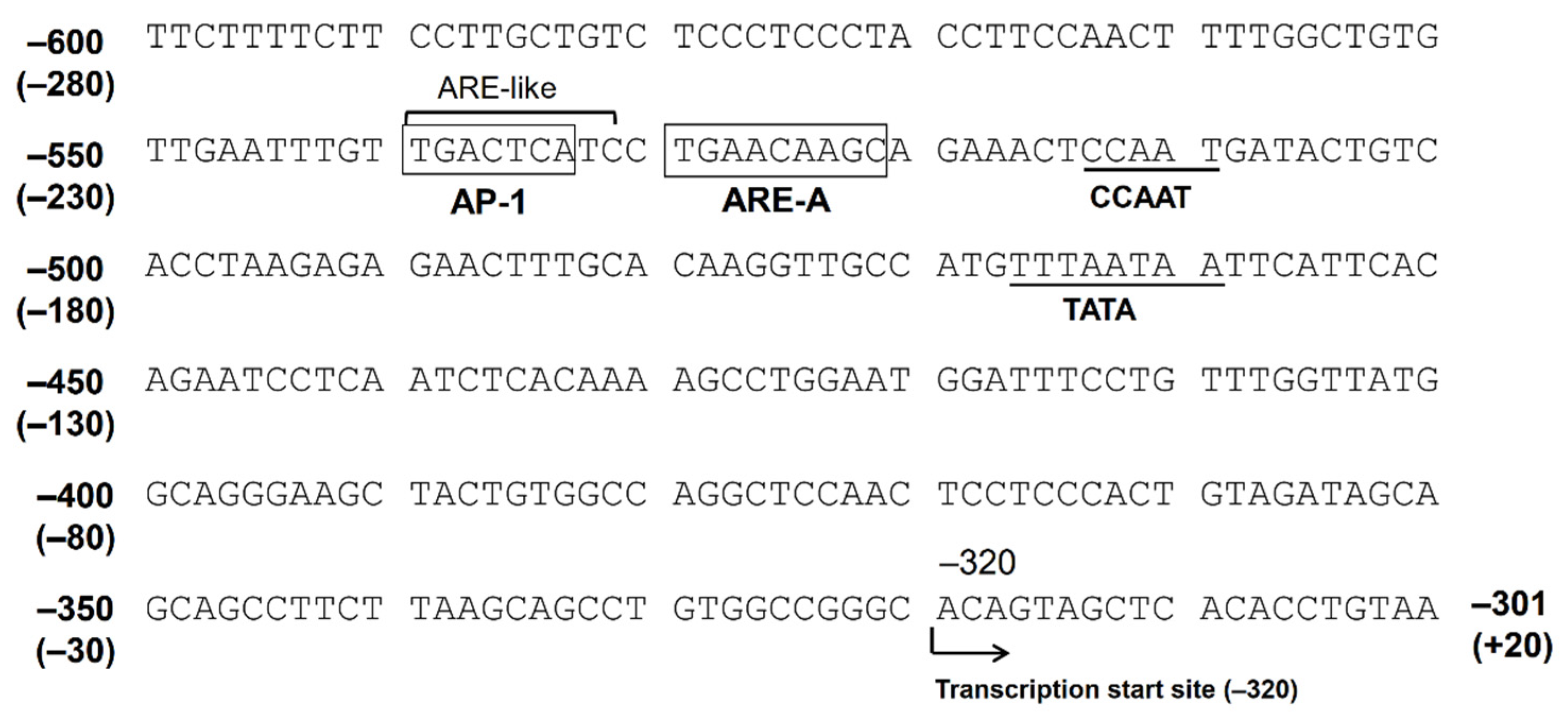
The Nrf2 ARE consensus sequence is 5′-TGACnnnGC-3′, and there are at least five potential AREs found in the 5′-flanking region, up to −3282 bp from the translation start site (−2962 bp from the putative transcription start site). We indicated that only AREs located between −530 bp and −522 bp (between −210 bp and −202 bp from the transcription start site, ARE-A) are functional among these five AREs [31]. There is an AP-1 site (between −540 bp and −532 bp) just upstream of this ARE. The consensus sequence of AP-1 is 5′-TGACTCA-3′, and the corresponding nucleotide sequence of the AKR1B10 gene, 5′-TGACTCATC-3′, resembles the ARE. In fact, the introduction of a mutation in the AP-1 site resulted in a reduced response to Nrf2 in the gene reporter assay. Accordingly, this AP-1 site may function as an ARE, and this tandem repeat of AREs may be important for a strong response to stimulations targeting Nrf2 [31] (Figure 2).
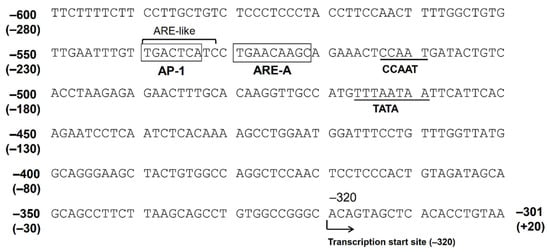
Figure 2.
The nucleotide sequence of the 5′-flanking region of the AKR1B10 gene. The numbers represent the nucleotide position when +1 is defined as the translation start site. The numbers in parentheses represent the position when +1 is defined as the putative transcription start site.
2.3. The Function of AP-1 Protein in AKR1B10 Gene Regulation
The induction of AKR1B10 by EGF and insulin in HepG2 cells also shows the importance of the AP-1 site [19]. The AP-1 protein consists of a heterodimer of c-Jun and c-Fos proteins binding to the AP-1 site [32]. Cell treatment with EGF or insulin significantly increases the amount of AP-1 protein. The introduction of c-Jun and c-Fos proteins also augments transcriptional activity in gene reporter assays. Interestingly, the injection of c-Fos shRNA to HepG2-inoculated mice to knockdown c-Fos results in the loss of AKR1B10 expression, suggesting that endogenously expressing c-Fos is an important regulator in the expression of AKR1B10. Furthermore, Cheng et al. [33] have proposed that AP-1 is involved in the induction of AKR1B10 in HCC cells. They show that the knockdown of interleukin-1 associated kinase 1 (IRAK1) leads to a decrease in AKR1B10 expression possibly mediated through regulating AP-1 signal transduction.
On the other hand, the c-Jun component of AP-1 protein has the opposite function on AKR1B10 gene regulation. We have shown that TPA downregulates AKR1B10 expression through the induction of c-Jun protein in A549 cells [15]. In addition, the forced expression of c-Jun protein in A549 cells resulted in decreased AKR1B10 expression, and the introduction of c-Jun in gene reporter assays led to the suppression of transcriptional activity in an AP-1 site-independent manner. The leucine zipper domain is necessary for c-Jun to exhibit its suppressive action, indicating that c-Jun needs to interact with other proteins. Hence, c-Jun also acts as a negative regulator. Although it is not clear how c-Jun acts differently, the variations of other protein factors existing in the cells, such as c-Fos protein, may determine the action of c-Jun.
2.4. Signal Transduction
The cAMP-dependent signaling pathway may take part in the regulation of AKR1B10, because a phosphodiesterase inhibitor, IBMX, with BMP2, is able to induce AKR1B10 expression [21]. Since a potential cAMP-response element (CRE) has not been identified in the AKR1B10 gene promoter region to date, it could be that this pathway might modify the other signaling cascades. Extracellular signal-regulated kinase (ERK), a mitogen-activated protein kinase (MAPK), is an important signal molecule in AKR1B10 gene regulation. The induction of AKR1B10 by EGF and insulin is mediated through the activation of ERK in HepG2 cells, and totally abolished by the inhibition of its upstream kinase, MAPK/ERK kinase (MEK) [19]. Thus, the receptor tyrosine kinase/ERK cascade is one of the primary signaling pathways involved in the induction of AKR1B10. In addition, ERK regulates the basal expression of AKR1B10 in this cell line as well as in A549 cells, which is suggested by studies using MEK inhibition [15]. On the other hand, ERK activation is also a key event in the TPA-mediated down-regulation of AKR1B10. The inhibition of MEK resulted in decreased c-Jun induction, leading to the loss of TPA’s action. It is very interesting that ERK activation is involved in both up-regulation and down-regulation of AKR1B10 expression as seen in the case of c-Jun.
Recently, p53 protein, a tumor suppressor gene product, was shown to play a key role in determining whether AKR1B10 expression is upregulated or down-regulated. In CRC HT29 cells possessing mutant p53, treatment with 5-FU or L-OHP reduced AKR1B10 expression. Alternatively, expression was induced by these agents in c CRC HCT116 cells having wild-type p53. When the p53 gene in HCT116 cells is knocked-down, 5-FU can no longer induce AKR1B10 [16]. Thus, p53 protein might act as a switch to determine the direction of AKR1B10 expression, suggesting that the level and status of p53 might be an important factor.
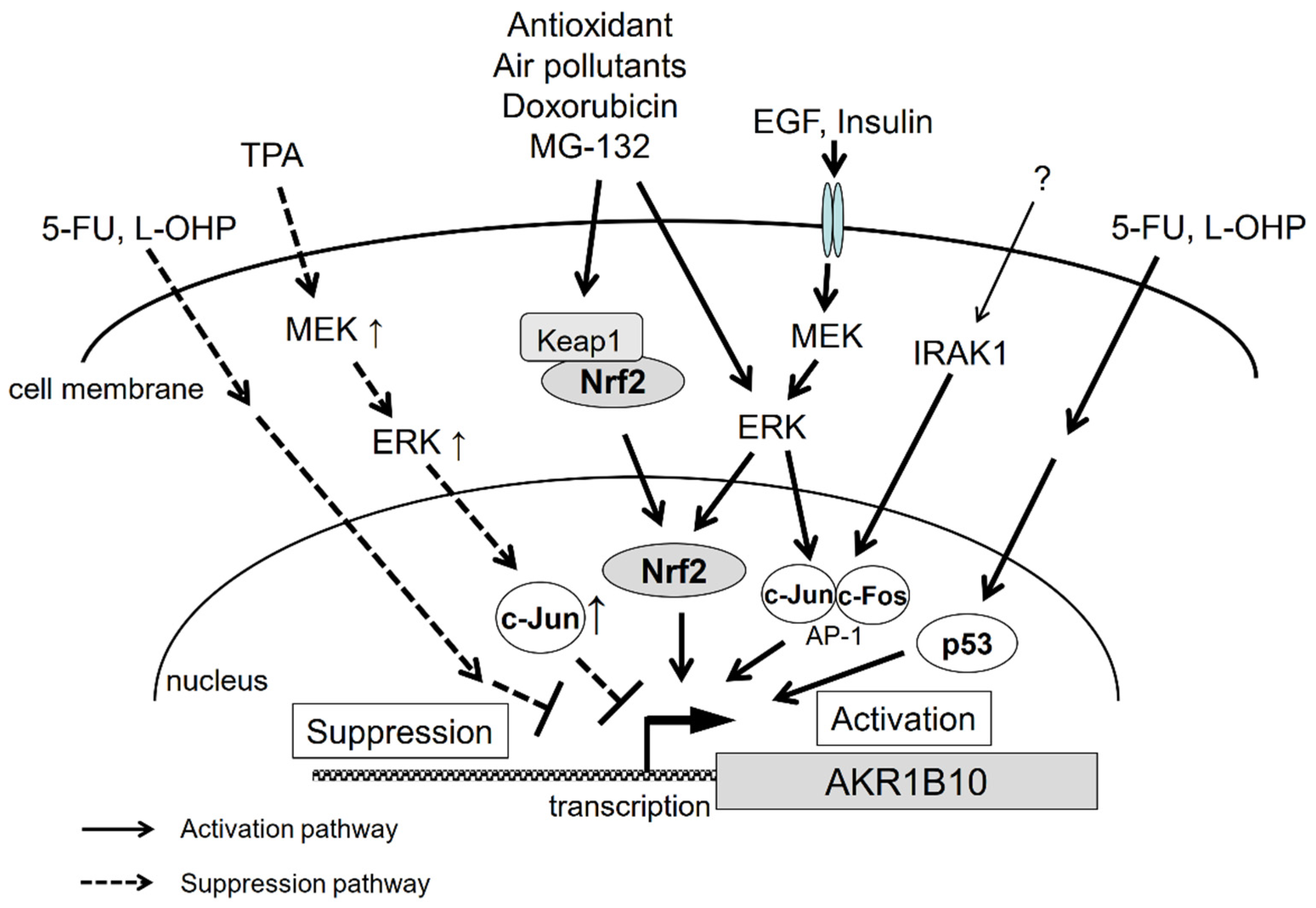
The gene regulation pathways for AKR1B10 are summarized in Figure 3. As described above, we still have little knowledge of the mechanisms underlying AKR1B10 gene regulation. The regulation pathways appear to be complex, since, for example, the same factors can produce opposing outcomes. There are likely more factors involved, and therefore, further investigations are necessary to solve the puzzle of gene regulation of AKR1B10 and its physiological significance.
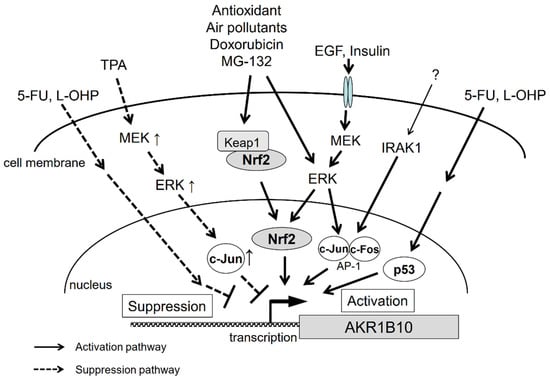
Figure 3.
Signal transduction pathways that regulate AKR1B10 gene expression.
3. AKR1B10 as a Multifunctional NADPH-Dependent Reductase
AKR1B10 reduces a variety of endogenous carbonyl compounds, such as retinals, lipid peroxidation-derived cytotoxic aldehydes, and isoprenyl aldehydes. It also metabolizes xenobiotics, including several drugs and polycyclic aromatic hydrocarbon (PAH) derivatives found in cigarette smoke and the environment. Like all AKR1 subfamily enzymes, AKR1B10 catalyzes a sequential-ordered Bi-Bi kinetic mechanism, with NADPH binding occurring before substrate binding [4]. The active site of AKR1B10 is located above its α/β-barrel tertiary structure consisting of eight α-helices and β-sheets (Figure 4a).
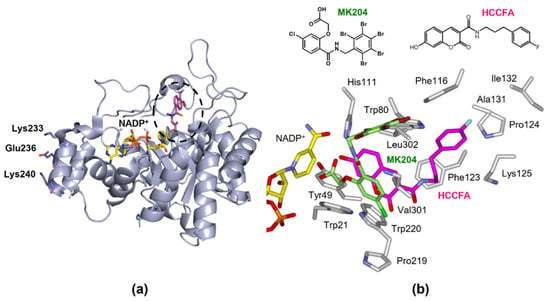
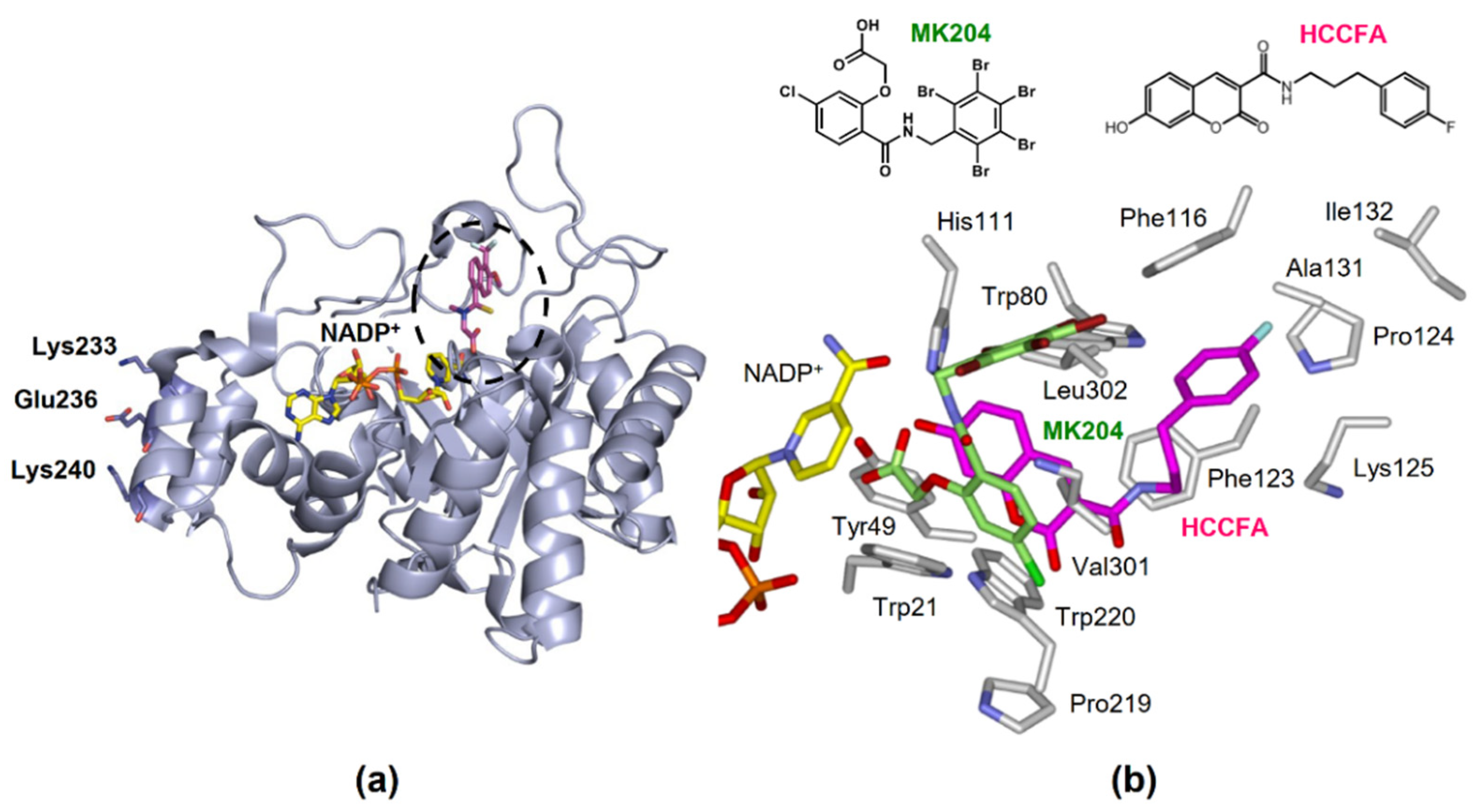
Figure 4.
The structure of AKR1B10. (a) Tertiary structure complexed with NADP+ and an inhibitor tolrestat (PDB 1ZUA), showing the active site (dotted circle) and residues involved in binding to HSP90α. (b) The orientations of HCCFA (pink) and MK204 (light green) in the active site.
3.1. Retinoid Metabolism
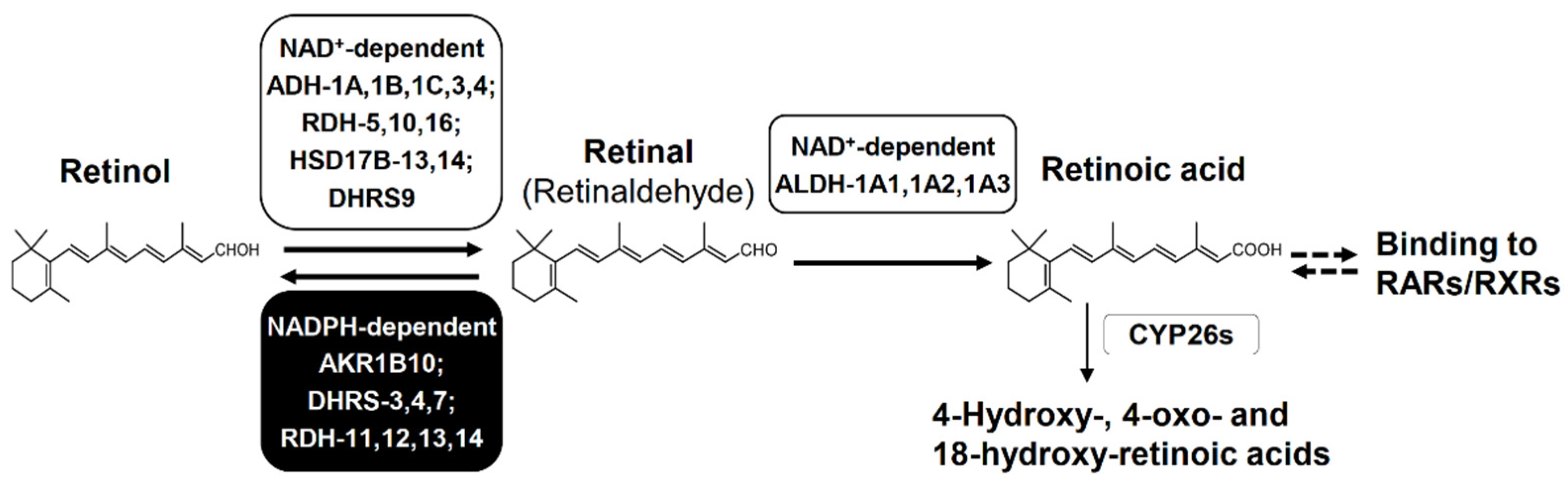
AKR1B10 efficiently catalyzes the reduction of retinal (i.e., all-trans-retinaldehyde) to retinol, which is the first reversible step of retinoid metabolism producing retinoic acid (i.e., all-trans-retinoic acid) that plays a pivotal role in proliferation, differentiation and morphogenesis of many cell types through binding to retinoic acid receptors or retinoid X receptors [34,35] (Figure 5). The reduction of retinal is mediated by other enzymes including AKRs (1B1, 1B15, 1C3 and 1C4), retinol dehydrogenases (RDHs) and dehydrogenase/reductase (SDR family) members (DHRSs). The kcat/Km value for retinal (45 min−1 μM−1) of AKR1B10 is much higher than those (0.14–5.3 min−1 μM−1) of the other AKRs [5,34], DHRS4 [36], DHRS7 [37], and RDH13 [38], although its Km value for retinal (0.6 μM) is higher than those of RDH11, RDH12 and RDH14 (0.04–0.15 μM) [38] and DHRS3 [39]. In addition, the Km and kcat/Km values of AKR1B10 are much lower and higher, respectively, than Km (2.4–11 μM) and kcat/Km values (1.6–7.6 min−1 μM−1) of aldehyde dehydrogenases (ALDHs: 1A1, 1A2 and 1A3) [40], which catalyze the oxidation of retinal to retinoic acid. Thus, AKR1B10 acts as one of the enzymes that control the cellular concentration of retinal, and contributes to maintain the retinoid homeostasis in normal gastrointestinal tissues with its high-expression. In extra-gastrointestinal tissues with little or no expression of AKR1B10, the retinoid metabolism proceeds normally by actions of other enzymes. However, the down-regulation of AKR1B10 in gastrointestinal cancers and its high expression in other cancers may disturb the retinoid homeostasis as described in Section 5 and Section 6.
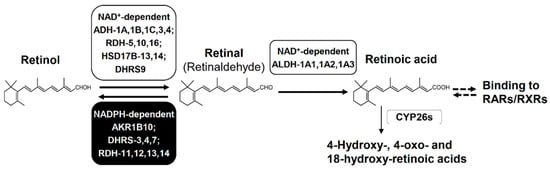
Figure 5.
Biosynthesis and degradation of retinoic acid. Retinol is oxidized to retinal by NAD+-dependent enzymes, but this step is reversible by NADPH-dependent reductases. Retinal is oxidized irreversibly to retinoic acid by NAD+-dependent enzymes. Retinoic acid activates RAR and RXR to initiate the transcription of target genes that regulate differentiation, apoptosis, and cell cycle arrest. Retinoic acid is also eliminated by conversion to its hydroxy- and/or oxo-metabolites. Abbreviations: ADH, alcohol dehydrogenase; RDH, retinol dehydrogenase; HSD17B, 17β-hydroxysteroid dehydrogenase; DHRS, dehydrogenase/reductase (SDR family) member; ALDH, aldehyde dehydrogenase; CYP26s, cytochrome P450 family 26 enzymes, RAR, retinoic acid receptor; and RXR, retinoid X receptor. DHRS9 is called RDHL, and ALDH-1A1, 1A2, and 1A3 are also called retinal dehydrogenase (RALDH)-1, 2, and 3, respectively.
3.2. Detoxification of Reactive Carbonyl Species (RCS)
The continuous oxidation of carbohydrates and lipids generates RCSs, such as methylglyoxal, 3-deoxyglucosone, hexanal, acrolein, 4-hydroxynonenal, 4-oxo-2-nonenal, and phospholipid aldehydes. These RCS are metabolized by several members of the AKR superfamily, as well as by ALDHs and alcohol dehydrogenases [41,42]. AKR1B10 reduces the above RCSs to their less-toxic alcohol [4,43,44,45,46,47,48], of which highly cytotoxic 4-hydroxynonenal and 4-oxo-2-nonenal are most efficiently reduced to their less-toxic alcohols by AKR1B10 among the members of the AKR superfamily. Kinetic constants for 4-hydroxynonenal are shown in Table 2, and AKR1B10 also exhibits high catalytic efficiency for 4-oxo-2-nonenal (kcat/Km = 186 min−1 μM−1) [49]. Furthermore, the catalytic efficiency for 4-hydroxynonenal of AKR1B10 is higher than those of the ALDHs (1A1 and 1A3) that oxidize to 4-hydroxy-2-nonenoic acid (Table 2). Thus, AKR1B10 plays a key role in the detoxification of lipid peroxidation derived RCSs.

Table 2.
Kinetic constants for 4-hydroxynonenal of human AKRs and ALDHs.
3.3. Isoprenoid Metabolism
AKR1B10 efficiently reduces farnesal and geranylgeranial into their alcohols (farnesol and geranylgeraniol) with low Km (2.5 μM and 0.9 μM, respectively) and high kcat/Km values (9.1 min−1 μM−1 and 8.3 min−1 μM−1, respectively) [4]. The catalytic efficiency of AKR1B10 is superior to other AKRs, although for geranylgeranial, it is similar to that of AKR1C3 [52]. Farnesol and geranylgeraniol are phosphorylated to their pyrophosphates, which are required for the transformational activity of many oncogenic proteins, including some RAS family members. The role of AKR1B10 in modulating the Kras-E-cadherin pathway is further supported by the overexpression, knockdown, and inhibition of the enzyme in pancreatic cancer CD18/HPAF and Panc10.05 cells and xenograft tumors [53,54]. It has also been suggested that the proliferative capacity of oxaliplatin-resistant CRC HT29 cells is affected by the modulation of isoprenoid metabolism by AKR1B10 [55].
3.4. Xenobiotic Metabolism
The substrate specificity of AKR1B10 is broad, reducing xenobiotic aromatic aldehydes [2,4], quinones [20,56], and ketones, including several drugs [57,58,59,60]. The drug substrates include dolasteron, oracin, daunorubicin [57], doxorubicin [58], nabumetone [59], and tiaprofenic acid [60], which are also reduced by other AKRs and carbonyl reductase 1. Among these drugs, dolasteron and nabumetone are efficiently reduced by AKR1B10, whose kcat/Km values are higher than (or comparable to) those of the other reductases [57,59,61]. Since AKR1B10 is expressed abundantly in the intestine, it may play a significant role in the intestinal first-pass reductive metabolism of orally administered dolasteron and nabumetone. In addition, AKR1B10 reduces redox-active 9,10-phenanthrenequinone in diesel exhaust [20] and PAH o-quinones derived from the oxidation of PAH trans-dihydrodiols by the enzyme [62]. Furthermore, the gut microbiota produces various carbonyl compounds [63], several of which are reduced by AKR1B10 [2,4,5,43,44,45,47].
4. Moonlighting Functions of AKR1B10
AKR1B10 has three moonlighting functions that are independent of its enzyme activity.
- (1)
- Fatty acid/lipid synthesis. In breast cancer RAO-3 cells, AKR1B10 interacts with acetyl-CoA carboxylase-α (a rate-limiting enzyme of de novo fatty acid synthesis), preventing acetyl-CoA carboxylase-α ubiquitination and proteolysis, and thereby promoting fatty acid/lipid synthesis [64].
- (2)
- Interaction with heat shock protein (HSP) 90α in AKR1B10 secretion. Cytosolic AKR1B10 is secreted from cells through a lysosome-mediated nonclassical pathway, increasing its presence in breast cancer patients’ serum [65]. The AKR1B10 secretion is mediated by interaction with HSP90α, which binds to Lys-233, Glu-236, and Lys-240 in AKR1B10 [66] (Figure 4a).
- (3)
- Interaction with glyceraldehyde-3-phosphate dehydrogenase (GAPDH). A recent report shows that AKR1B10 interacts with GAPDH in CRC HT29 cells [67]. The interaction inhibits the nuclear import of GAPDH, and subsequently results in autophagy repression, for which AKR1B10 reductase activity is likely to be important.
5. AKR1B10 in the Gastrointestinal Tract and Cancer
AKR1B10 is abundant primarily in human stomach, small intestine, and colon [2,8]. Immunohistochemical analysis has revealed that AKR1B10 is localized in the rapidly renewing epithelial cells of the colon [68] and stomach [69]. In contrast, AKR1B10 is markedly decreased or undetectable in cancerous lesions of the colon [68,70,71,72,73,74,75,76] and stomach [69,77,78,79] (Table 3), as well as in precancerous lesions of the colon (including ulcerative colitis), Crown’s disease, and adenomatous polyps [68,73]. Zu et al. [73] reported that AKR1B10 is critical for protecting host cells from DNA damage induced by electrophilic carbonyl compounds, which are ingested and/or metabolically formed in the intestine. The authors suggest the enzymatic activity (i.e., detoxification of RCS) of AKR1B10 is more important than its promotion of fatty acid synthesis for maintaining homeostasis of colon epithelium. However, whether other AKR1B10 functions (as described in Section 4) are involved in intestinal physiology remains to be elucidated. Additionally, since it activates ERK signaling (as described in Section 6), AKR1B10 might participate in the renewal of epithelial cells through the mitogen-activated protein kinase (MAPK) pathway that regulates the proliferation and differentiation of gastrointestinal epithelial cells [80].

Table 3.
AKR1B10 expression in extra-hepatic cancers and its prognostic impact.
The expression of AKR1B10 is decreased in patients with CRC and the low expression correlates with poor prognosis and reduced survival [16,71,74,75,76]. It has been suggested that AKR1B10 down-regulation results from mutation of the tumor suppressor p53 gene: Wild-type p53 is a transcriptional activator of AKR1B10, but mutant p53 acts as a repressor [16]. Recently, Li et al. [67] reported that AKR1B10 expression is negatively regulated by a membrane receptor neuropilin1 (NRP1), which was upregulated in CRC HT29 cells that underwent autophagy following glucose deprivation. This study also showed that AKR1B10 inhibits glucose-deprivation autophagy by interacting with GAPDH, and suggested that low AKR1B10 expression in CRC promotes tumor development by upregulated autophagy.
Several studies suggest that decreased AKR1B10 expression mediates tumorigenesis. Ohashi et al. [71] reported that the decreased expression of AKR1B10 disrupts the tumor-suppressive function of p53. Kropotova et al. [72] observed a decrease in the expression of mRNAs for the retinoid metabolizing enzymes (AKR1B10, ADH1B, ADH1C, DHRS9, RDH5, and ALDH1A), and suggested that the resultant disturbance to retinoid homeostasis contributes to the progression of CRC. Zu et al. [73] suggest that AKR1B10 deficiency may leave the colonic HCT-8 cells vulnerable to electrophilic carbonyl lesions, leading to DNA damage and carcinogenesis. Recently, Yao et al. [75] reported that AKR1B10 depletion promotes the progression of CRC by down-regulating fibroblast growth factor (FGF)-1, which is related to the growth and migration of CRC and gastric cancer [100,101,102]. This finding implies that FGF-1 expression is low in a normal colon with high levels of AKR1B10, in contrast to the high levels of FGF-1 due to low AKR1B10 expression in CRC. Further studies of the underlying mechanism by which FGF-1 is suppressed by AKR1B10 might lead to the identification of a novel moonlighting function of AKR1B10.
A poor prognosis in gastric cancer is suggested by the down-regulation of AKR1B10 expression [78]. By contrast, patients who underwent neoadjuvant chemotherapy showed that high AKR1B10 expression was associated with lymph node metastasis and a poorer prognosis, along with a weak response to neoadjuvant chemotherapy [79]. In addition, the overexpression of AKR1B10 in gastric cancer MKN46 cells stimulates migration [18] and down-regulates peroxisome proliferator-activated receptor-γ (PPARγ) that is closely linked to growth suppression and death of cancer cells [103]. However, there has been no mechanistic study relating AKR1B10 with gastric cancer progression, with the exception of Kropotova et al. [77], who suggested that retinoic acid synthesis was dysregulated due to significant decreases in the mRNAs for AKR1B10 and other retinoid-metabolizing enzymes (ADH4, ADH1B, ADH1C, DHRS9, and RDH12). Further studies to elucidate whether the low expression of AKR1B10 is associated with carcinogenesis are warranted.
6. Diseases Associated with AKR1B10 Elevation
6.1. Hepatocellular Carcinoma (HCC)
The overexpression of AKR1B10 in HCC was first identified by Scuric et al. [12] in 1998, which alluded to a role of AKR1B10 in liver carcinogenesis. Subsequent literature (through 2019) supporting the diagnostic and prognostic utility of AKR1B10 as a potential biomarker for HCC has been reviewed by DiStefano and Davis [104]. They summarize the roles of the enzyme in HCC development and progression through lipogenesis, oxidative stress, detoxification of cytotoxic RCS, and the regulation of sphingosine-1 phosphate and retinoic acid. Although this review also includes the underlying upregulation mechanisms of AKR1B10 in the process of hepatocarcinogenesis by microRNA (miR-383-5p), 14-3-3ε, and AP1 downstream of IRAK1 or EGFR signaling, a recent study reported that AKR1B10 elevation results from compensatory upregulation to protect hepatocytes against oxidative stress during hepatocarcinogenesis [105]. Thus, AKR1B10 is emerging as a promising biomarker for HCC. In addition to AKR1B10, HSP90α is also a potential serum biomarker for diagnosing α-fetoprotein negative HCC [106], as it interacts with AKR1B10 [66].
6.2. Nonalcoholic Fatty Liver Disease (NAFLD)
NAFLD encompasses a spectrum of liver pathologies which involve an accumulation of triglycerides in the hepatocytes, hepatocyte apoptosis, liver inflammation and fibrosis termed as non-alcoholic steatohepatitis (NASH), and, in extreme cases, it can progress to cirrhosis and HCC [107]. Since a significant increase of AKR1B10 in steatohepatitis was first found in 2012 [108], it has been confirmed in biopsy samples and sera from NAFLD patients [109,110,111,112,113,114,115,116], indicating that AKR1B10 is a consistent marker for the progression of NAFLD/NASH. Oxidative and endoplasmic reticulum (ER) stresses mediated by chronic lipotoxicity play a major role in the cellular mechanisms responsible for NAFLD/NASH pathogenesis [107] and induces Nrf2 transcriptional activity [117,118]. The upregulation of the Nrf2-target AKR1B10 gene in NAFLD is, thus, due to oxidative and ER stresses [115]. Furthermore, it may also be due to a reduction in pregnane X receptor (PXR) that negatively regulates the enzyme’s expression [110]. Several roles have been proposed for the upregulated AKR1B10 in NAFLD/NASH progression. This enzyme contributes to lipogenesis through the stabilization of acetyl-CoA carboxylase [110], in addition to detoxification of RCS derived from lipid peroxidation. This AKR1B10 overexpression, together with the underexpression of ALDHs (1A2 and 1A3) [111] and HSD17B14 [115], may reduce hepatic retinal levels, and hence lead to a decrease in retinoic acid, favoring progression of NASH to HCC. Furthermore, the immune deregulations associated with the progression of NAFLD may be related to AKR1B10 [116]. Two studies [113,115] suggest that AKR1B10 is a useful serum biomarker for advanced liver fibrosis and, thereby, the diagnosis of NAFLD/NASH. These studies recommend combining AKR1B10 with other markers such as Wisteria floribunda agglutinin-positive Mac-2-binding protein (WFA+-M2BP) [112] and growth/differentiation factor 15 (GDF15) [115].
6.3. Lung Cancer
There are two major types of lung cancer: small cell lung carcinoma and non-small cell lung carcinoma (NSCLC). Subclassifications of NSCLC include squamous cell carcinoma (SCC), adenocarcinoma (ADC), and large cell carcinoma. In 2005, Fukumoto et al. [8] were the first to report the overexpression of AKR1B10 in SCC and ADC. They proposed that AKR1B10 could be a potential diagnostic marker specific to NSCLC in smokers. The overexpression of AKR1B10 in NSCLC at the mRNA and/or protein level has been observed in many studies [81,82,83,84,85,86,87,88,89,90] (Table 3), which have shown SCC to have higher AKR1B10 expression than ADC [8,81,84,87]. Higher AKR1B10 expression may be due to the Nrf2 mutations that occur primarily in SCC [84]. The overexpression of AKR1B10 is a prognostic factor for NSCLC [82,89] and an indicator for poor recurrence-free survival in patients with resected ADC [88]. The upregulation of AKR1B10 may be involved in the carcinogenicity and metastasis of NSCLC by activating the ERK signaling pathway [87,89,90], as well as by the detoxification of RCS derived from lipid peroxidation and suppressing the synthesis of retinoic acid [85].
Similar to HCC and NAFLD, the oxidative stress-dependent activation of Nrf2 in NSCLC cells is a major factor responsible for upregulating AKR1B10 expression. However, a recent study with lung ADC A549 cells suggests another mechanism of AKR1B10 upregulation via a long non-coding RNA linc00665, which exerts its oncogenic role by acting as a competing endogenous RNA for miR-98 and subsequently increasing enzyme expression [89]. In addition, comparative analyses of lung specimens between healthy smokers and nonsmokers, and between smoking and non-smoking NSCLC patients, have shown that smoking mediates the upregulation of AKR1B10 expression [8,23,82,83,87,119]. AKR1B10 is also frequently overexpressed in interstitial pneumonia, a disease that increases the risk of lung cancer in smokers [120]. Thus, AKR1B10 is a diagnostic marker of NSCLC in smokers, as its smoking-induced upregulation may be an early indicator among the multiple events leading to lung cancer. AKR1B10 metabolizes PAH trans-dihydrodiols to redox-active o-quinones, enhancing the production of reactive oxygen species (ROS) and leading to further overexpression of Nrf2-target genes, including AKR1B10 [62]. Indeed, a redox-active PAH o-quinone, 9,10-phenanthrenequinone, is reduced by AKR1B10. Its treatment of A549 cells induces the expression of AKR1B10 through ROS- and Nrf2-dependent mechanisms, leading to further progression of malignant lung cancer cells through activating the ERK pathway [22]. Thus, AKR1B10 may be involved in the initial steps of carcinogenesis caused by smoking.
6.4. Breast Cancer
Breast cancer RAO-3 cells exhibit elevated AKR1B10 expression [64], and the high expression is observed in situ and in the serum of the patients, suggesting that AKR1B10 expression may be a diagnostic and therapeutic marker [91,92,93,94,95] as well as a poor prognostic factor for breast cancer [91,95] (Table 3). Notably, AKR1B10 expression is significant in breast cancers that are positive for human epidermal growth-factor receptor type 2 (HER2) [92,96]. Several roles have been proposed for AKR1B10 in cancer growth and metastasis. AKR1B10 may support breast cancer cell survival by releasing carbonyl stress and promoting fatty acid/lipid synthesis [91]. In addition, AKR1B10 promotes cancer metastasis through activation of the ERK signaling pathway, which stimulates the downstream integrin α5/δ-catenin mediated FAK/Src/Rac1 signaling pathway [93] and increases the expressions of matrix metalloproteinase-2 (MMP2) and vimentin [95]. The proposed mechanism underlying the activation of the ERK signaling pathway by AKR1B10 is as follows: AKR1B10 promotes lipogenesis, which leads to increases in second messengers (PIP2, DAG, and IP3) of the PKC-mediated ERK signaling cascade [121]. However, van Weverwijk et al. [96] show that lipogenesis is significantly reduced in breast cancer cells with high AKR1B10 expression. Instead, fatty acid oxidation is sustained by AKR1B10-mediated limitation of the toxic side effects of oxidative stress. Thus, further research on lipid metabolism in breast cancer is warranted.
6.5. Pancreatic Cancer
In pancreatic cancer, Keap1–Nrf2 signaling is dysregulated and Nrf2-ARE-driven genes are frequently upregulated [122]. As an Nrf2-ARE effector protein, AKR1B10 is overexpressed in pancreatic adenocarcinoma, intraepithelial neoplasia [54], and mucinous cystic pancreatic tumors [97] (Table 3). AKR1B10 efficiently reduces isoprenyl aldehydes (farnesal and geranylgeranial) to protein prenylation precursors (farnesol and geranylgeraniol) [52]. Mechanistic studies show that the upregulated enzyme promotes cancer cell growth by activating ERK signaling via modulating the prenylation of Kras protein [53,54].
6.6. Oral Cancer
More than 90% of oral cancers are squamous cell carcinomas (OSCC), in which AKR1B10 is overexpressed (Table 3). As such, AKR1B10 is a prognostic biomarker for predicting recurrence and survival in OSCC [98,99]. In addition, patient salivary levels of AKR1B10 are high and correlated with poor prognosis and elevated tumor recurrence incidence in OSCC [123]. AKR1B10 is thought to increase the prenylation of Ras protein that is critical in OSCC [98], as described in the pancreatic cancer section (Section 6.5). Several environmental factors increase the risk of OSCC, the greatest of which is cigarette smoking. Cigarette smoke condensate induces AKR1B10 expression in oral dysplasia (Leuk1 and Leuk2) and OSCC 101A cells [124], in which AKR1B10 may activate pro-carcinogen PAHs, promoting carcinogenesis, as described in Section 6.4.
6.7. Other Cancers
Uterine cancer. Immunohistochemical analysis has detected AKR1B10 expression in 20.0% and 15.8% of cervical and endometrial cancer cases, respectively. Tumor recurrence was associated with AKR1B10 expression after surgery for cervical cancer [125]. In subsequent studies on endometrial cancer [126,127], the expression of AKR1B10 at the mRNA level was significantly increased, while there were significantly decreased protein levels. Differences in the expression of AKR1B10 among these studies might be explained by the type of cancer cells, as cervical and endometrial cancers are squamous cell carcinomas and adenocarcinomas, respectively.
Renal cancer. In the Human Protein Atlas database (https://www.proteinatlas.org, [accessed on 2 February 2021]), AKR1B10 levels in renal cancer are significantly high and unfavorable for prognosis. There is only one report in the literature showing that AKR1B10 is highly expressed in specimens of hereditary and sporadic type 2 papillary renal cell carcinoma, in which Nrf2 is activated by fumarate hydratase mutations [128].
Nasopharyngeal carcinoma (NPC). AKR1B10 expression is increased in NPC tissues (compared to the normal tissues), in which squamous cell carcinomas show higher AKR1B10 expression than basal cell carcinomas, adenoid cystic carcinomas, adenocarcinomas, and undifferentiated carcinomas. However, AKR1B10 expression levels in NPC tissues are lower than those in nasopharyngeal hyperplasia and benign tumors, where AKR1B10 was predominantly present in nuclei [129]. Guo et al. also reported the lower expression of AKR1B10 in NPC than benign tumors, and they suggested that AKR1B10 may be involved in regulating the proliferation and migration of NPC cells [130]. In contrast, AKR1B10 was overexpressed in NPC tissues from patients resistant to radiotherapy, thereby suggesting that AKR1B10 confers radio-resistance [131].
Esophageal carcinoma. AKR1B10 expression is increased in epithelium specimens from patients with erosive gastro-esophageal reflux disease and Barrett’s esophagus but not in those with esophageal adenocarcinoma [132,133]. Since high expression of AKR1B10 has been observed in an esophageal squamous cell carcinoma cell line, OE-21 [132], it would be interesting to analyze the expression of AKR1B10 in specimens from patients with squamous cell carcinoma, which is the predominant form of esophageal carcinoma worldwide.
6.8. Non-Neoplastic Skin Diseases
The up-regulation of AKR1B10 has been observed in skin lesions of patients with psoriasis [134,135], keloids [136,137], atopic dermatitis [138], and type 2 reaction leprosy [139]. Psoriasis and keloid lesions show extremely high AKR1B10 expression levels (23-86-fold higher than normal skin) [134,135,136,137]. Although the role of AKR1B10 in the pathogenesis of atopic dermatitis and type 2 reaction leprosy remains unknown, its overexpression is related to the proliferation and migration of keratinocytes in psoriasis [135] and causes the dysregulation of retinoid metabolism, one of proposed mechanisms contributing to keloid disease [137,138]. The AKR1B10-overexpressing keratinocytes also secrete paracrine signals that enhance the fibrogenic activity of dermal fibroblasts and contribute to keloid pathobiology [136,137].
7. AKR1B10 in Anti-Cancer Drug Resistance
Chemotherapy is a common therapeutic approach for treating many cancers. However, primary or acquired drug resistance is a major problem during chemotherapy, which consequently leads to malignant transformation and metastasis. Human AKRs (1B1, 1B10, 1C1, 1C2, and 1C3) are regarded as novel factors in chemoresistance, along with drug transporters, growth factor receptors, and the ubiquitin-proteasome system [55,140,141]. Among the five AKRs, AKR1B10 may play a crucial role in acquiring resistance to several anti-cancer drugs. In clinical studies, bladder cancer specimens receiving carboplatin-gemcitabine combination chemotherapy displayed higher levels of AKR1B10 expression compared to the pre-chemotherapy group [142]. A study on patients with gastric cancer who underwent neoadjuvant chemotherapy showed that high AKR1B10 expression was associated with lymph node metastasis and a poor response to neoadjuvant chemotherapy [79]. These two studies also indicate that AKR1B10 upregulation is associated with a poorer prognosis for patients receiving chemotherapy. In addition, the upregulation of AKR1B10 has been observed in several cancer cell variants established by continuous exposure to anti-cancer drugs, as summarized in Table 4. In these resistant cells, chemoresistance was overcome by the addition of an AKR1B10 inhibitor or siRNA-mediated silencing [14,48,55,103,143,144,145]. Thus, AKR1B10 is considered a key factor responsible for the development of chemoresistance in these cancer cells.

Table 4.
AKR1B10-upregulated chemoresistant cancer cells.
The most accepted mechanism underlying the upregulation of AKR1B10 is the constitutive activation of Nrf2, a transcription factor involved in the gene expression of AKR1B10, as described in Section 2. Although specific details of the mechanism(s) of constitutive Nrf2 activation remain unclear, some researches propose an association between the development of chemoresistance and somatic mutations in Nrf2/Keap1, based on results showing a loss of the Nrf2-Keap1 interaction in cancer cell lines showing chemoresistance [148,149,150]. Another study also concluded that EGF induces AKR1B10 expression through AP-1 signaling in HCC HepG2 and Hep3B cells [18]. Moreover, recent transcriptome sequencing of HCC samples found a significant upregulation of IRAK1, which is likely to confer chemoresistance through a signaling pathway dependent on AP-1 and AKR1B10 [33]. Thus, AKR1B10 is considered to be upregulated through various processes, including Keap1 mutation, EGF exposure, and IRAK1 overexpression (Figure 3). Further studies are warranted to clarify the effect of AKR1B10 gene regulation on developing chemoresistance.
Studies with chemoresistant cells suggest that the role of AKR1B10 in chemoresistance development is dependent on the anti-cancer drugs or cancer cells used. While AKR1B10 may contribute to oxaliplatin-resistant HT29 cells through modulating isoprenoid metabolism [55], its role in chemoresistance to anthracycline drugs is performed by modulating the metabolism of the drugs to less toxic metabolites by reducing the C13 ketone group [58,146,151]. Exposing doxorubicin, docetaxel, cisplatin, and mitomycin c to resistant cancer cells generates ROS, which causes lipid peroxidation and subsequent increases in RCS, including highly cytotoxic 4-hydroxynonenal and 4-oxo-2-nonenal [18,48,55,143,144,145]. As described in Section 3.2, AKR1B10 efficiently detoxifies the two cytotoxic aldehydes. In addition, knockdown of the AKR1B10 gene by siRNAs sensitizes CRC HCT-8 cells to acrolein and crotonaldehyde [152]. Thus, the detoxification of RCS by AKR1B10 is the most critical step in developing resistance to these anti-cancer drugs.
In the doxorubicin-resistant MKN45 cells, it has been suggested that AKR1B10 facilitates the induction and secretion of MMP2 through activation of the ERK signaling pathway and the above-mentioned isoprenoid metabolism with consequent activation of small-G protein [18]. In cisplatin-resistant MKN45 and LoVo cells, AKR1B10 is proposed to be involved in the down-regulation of PPARγ through activation of the isoprenoid-dependent signaling pathway and/or the catalysis of PPARγ substrates [103]. Further studies are needed to elucidate the detailed mechanism(s) underlying the induction of MMP2 and down-regulation of PPARγ.
8. AKR1B10 Inhibitors
Inhibitors of AKR1B10 are considered promising candidates for treating extra-gastrointestinal cancers and chemoresistance described in Section 6 and Section 7. Since aldose reductase (AKR1B1) has a similar structure to AKR1B10 and is involved in glucose and prostaglandin metabolism, developing inhibitors as anti-cancer drugs or adjuvant therapies for chemotherapeutic drug resistance must ensure selective inhibition of AKR1B10. Previously, Hung et al. [153] reviewed various synthetic and natural AKR1B10 inhibitors reported until 2015 [153]. Among them, HAHE [154] was the most potent, and oleanolic acid was the most selective to AKR1B10 compared to AKR1B1 [155]. Table 5 shows the inhibitory potency and selectivity (ratio of AKR1B10 to AKR1B1) of the inhibitors reported only from 2016 onwards, in addition to those of 3-(4-hydroxy-2-methoxyphenyl)acrylic acid 3-(3-hydroxyphenyl)propyl ester (HAHE), oleanolic acid, epalrestat, and glycyrrhetinic acid [156], which were used in studies on cellular AKR1B10-mediated metabolism. The natural inhibitors include flavonoid emodin [157], unsaturated fatty acid arachidonic acid [158], cohumulone, and 8-prenylnaringenin, the latter two of which are plant hop-derived α-bitter acid and prenylflavanone, respectively [159,160]. Although cohumulone is highly selective, emodin potently inhibits human carbonyl reductase 1 [157] with S-nitroso-glutathione reductase activity [161]. Furthermore, unsaturated fatty acids, including arachidonic acid, are similarly inhibitory to both carbonyl reductase 1 and human four AKRs (1C1-1C4) with hydroxysteroid dehydrogenase activity [162,163]. In contrast, the synthetic inhibitors, 7-Hydroxy-2-oxo-2H-chromene-3-carboxylic Acid [3-(4-Fluorophenyl)propyl]amide (HCCFA) [164], MK204 [165], and androst-3β,5α,6β,19-tetrol [166], show greater inhibitory potency and/or selectivity. HCCFA is the most potent and shows negligible or low inhibition to human carbonyl reductase 1 and AKRs (1A1, 1C1, 1C2, 1C3, and 1C4). HCCFA has greater chemical stability than HAHE. The inhibitors bind to the active site of AKR1B10 [164,165], in which their hydroxyl or carbonyl group interacts with the catalytically important residues, Tyr49 and His118. However, the orientations of their other parts are different, reflecting their chemical structures (Figure 4b). The residue differences between AKR1B10 and AKR1B1 at positions 114, 125, 301, and/or 304 may contribute to selective inhibition [154,155,164,165].

Table 5.
Recently identified inhibitors of AKR1B10.
Among the above inhibitors, only HCCFA significantly suppresses not only migration, proliferation and metastasis of lung cancer A549 cells, but also metastatic and invasive potentials of cisplatin-resistant A549 cells [164]. Of the previously discovered inhibitors, HAHE and oleanolic acid suppress the proliferative potential of gastric carcinoma MNK45 and CRC Lovo cells that are resistant to cisplatin [103] and doxorubicin [144]. Oleanolic acid also increases the drug sensitivity of docetaxel-resistant A549 and prostate cancer Du145 cells [145]. Although epalrestat may be harmful to CRC HCT-8 cells by promoting DNA damage [73], it enhances the efficacy of sorafenib (an anti-cancer drug) on HCC by inhibiting AKR1B10 [167]. In addition, glycyrrhetinic acid may attenuate disturbed vitamin A metabolism in NAFLD and/or NASH through AKR1B10 inhibition [168]. Since HCCFA, MK204, and oleanolic acid are commercially available, they can be used for elucidating the mechanisms underlying tumorigenesis of AKR1B10-overexpressing cancers and chemotherapy resistance, which would precede AKR1B10-based therapies in clinical cancer management.
9. Conclusions
Functional studies reveal that AKR1B10 physiologically contributes to the maintenance of cellular homeostasis through the detoxification of cytotoxic RCS and regulating the metabolism of isoprenoid, retinoid, and lipid. AKR1B10 expression is constitutively high in the epithelial cells of the gastrointestinal tract, where the enzyme plays a critical role in epithelial cell renewal and the metabolism of several therapeutic drugs and carbonyl compounds produced by gut bacteria.
Clinical and basic research has proven that AKR1B10 is down-regulated in gastrointestinal cancers and several inflammatory bowel diseases, proposing that its low expression is correlated with poor prognosis and decreased survival for patients with CRC and gastric cancer. By contrast, in other tissues with normally low or no expression, the evidence is mounting that AKR1B10 is upregulated and involved in several forms of cancer and inflammatory disease. In addition, AKR1B10 is increased and implicated in the acquisition of anti-cancer drug resistance. In liver, lung, and breast cancers, AKR1B10 may be upregulated through the activation of stress-induced signal transduction pathways (such as Nrf2, AP-1, and MAPK pathways) and related to the proliferation and migration of cancer cells. However, the mechanisms underlying AKR1B10 down-regulation and its association with gastrointestinal diseases have been poorly elucidated. Thus, AKR1B10 acts as a double-edged sword depending on the type of cancer cells, and further understanding its role in the onset and progression of these diseases should be pursued.
A number of AKR1B10 inhibitors have been developed, of which HCCFA, the most potent, significantly inhibits not only the migration, proliferation, and metastasis of lung cancer A549 cells, but also the metastatic and invasive abilities of cisplatin-resistant A549 cells. Therefore, utilizing AKR1B10 inhibitors for cancers that highly express AKR1B10 is expected in clinical practice. Since no genes encoding homologues of human AKR1B10 were identified in experimental animals such as rats and mice [169,170], the efficacy and pharmacokinetics of the AKR1B10 inhibitors in vivo are unclear, and as such, further studies are needed to develop a more clinically relevant approach.
Author Contributions
Conceptualization, S.E.; writing—original draft preparation, S.E., T.M., and T.N.; writing—review and editing, S.E., T.M., and T.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
We wish to acknowledge Akira Hara, Gifu Pharmaceutical University, for his advice and suggestions while writing the review.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Penning, T.M. The aldo-keto reductases (AKRs): Overview. Chem. Biol. Interact. 2015, 234, 236–246. [Google Scholar] [CrossRef]
- Cao, D.; Fan, S.T.; Chung, S.S. Identification and characterization of a novel human aldose reductase-like gene. J. Biol. Chem. 1998, 273, 11429–11435. [Google Scholar] [CrossRef] [PubMed]
- Weber, S.; Salabei, J.K.; Moller, G.; Kremmer, E.; Bhatnagar, A.; Adamski, J.; Barski, O.A. Aldo-keto Reductase 1B15 (AKR1B15): A mitochondrial human aldo-keto reductase with activity toward steroids and 3-keto-acyl-CoA conjugates. J. Biol. Chem. 2015, 290, 6531–6545. [Google Scholar] [CrossRef] [PubMed]
- Endo, S.; Matsunaga, T.; Mamiya, H.; Ohta, C.; Soda, M.; Kitade, Y.; Tajima, K.; Zhao, H.T.; El-Kabbani, O.; Hara, A. Kinetic studies of AKR1B10, human aldose reductase-like protein: Endogenous substrates and inhibition by steroids. Arch. Biochem. Biophys. 2009, 487, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gimenez-Dejoz, J.; Kolar, M.H.; Ruiz, F.X.; Crespo, I.; Cousido-Siah, A.; Podjarny, A.; Barski, O.A.; Fanfrlik, J.; Pares, X.; Farres, J.; et al. Substrate Specificity, Inhibitor Selectivity and Structure-Function Relationships of Aldo-Keto Reductase 1B15: A Novel Human Retinaldehyde Reductase. PLoS ONE 2015, 10, e0134506. [Google Scholar] [CrossRef]
- Gimenez-Dejoz, J.; Weber, S.; Fernandez-Pardo, A.; Moller, G.; Adamski, J.; Porte, S.; Pares, X.; Farres, J. Engineering aldo-keto reductase 1B10 to mimic the distinct 1B15 topology and specificity towards inhibitors and substrates, including retinoids and steroids. Chem. Biol. Interact. 2019, 307, 186–194. [Google Scholar] [CrossRef]
- Kabututu, Z.; Manin, M.; Pointud, J.C.; Maruyama, T.; Nagata, N.; Lambert, S.; Lefrancois-Martinez, A.M.; Martinez, A.; Urade, Y. Prostaglandin F2{alpha} Synthase Activities of Aldo-Keto Reductase 1B1, 1B3 and 1B7. J. Biochem. 2009, 145, 161–168. [Google Scholar] [CrossRef]
- Fukumoto, S.; Yamauchi, N.; Moriguchi, H.; Hippo, Y.; Watanabe, A.; Shibahara, J.; Taniguchi, H.; Ishikawa, S.; Ito, H.; Yamamoto, S.; et al. Overexpression of the aldo-keto reductase family protein AKR1B10 is highly correlated with smokers’ non-small cell lung carcinomas. Clin. Cancer Res. 2005, 11, 1776–1785. [Google Scholar] [CrossRef]
- Hyndman, D.J.; Flynn, T.G. Sequence and expression levels in human tissues of a new member of the aldo-keto reductase family. Biochim. Biophys. Acta 1998, 1399, 198–202. [Google Scholar] [CrossRef]
- Oates, P.J. Aldose reductase, still a compelling target for diabetic neuropathy. Curr. Drug Targets 2008, 9, 14–36. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.C.; Petrash, J.M. Aldo-Keto Reductases: Multifunctional Proteins as Therapeutic Targets in Diabetes and Inflammatory Disease. Adv. Exp. Med. Biol. 2018, 1032, 173–202. [Google Scholar] [CrossRef] [PubMed]
- Scuric, Z.; Stain, S.C.; Anderson, W.F.; Hwang, J.J. New member of aldose reductase family proteins overexpressed in human hepatocellular carcinoma. Hepatology 1998, 27, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhong, L.; Krishack, P.A.; Robbins, S.; Cao, J.X.; Zhao, Y.; Chung, S.; Cao, D. Structure and promoter characterization of aldo-keto reductase family 1 B10 gene. Gene 2009, 437, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Nishinaka, T.; Miura, T.; Okumura, M.; Nakao, F.; Nakamura, H.; Terada, T. Regulation of aldo-keto reductase AKR1B10 gene expression: Involvement of transcription factor Nrf2. Chem. Biol. Interact. 2011, 191, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Nishinaka, T.; Miura, T.; Sakou, M.; Hidaka, C.; Sasaoka, C.; Okamura, A.; Okamoto, A.; Terada, T. Down-regulation of aldo-keto reductase AKR1B10 gene expression by a phorbol ester via the ERK/c-Jun signaling pathway. Chem. Biol. Interact. 2015, 234, 274–281. [Google Scholar] [CrossRef]
- Zinovieva, O.L.; Grineva, E.N.; Krasnov, G.S.; Karpov, D.S.; Zheltukhin, A.O.; Snezhkina, A.V.; Kudryavtseva, A.V.; Mashkova, T.D.; Lisitsyn, N.A. Treatment of cancer cells with chemotherapeutic drugs results in profound changes in expression of genes encoding aldehyde-metabolizing enzymes. J. Cancer 2019, 10, 4256–4263. [Google Scholar] [CrossRef]
- Ebert, B.; Kisiela, M.; Wsol, V.; Maser, E. Proteasome inhibitors MG-132 and bortezomib induce AKR1C1, AKR1C3, AKR1B1, and AKR1B10 in human colon cancer cell lines SW-480 and HT-29. Chem. Biol. Interact. 2011, 191, 239–249. [Google Scholar] [CrossRef]
- Morikawa, Y.; Kezuka, C.; Endo, S.; Ikari, A.; Soda, M.; Yamamura, K.; Toyooka, N.; El-Kabbani, O.; Hara, A.; Matsunaga, T. Acquisition of doxorubicin resistance facilitates migrating and invasive potentials of gastric cancer MKN45 cells through up-regulating aldo-keto reductase 1B10. Chem. Biol. Interact. 2015, 230, 30–39. [Google Scholar] [CrossRef]
- Liu, Z.; Yan, R.; Al-Salman, A.; Shen, Y.; Bu, Y.; Ma, J.; Luo, D.X.; Huang, C.; Jiang, Y.; Wilber, A.; et al. Epidermal growth factor induces tumour marker AKR1B10 expression through activator protein-1 signalling in hepatocellular carcinoma cells. Biochem. J. 2012, 442, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Shaw, N.; Yang, B.; Millward, A.; Demaine, A.; Hodgkinson, A. AKR1B10 is induced by hyperglycaemia and lipopolysaccharide in patients with diabetic nephropathy. Cell Stress Chaperones 2014, 19, 281–287. [Google Scholar] [CrossRef]
- Van Zoelen, E.J.; Duarte, I.; Hendriks, J.M.; van der Woning, S.P. TGFbeta-induced switch from adipogenic to osteogenic differentiation of human mesenchymal stem cells: Identification of drug targets for prevention of fat cell differentiation. Stem Cell Res. Ther. 2016, 7, 123. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, T.; Morikawa, Y.; Haga, M.; Endo, S.; Soda, M.; Yamamura, K.; El-Kabbani, O.; Tajima, K.; Ikari, A.; Hara, A. Exposure to 9,10-phenanthrenequinone accelerates malignant progression of lung cancer cells through up-regulation of aldo-keto reductase 1B10. Toxicol. Appl. Pharmacol. 2014, 278, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, G.; Ricard, M.J.; Ferris, B.; Strulovici-Barel, Y.; Salit, J.; Hackett, N.R.; Gudas, L.J.; Crystal, R.G. Smoking-induced upregulation of AKR1B10 expression in the airway epithelium of healthy individuals. Chest 2010, 138, 1402–1410. [Google Scholar] [CrossRef]
- Itoh, K.; Igarashi, K.; Hayashi, N.; Nishizawa, M.; Yamamoto, M. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol. Cell. Biol. 1995, 15, 4184–4193. [Google Scholar] [CrossRef] [PubMed]
- Rushmore, T.H.; Morton, M.R.; Pickett, C.B. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 1991, 266, 11632–11639. [Google Scholar] [CrossRef]
- Kobayashi, A.; Kang, M.I.; Okawa, H.; Ohtsuji, M.; Zenke, Y.; Chiba, T.; Igarashi, K.; Yamamoto, M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 2004, 24, 7130–7139. [Google Scholar] [CrossRef]
- Tong, K.I.; Padmanabhan, B.; Kobayashi, A.; Shang, C.; Hirotsu, Y.; Yokoyama, S.; Yamamoto, M. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol. Cell. Biol. 2007, 27, 7511–7521. [Google Scholar] [CrossRef]
- Mimura, J.; Inose-Maruyama, A.; Taniuchi, S.; Kosaka, K.; Yoshida, H.; Yamazaki, H.; Kasai, S.; Harada, N.; Kaufman, R.J.; Oyadomari, S.; et al. Concomitant Nrf2- and ATF4-activation by Carnosic Acid Cooperatively Induces Expression of Cytoprotective Genes. Int. J. Mol. Sci. 2019, 20, 1706. [Google Scholar] [CrossRef]
- MacLeod, A.K.; McMahon, M.; Plummer, S.M.; Higgins, L.G.; Penning, T.M.; Igarashi, K.; Hayes, J.D. Characterization of the cancer chemopreventive NRF2-dependent gene battery in human keratinocytes: Demonstration that the KEAP1-NRF2 pathway, and not the BACH1-NRF2 pathway, controls cytoprotection against electrophiles as well as redox-cycling compounds. Carcinogenesis 2009, 30, 1571–1580. [Google Scholar] [CrossRef]
- Rooney, J.P.; Chorley, B.; Hiemstra, S.; Wink, S.; Wang, X.; Bell, D.A.; van de Water, B.; Corton, J.C. Mining a human transcriptome database for chemical modulators of NRF2. PLoS ONE 2020, 15, e0239367. [Google Scholar] [CrossRef]
- Nishinaka, T.; Miura, T.; Shimizu, K.; Terada, T. Identification and characterization of functional antioxidant response elements in the promoter of the aldo-keto reductase AKR1B10 gene. Chem. Biol. Interact. 2017. [Google Scholar] [CrossRef] [PubMed]
- Chiu, R.; Boyle, W.J.; Meek, J.; Smeal, T.; Hunter, T.; Karin, M. The c-Fos protein interacts with c-Jun/AP-1 to stimulate transcription of AP-1 responsive genes. Cell 1988, 54, 541–552. [Google Scholar] [CrossRef]
- Cheng, B.Y.; Lau, E.Y.; Leung, H.W.; Leung, C.O.; Ho, N.P.; Gurung, S.; Cheng, L.K.; Lin, C.H.; Lo, R.C.; Ma, S.; et al. IRAK1 Augments Cancer Stemness and Drug Resistance via the AP-1/AKR1B10 Signaling Cascade in Hepatocellular Carcinoma. Cancer Res. 2018, 78, 2332–2342. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, F.X.; Porte, S.; Pares, X.; Farres, J. Biological role of aldo-keto reductases in retinoic Acid biosynthesis and signaling. Front. Pharmacol. 2012, 3, 58. [Google Scholar] [CrossRef] [PubMed]
- Kedishvili, N.Y. Retinoic Acid Synthesis and Degradation. Subcell. Biochem. 2016, 81, 127–161. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, T.; Endo, S.; Maeda, S.; Ishikura, S.; Tajima, K.; Tanaka, N.; Nakamura, K.T.; Imamura, Y.; Hara, A. Characterization of human DHRS4: An inducible short-chain dehydrogenase/reductase enzyme with 3beta-hydroxysteroid dehydrogenase activity. Arch. Biochem. Biophys. 2008, 477, 339–347. [Google Scholar] [CrossRef]
- Stambergova, H.; Zemanova, L.; Lundova, T.; Malcekova, B.; Skarka, A.; Safr, M.; Wsol, V. Human DHRS7, promising enzyme in metabolism of steroids and retinoids? J. Steroid Biochem. Mol. Biol. 2016, 155, 112–119. [Google Scholar] [CrossRef]
- Belyaeva, O.V.; Adams, M.K.; Popov, K.M.; Kedishvili, N.Y. Generation of Retinaldehyde for Retinoic Acid Biosynthesis. Biomolecules 2019, 10, 5. [Google Scholar] [CrossRef]
- Adams, M.K.; Belyaeva, O.V.; Wu, L.; Kedishvili, N.Y. The retinaldehyde reductase activity of DHRS3 is reciprocally activated by retinol dehydrogenase 10 to control retinoid homeostasis. J. Biol. Chem. 2014, 289, 14868–14880. [Google Scholar] [CrossRef]
- Pequerul, R.; Vera, J.; Gimenez-Dejoz, J.; Crespo, I.; Coines, J.; Porte, S.; Rovira, C.; Pares, X.; Farres, J. Structural and kinetic features of aldehyde dehydrogenase 1A (ALDH1A) subfamily members, cancer stem cell markers active in retinoic acid biosynthesis. Arch. Biochem. Biophys. 2020, 681, 108256. [Google Scholar] [CrossRef]
- Conklin, D.; Prough, R.; Bhatanagar, A. Aldehyde metabolism in the cardiovascular system. Mol. Biosyst. 2007, 3, 136–150. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Kapoor, A.; Bhatnagar, A. Oxidative and reductive metabolism of lipid-peroxidation derived carbonyls. Chem. Biol. Interact. 2015, 234, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Spite, M.; Baba, S.P.; Ahmed, Y.; Barski, O.A.; Nijhawan, K.; Petrash, J.M.; Bhatnagar, A.; Srivastava, S. Substrate specificity and catalytic efficiency of aldo-keto reductases with phospholipid aldehydes. Biochem. J. 2007, 405, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Baba, S.P.; Barski, O.A.; Ahmed, Y.; O’Toole, T.E.; Conklin, D.J.; Bhatnagar, A.; Srivastava, S. Reductive metabolism of AGE precursors: A metabolic route for preventing AGE accumulation in cardiovascular tissue. Diabetes 2009, 58, 2486–2497. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Liu, Z.; Yan, R.; Johnson, S.; Fang, X.; Cao, D. Aldo-keto reductase family 1 B10 protein detoxifies dietary and lipid-derived alpha, beta-unsaturated carbonyls at physiological levels. Biochem. Biophys. Res. Commun. 2009, 387, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.J.; Maser, E. Role of human aldo-keto-reductase AKR1B10 in the protection against toxic aldehydes. Chem. Biol. Interact. 2009, 178, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhong, L.; Johnson, S.; Cao, D. Human aldo-keto reductases 1B1 and 1B10: A comparative study on their enzyme activity toward electrophilic carbonyl compounds. Chem. Biol. Interact. 2011, 191, 192–198. [Google Scholar] [CrossRef]
- Matsunaga, T.; Yamaji, Y.; Tomokuni, T.; Morita, H.; Morikawa, Y.; Suzuki, A.; Yonezawa, A.; Endo, S.; Ikari, A.; Iguchi, K.; et al. Nitric oxide confers cisplatin resistance in human lung cancer cells through upregulation of aldo-keto reductase 1B10 and proteasome. Free Radic. Res. 2014, 48, 1371–1385. [Google Scholar] [CrossRef]
- Vander Jagt, D.L.; Kolb, N.S.; Vander Jagt, T.J.; Chino, J.; Martinez, F.J.; Hunsaker, L.A.; Royer, R.E. Substrate specificity of human aldose reductase: Identification of 4-hydroxynonenal as an endogenous substrate. Biochim. Biophys. Acta 1995, 1249, 117–126. [Google Scholar] [CrossRef]
- Ramana, K.V.; Dixit, B.L.; Srivastava, S.; Balendiran, G.K.; Srivastava, S.K.; Bhatnagar, A. Selective recognition of glutathiolated aldehydes by aldose reductase. Biochemistry 2000, 39, 12172–12180. [Google Scholar] [CrossRef]
- Burczynski, M.E.; Sridhar, G.R.; Palackal, N.T.; Penning, T.M. The reactive oxygen species- and Michael acceptor-inducible human aldo-keto reductase AKR1C1 reduces the alpha,beta-unsaturated aldehyde 4-hydroxy-2-nonenal to 1,4-dihydroxy-2-nonene. J. Biol. Chem. 2001, 276, 2890–2897. [Google Scholar] [CrossRef] [PubMed]
- Endo, S.; Matsunaga, T.; Ohta, C.; Soda, M.; Kanamori, A.; Kitade, Y.; Ohno, S.; Tajima, K.; El-Kabbani, O.; Hara, A. Roles of rat and human aldo-keto reductases in metabolism of farnesol and geranylgeraniol. Chem. Biol. Interact. 2011, 191, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, H.; Yang, Y.; Liao, J.; Yang, G.Y. Knockdown or inhibition of aldo-keto reductase 1B10 inhibits pancreatic carcinoma growth via modulating Kras-E-cadherin pathway. Cancer Lett. 2014, 355, 273–280. [Google Scholar] [CrossRef]
- Chung, Y.T.; Matkowskyj, K.A.; Li, H.; Bai, H.; Zhang, W.; Tsao, M.S.; Liao, J.; Yang, G.Y. Overexpression and oncogenic function of aldo-keto reductase family 1B10 (AKR1B10) in pancreatic carcinoma. Mod. Pathol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, T.; Wada, Y.; Endo, S.; Soda, M.; El-Kabbani, O.; Hara, A. Aldo-Keto Reductase 1B10 and Its Role in Proliferation Capacity of Drug-Resistant Cancers. Front. Pharmacol. 2012, 3, 5. [Google Scholar] [CrossRef]
- Matsunaga, T.; Endo, S.; Takemura, M.; Soda, M.; Yamamura, K.; Tajima, K.; Miura, T.; Terada, T.; El-Kabbani, O.; Hara, A. Reduction of cytotoxic p-quinone metabolites of tert-butylhydroquinone by human aldo-keto reductase (AKR) 1B10. Drug Metab. Pharmacokinet. 2012, 27, 553–558. [Google Scholar] [CrossRef]
- Martin, H.J.; Breyer-Pfaff, U.; Wsol, V.; Venz, S.; Block, S.; Maser, E. Purification and characterization of akr1b10 from human liver: Role in carbonyl reduction of xenobiotics. Drug Metab. Dispos. 2006, 34, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Bains, O.S.; Grigliatti, T.A.; Reid, R.E.; Riggs, K.W. Naturally occurring variants of human aldo-keto reductases with reduced in vitro metabolism of daunorubicin and doxorubicin. J. Pharmacol. Exp. Ther. 2010, 335, 533–545. [Google Scholar] [CrossRef]
- Skarydova, L.; Nobilis, M.; Wsol, V. Role of carbonyl reducing enzymes in the phase I biotransformation of the non-steroidal anti-inflammatory drug nabumetone in vitro. Xenobiotica 2013, 43, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Malatkova, P.; Skarka, A.; Musilova, K.; Wsol, V. Reductive metabolism of tiaprofenic acid by the human liver and recombinant carbonyl reducing enzymes. Chem. Biol. Interact. 2017. [Google Scholar] [CrossRef]
- Breyer-Pfaff, U.; Nill, K. Carbonyl reduction of naltrexone and dolasetron by oxidoreductases isolated from human liver cytosol. J. Pharm. Pharmacol. 2004, 56, 1601–1606. [Google Scholar] [CrossRef] [PubMed]
- Quinn, A.M.; Harvey, R.G.; Penning, T.M. Oxidation of PAH trans-Dihydrodiols by Human Aldo-Keto Reductase AKR1B10. Chem. Res. Toxicol. 2008, 21, 2207–2215. [Google Scholar] [CrossRef]
- Conway, L.P.; Garg, N.; Lin, W.; Vujasinovic, M.; Lohr, J.M.; Globisch, D. Chemoselective probe for detailed analysis of ketones and aldehydes produced by gut microbiota in human samples. Chem. Commun. (Camb.) 2019, 55, 9080–9083. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Yan, R.; Zu, X.; Cheng, J.M.; Rao, K.; Liao, D.F.; Cao, D. Aldo-keto Reductase Family 1 B10 Affects Fatty Acid Synthesis by Regulating the Stability of Acetyl-CoA Carboxylase-{alpha} in Breast Cancer Cells. J. Biol. Chem. 2008, 283, 3418–3423. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.X.; Huang, M.C.; Ma, J.; Gao, Z.; Liao, D.F.; Cao, D. Aldo-keto reductase family 1, member B10 is secreted through a lysosome-mediated non-classical pathway. Biochem. J. 2011, 438, 71–80. [Google Scholar] [CrossRef]
- Luo, D.; Bu, Y.; Ma, J.; Rajput, S.; He, Y.; Cai, G.; Liao, D.F.; Cao, D. Heat shock protein 90-alpha mediates aldo-keto reductase 1B10 (AKR1B10) protein secretion through secretory lysosomes. J. Biol. Chem. 2013, 288, 36733–36740. [Google Scholar] [CrossRef]
- Li, W.; Liu, C.; Huang, Z.; Shi, L.; Zhong, C.; Zhou, W.; Meng, P.; Li, Z.; Wang, S.; Luo, F.; et al. AKR1B10 negatively regulates autophagy through reducing GAPDH upon glucose starvation in colon cancer. J. Cell Sci. 2021. [Google Scholar] [CrossRef]
- Shen, Y.; Ma, J.; Yan, R.; Ling, H.; Li, X.; Yang, W.; Gao, J.; Huang, C.; Bu, Y.; Cao, Y.; et al. Impaired self-renewal and increased colitis and dysplastic lesions in colonic mucosa of AKR1B8-deficient mice. Clin. Cancer Res. 2015, 21, 1466–1476. [Google Scholar] [CrossRef]
- Lee, H.J.; Nam, K.T.; Park, H.S.; Kim, M.A.; Lafleur, B.J.; Aburatani, H.; Yang, H.K.; Kim, W.H.; Goldenring, J.R. Gene expression profiling of metaplastic lineages identifies CDH17 as a prognostic marker in early stage gastric cancer. Gastroenterology 2010, 139, 213–225.e3. [Google Scholar] [CrossRef]
- Kropotova, E.S.; Tychko, R.A.; Zinov’eva, O.L.; Zyrianova, A.F.; Khankin, S.L.; Cherkes, V.L.; Aliev, V.A.; Beresten, S.F.; Oparina, N.; Mashkova, T.D. [Downregulation of AKR1B10 gene expression in colorectal cancer]. Mol. Biol. (Mosk.) 2010, 44, 216–222. [Google Scholar] [CrossRef]
- Ohashi, T.; Idogawa, M.; Sasaki, Y.; Suzuki, H.; Tokino, T. AKR1B10, a transcriptional target of p53, is downregulated in colorectal cancers associated with poor prognosis. Mol. Cancer Res. 2013, 11, 1554–1563. [Google Scholar] [CrossRef] [PubMed]
- Kropotova, E.S.; Zinovieva, O.L.; Zyryanova, A.F.; Dybovaya, V.I.; Prasolov, V.S.; Beresten, S.F.; Oparina, N.Y.; Mashkova, T.D. Altered expression of multiple genes involved in retinoic acid biosynthesis in human colorectal cancer. Pathol. Oncol. Res. 2014, 20, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Zu, X.; Yan, R.; Pan, J.; Zhong, L.; Cao, Y.; Ma, J.; Cai, C.; Huang, D.; Liu, J.; Chung, F.L.; et al. Aldo-keto reductase 1B10 protects human colon cells from DNA damage induced by electrophilic carbonyl compounds. Mol. Carcinog. 2017, 56, 118–129. [Google Scholar] [CrossRef]
- Taskoparan, B.; Seza, E.G.; Demirkol, S.; Tuncer, S.; Stefek, M.; Gure, A.O.; Banerjee, S. Opposing roles of the aldo-keto reductases AKR1B1 and AKR1B10 in colorectal cancer. Cell. Oncol. (Dordr.) 2017, 40, 563–578. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Wang, X.; Zhou, D.; Li, H.; Qian, H.; Zhang, J.; Jiang, L.; Wang, B.; Lin, Q.; Zhu, X. Loss of AKR1B10 promotes colorectal cancer cells proliferation and migration via regulating FGF1-dependent pathway. Aging (Albany N. Y.) 2020, 12, 13059–13075. [Google Scholar] [CrossRef]
- Dai, G.P.; Wang, L.P.; Wen, Y.Q.; Ren, X.Q.; Zuo, S.G. Identification of key genes for predicting colorectal cancer prognosis by integrated bioinformatics analysis. Oncol. Lett. 2020, 19, 388–398. [Google Scholar] [CrossRef]
- Kropotova, E.S.; Zinov’eva, O.L.; Zyrianova, A.F.; Choinzonov, E.L.; Afanas’ev, S.G.; Cherdyntseva, N.V.; Beresten, S.F.; Oparina, N.; Mashkova, T.D. [Expression of genes involved in retinoic acid biosynthesis in human gastric cancer]. Mol. Biol. (Mosk.) 2013, 47, 317–330. [Google Scholar] [CrossRef]
- Yao, H.B.; Xu, Y.; Chen, L.G.; Guan, T.P.; Ma, Y.Y.; He, X.J.; Xia, Y.J.; Tao, H.Q.; Shao, Q.S. AKR1B10, a good prognostic indicator in gastric cancer. Eur. J. Surg. Oncol. 2014, 40, 318–324. [Google Scholar] [CrossRef]
- Ahmed, S.M.U.; Jiang, Z.N.; Zheng, Z.H.; Li, Y.; Wang, X.J.; Tang, X. AKR1B10 expression predicts response of gastric cancer to neoadjuvant chemotherapy. Oncol. Lett. 2019, 17, 773–780. [Google Scholar] [CrossRef]
- Osaki, L.H.; Gama, P. MAPKs and signal transduction in the control of gastrointestinal epithelial cell proliferation and differentiation. Int. J. Mol. Sci. 2013, 14, 10143–10161. [Google Scholar] [CrossRef]
- Mashkova, T.D.; Oparina, N.; Zinov’eva, O.L.; Kropotova, E.S.; Dubovaia, V.I.; Poltaraus, A.B.; Fridman, M.V.; Kopantsev, E.P.; Vinogradova, T.V.; Zinov’eva, M.V.; et al. [Transcription TIMP3, DAPk1 and AKR1B10 genes in squamous cell lung cancer]. Mol. Biol. (Mosk.) 2006, 40, 1047–1054. [Google Scholar] [CrossRef]
- Kang, M.W.; Lee, E.S.; Yoon, S.Y.; Jo, J.; Lee, J.; Kim, H.K.; Choi, Y.S.; Kim, K.; Shim, Y.M.; Kim, J.; et al. AKR1B10 is associated with smoking and smoking-related non-small-cell lung cancer. J. Int. Med. Res. 2011, 39, 78–85. [Google Scholar] [CrossRef]
- Szymanowska-Narloch, A.; Jassem, E.; Skrzypski, M.; Muley, T.; Meister, M.; Dienemann, H.; Taron, M.; Rosell, R.; Rzepko, R.; Jarzab, M.; et al. Molecular profiles of non-small cell lung cancers in cigarette smoking and never-smoking patients. Adv. Med. Sci. 2013, 58, 196–206. [Google Scholar] [CrossRef]
- MacLeod, A.K.; Acosta-Jimenez, L.; Coates, P.J.; McMahon, M.; Carey, F.A.; Honda, T.; Henderson, C.J.; Wolf, C.R. Aldo-keto reductases are biomarkers of NRF2 activity and are co-ordinately overexpressed in non-small cell lung cancer. Br. J. Cancer 2016, 115, 1530–1539. [Google Scholar] [CrossRef]
- Kuznetsova, E.S.; Zinovieva, O.L.; Oparina, N.Y.; Prokofjeva, M.M.; Spirin, P.V.; Favorskaya, I.A.; Zborovskaya, I.B.; Lisitsyn, N.A.; Prassolov, V.S.; Mashkova, T.D. [Abnormal expression of genes that regulate retinoid metabolism and signaling in non-small-cell lung cancer]. Mol. Biol. (Mosk.) 2016, 50, 255–265. [Google Scholar] [CrossRef]
- Ludovini, V.; Bianconi, F.; Siggillino, A.; Piobbico, D.; Vannucci, J.; Metro, G.; Chiari, R.; Bellezza, G.; Puma, F.; Della Fazia, M.A.; et al. Gene identification for risk of relapse in stage I lung adenocarcinoma patients: A combined methodology of gene expression profiling and computational gene network analysis. Oncotarget 2016, 7, 30561–30574. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zhao, Y.; Gu, L.; Niu, X.; Lu, S. Inhibiting proliferation and migration of lung cancer using small interfering RNA targeting on Aldo-keto reductase family 1 member B10. Mol. Med. Rep. 2018, 17, 2153–2160. [Google Scholar] [CrossRef]
- Hung, J.J.; Yeh, Y.C.; Hsu, W.H. Prognostic significance of AKR1B10 in patients with resected lung adenocarcinoma. Thorac. Cancer 2018, 9, 1492–1499. [Google Scholar] [CrossRef]
- Cong, Z.; Diao, Y.; Xu, Y.; Li, X.; Jiang, Z.; Shao, C.; Ji, S.; Shen, Y.; De, W.; Qiang, Y. Long non-coding RNA linc00665 promotes lung adenocarcinoma progression and functions as ceRNA to regulate AKR1B10-ERK signaling by sponging miR-98. Cell Death Dis. 2019, 10, 84. [Google Scholar] [CrossRef]
- Liu, W.; Song, J.; Du, X.; Zhou, Y.; Li, Y.; Li, R.; Lyu, L.; He, Y.; Hao, J.; Ben, J.; et al. AKR1B10 (Aldo-keto reductase family 1 B10) promotes brain metastasis of lung cancer cells in a multi-organ microfluidic chip model. Acta Biomater. 2019, 91, 195–208. [Google Scholar] [CrossRef]
- Ma, J.; Luo, D.X.; Huang, C.; Shen, Y.; Bu, Y.; Markwell, S.; Gao, J.; Liu, J.; Zu, X.; Cao, Z.; et al. AKR1B10 overexpression in breast cancer: Association with tumor size, lymph node metastasis and patient survival and its potential as a novel serum marker. Int. J. Cancer 2012, 131, E862–E871. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; He, X.; Xing, S.; Ni, J.; Zhang, W.; Xu, X.; Gao, F.; Gai, J.; Zhao, Z.; Li, J.; et al. Overexpression of Aldo-keto reductase family 1 B10 protein in ductal carcinoma in situ of the breast correlates with HER2 positivity. Cancer Biomark. 2013, 13, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Verhulst, S.; Shen, Y.; Bu, Y.; Cao, Y.; He, Y.; Wang, Y.; Huang, D.; Cai, C.; Rao, K.; et al. AKR1B10 promotes breast cancer metastasis through integrin alpha5/delta-catenin mediated FAK/Src/Rac1 signaling pathway. Oncotarget 2016. [Google Scholar] [CrossRef]
- Reddy, K.A.; Kumar, P.U.; Srinivasulu, M.; Triveni, B.; Sharada, K.; Ismail, A.; Reddy, G.B. Overexpression and enhanced specific activity of aldoketo reductases (AKR1B1 & AKR1B10) in human breast cancers. Breast 2017, 31, 137–143. [Google Scholar] [CrossRef]
- Li, J.; Guo, Y.; Duan, L.; Hu, X.; Zhang, X.; Hu, J.; Huang, L.; He, R.; Hu, Z.; Luo, W.; et al. AKR1B10 promotes breast cancer cell migration and invasion via activation of ERK signaling. Oncotarget 2017, 8, 33694–33703. [Google Scholar] [CrossRef]
- Van Weverwijk, A.; Koundouros, N.; Iravani, M.; Ashenden, M.; Gao, Q.; Poulogiannis, G.; Jungwirth, U.; Isacke, C.M. Metabolic adaptability in metastatic breast cancer by AKR1B10-dependent balancing of glycolysis and fatty acid oxidation. Nat. Commun. 2019, 10, 2698. [Google Scholar] [CrossRef]
- Connor, J.P.; Esbona, K.; Matkowskyj, K.A. AKR1B10 expression by immunohistochemistry in surgical resections and fine needle aspiration cytology material in patients with cystic pancreatic lesions; potential for improved nonoperative diagnosis. Hum. Pathol. 2017, 70, 77–83. [Google Scholar] [CrossRef]
- Fang, C.Y.; Lin, Y.H.; Chen, C.L. Overexpression of AKR1B10 predicts tumor recurrence and short survival in oral squamous cell carcinoma patients. J. Oral Pathol. Med. 2019, 48, 712–719. [Google Scholar] [CrossRef]
- Ko, H.H.; Cheng, S.L.; Lee, J.J.; Chen, H.M.; Kuo, M.Y.; Cheng, S.J. Expression of AKR1B10 as an independent marker for poor prognosis in human oral squamous cell carcinoma. Head Neck 2017, 39, 1327–1332. [Google Scholar] [CrossRef]
- El-Hariry, I.; Pignatelli, M.; Lemoine, N. Fibroblast growth factor 1 and fibroblast growth factor 2 immunoreactivity in gastrointestinal tumours. J. Pathol. 1997, 181, 39–45. [Google Scholar] [CrossRef]
- Henriksson, M.L.; Edin, S.; Dahlin, A.M.; Oldenborg, P.A.; Oberg, A.; Van Guelpen, B.; Rutegard, J.; Stenling, R.; Palmqvist, R. Colorectal cancer cells activate adjacent fibroblasts resulting in FGF1/FGFR3 signaling and increased invasion. Am. J. Pathol. 2011, 178, 1387–1394. [Google Scholar] [CrossRef]
- Liu, N.; Zhang, J.; Sun, S.; Yang, L.; Zhou, Z.; Sun, Q.; Niu, J. Expression and clinical significance of fibroblast growth factor 1 in gastric adenocarcinoma. Onco Targets Ther. 2015, 8, 615–621. [Google Scholar] [CrossRef]
- Matsunaga, T.; Suzuki, A.; Kezuka, C.; Okumura, N.; Iguchi, K.; Inoue, I.; Soda, M.; Endo, S.; El-Kabbani, O.; Hara, A.; et al. Aldo-keto reductase 1B10 promotes development of cisplatin resistance in gastrointestinal cancer cells through down-regulating peroxisome proliferator-activated receptor-gamma-dependent mechanism. Chem. Biol. Interact. 2016, 256, 142–153. [Google Scholar] [CrossRef]
- DiStefano, J.K.; Davis, B. Diagnostic and Prognostic Potential of AKR1B10 in Human Hepatocellular Carcinoma. Cancers 2019, 11, 486. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Liu, H.; Guan, G.; Zhang, T.; Wang, L.; Qi, X.; Zheng, H.; Chen, C.C.; Liu, J.; et al. Compensatory upregulation of aldo-keto reductase 1B10 to protect hepatocytes against oxidative stress during hepatocarcinogenesis. Am. J. Cancer Res. 2019, 9, 2730–2748. [Google Scholar]
- Wang, T.; Zhang, K.H. New Blood Biomarkers for the Diagnosis of AFP-Negative Hepatocellular Carcinoma. Front. Oncol. 2020, 10, 1316. [Google Scholar] [CrossRef]
- Raza, S.; Rajak, S.; Anjum, B.; Sinha, R.A. Molecular links between non-alcoholic fatty liver disease and hepatocellular carcinoma. Hepatoma Res. 2019, 5, 42. [Google Scholar] [CrossRef]
- Starmann, J.; Falth, M.; Spindelbock, W.; Lanz, K.L.; Lackner, C.; Zatloukal, K.; Trauner, M.; Sultmann, H. Gene expression profiling unravels cancer-related hepatic molecular signatures in steatohepatitis but not in steatosis. PLoS ONE 2012, 7, e46584. [Google Scholar] [CrossRef]
- Tsuzura, H.; Genda, T.; Sato, S.; Murata, A.; Kanemitsu, Y.; Narita, Y.; Ishikawa, S.; Kikuchi, T.; Mori, M.; Hirano, K.; et al. Expression of aldo-keto reductase family 1 member b10 in the early stages of human hepatocarcinogenesis. Int. J. Mol. Sci. 2014, 15, 6556–6568. [Google Scholar] [CrossRef]
- Bitter, A.; Rummele, P.; Klein, K.; Kandel, B.A.; Rieger, J.K.; Nussler, A.K.; Zanger, U.M.; Trauner, M.; Schwab, M.; Burk, O. Pregnane X receptor activation and silencing promote steatosis of human hepatic cells by distinct lipogenic mechanisms. Arch. Toxicol. 2015, 89, 2089–2103. [Google Scholar] [CrossRef]
- Pettinelli, P.; Arendt, B.M.; Teterina, A.; McGilvray, I.; Comelli, E.M.; Fung, S.K.; Fischer, S.E.; Allard, J.P. Altered hepatic genes related to retinol metabolism and plasma retinol in patients with non-alcoholic fatty liver disease. PLoS ONE 2018, 13, e0205747. [Google Scholar] [CrossRef]
- Arendt, B.M.; Teterina, A.; Pettinelli, P.; Comelli, E.M.; Ma, D.W.L.; Fung, S.K.; McGilvray, I.D.; Fischer, S.E.; Allard, J.P. Cancer-related gene expression is associated with disease severity and modifiable lifestyle factors in non-alcoholic fatty liver disease. Nutrition 2019, 62, 100–107. [Google Scholar] [CrossRef]
- Kanno, M.; Kawaguchi, K.; Honda, M.; Horii, R.; Takatori, H.; Shimakami, T.; Kitamura, K.; Arai, K.; Yamashita, T.; Sakai, Y.; et al. Serum aldo-keto reductase family 1 member B10 predicts advanced liver fibrosis and fatal complications of nonalcoholic steatohepatitis. J. Gastroenterol. 2019, 54, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Li, X.P.; Niu, C.Y.; Liu, M.L.; Yan, Q.Q.; Fan, L.P.; Li, Y.; Zhang, K.L.; Gao, J.; Qian, M.R.; et al. Bioinformatics analysis reveals novel core genes associated with nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Gene 2020, 742, 144549. [Google Scholar] [CrossRef]
- Govaere, O.; Cockell, S.; Tiniakos, D.; Queen, R.; Younes, R.; Vacca, M.; Alexander, L.; Ravaioli, F.; Palmer, J.; Petta, S.; et al. Transcriptomic profiling across the nonalcoholic fatty liver disease spectrum reveals gene signatures for steatohepatitis and fibrosis. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Zhang, Y.; Han, X.; Zeng, M.; Gao, Y.; Weng, J. Predicting Non-Alcoholic Fatty Liver Disease Progression and Immune Deregulations by Specific Gene Expression Patterns. Front. Immunol. 2020, 11, 609900. [Google Scholar] [CrossRef] [PubMed]
- Tebay, L.E.; Robertson, H.; Durant, S.T.; Vitale, S.R.; Penning, T.M.; Dinkova-Kostova, A.T.; Hayes, J.D. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic. Biol. Med. 2015, 88, 108–146. [Google Scholar] [CrossRef]
- Cullinan, S.B.; Zhang, D.; Hannink, M.; Arvisais, E.; Kaufman, R.J.; Diehl, J.A. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol. Cell. Biol. 2003, 23, 7198–7209. [Google Scholar] [CrossRef]
- Cubillos-Angulo, J.M.; Fukutani, E.R.; Cruz, L.A.B.; Arriaga, M.B.; Lima, J.V.; Andrade, B.B.; Queiroz, A.T.L.; Fukutani, K.F. Systems biology analysis of publicly available transcriptomic data reveals a critical link between AKR1B10 gene expression, smoking and occurrence of lung cancer. PLoS ONE 2020, 15, e0222552. [Google Scholar] [CrossRef]
- Li, C.P.; Goto, A.; Watanabe, A.; Murata, K.; Ota, S.; Niki, T.; Aburatani, H.; Fukayama, M. AKR1B10 in usual interstitial pneumonia: Expression in squamous metaplasia in association with smoking and lung cancer. Pathol. Res. Pract. 2008, 204, 295–304. [Google Scholar] [CrossRef]
- Huang, C.; Cao, Z.; Ma, J.; Shen, Y.; Bu, Y.; Khoshaba, R.; Shi, G.; Huang, D.; Liao, D.F.; Ji, H.; et al. AKR1B10 activates diacylglycerol (DAG) second messenger in breast cancer cells. Mol. Carcinog. 2018, 57, 1300–1310. [Google Scholar] [CrossRef]
- Hayes, A.J.; Skouras, C.; Haugk, B.; Charnley, R.M. Keap1-Nrf2 signalling in pancreatic cancer. Int. J. Biochem. Cell Biol. 2015, 65, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.H.; Peng, H.H.; Cheng, S.J.; Kuo, M.Y. Increased salivary AKR1B10 level: Association with progression and poor prognosis of oral squamous cell carcinoma. Head Neck 2018, 40, 2642–2647. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, N.S.; Beckers, S.; Mensah, J.K.; Waigel, S.; Vigneswaran, N.; Zacharias, W. Cigarette smoke condensate induces cytochromes P450 and aldo-keto reductases in oral cancer cells. Toxicol. Lett. 2006, 165, 182–194. [Google Scholar] [CrossRef]
- Yoshitake, H.; Takahashi, M.; Ishikawa, H.; Nojima, M.; Iwanari, H.; Watanabe, A.; Aburatani, H.; Yoshida, K.; Ishi, K.; Takamori, K.; et al. Aldo-keto reductase family 1, member B10 in uterine carcinomas: A potential risk factor of recurrence after surgical therapy in cervical cancer. Int. J. Gynecol. Cancer 2007, 17, 1300–1306. [Google Scholar] [CrossRef]
- Hevir, N.; Sinkovec, J.; Lanisnik Rizner, T. Decreased levels of AKR1B1 and AKR1B10 in cancerous endometrium compared to adjacent non-cancerous tissue. Chem. Biol. Interact. 2013, 202, 226–233. [Google Scholar] [CrossRef]
- Sinreih, M.; Stupar, S.; Cemazar, L.; Verdenik, I.; Frkovic Grazio, S.; Smrkolj, S.; Rizner, T.L. STAR and AKR1B10 are down-regulated in high-grade endometrial cancer. J. Steroid Biochem. Mol. Biol. 2017, 171, 43–53. [Google Scholar] [CrossRef]
- Ooi, A.; Wong, J.C.; Petillo, D.; Roossien, D.; Perrier-Trudova, V.; Whitten, D.; Min, B.W.; Tan, M.H.; Zhang, Z.; Yang, X.J.; et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell 2011, 20, 511–523. [Google Scholar] [CrossRef]
- He, Y.C.; Shen, Y.; Cao, Y.; Tang, F.Q.; Tian, D.F.; Huang, C.F.; Tao, H.; Zhou, F.L.; Zhang, B.; Song, L.; et al. Overexpression of AKR1B10 in nasopharyngeal carcinoma as a potential biomarker. Cancer Biomark. 2016, 16, 127–135. [Google Scholar] [CrossRef]
- Guo, Y.; Luo, W.; Hu, Z.; Li, J.; Li, X.; Cao, H.; Li, J.; Wen, B.; Zhang, J.; Cheng, H.; et al. Low expression of Aldo-keto reductase 1B10 is a novel independent prognostic indicator for nasopharyngeal carcinoma. Cell Biosci. 2016, 6, 18. [Google Scholar] [CrossRef]
- Liu, X.; Hu, Z.; Qu, J.; Li, J.; Gong, K.; Wang, L.; Jiang, J.; Li, X.; He, R.; Duan, L.; et al. AKR1B10 confers resistance to radiotherapy via FFA/TLR4/NF-κB axis in nasopharyngeal carcinoma. Int. J. Biol. Sci. 2021, 17, 756–767. [Google Scholar] [CrossRef] [PubMed]
- Breton, J.; Gage, M.C.; Hay, A.W.; Keen, J.N.; Wild, C.P.; Donnellan, C.; Findlay, J.B.; Hardie, L.J. Proteomic screening of a cell line model of esophageal carcinogenesis identifies cathepsin D and aldo-keto reductase 1C2 and 1B10 dysregulation in Barrett’s esophagus and esophageal adenocarcinoma. J. Proteome Res. 2008, 7, 1953–1962. [Google Scholar] [CrossRef] [PubMed]
- Nancarrow, D.J.; Clouston, A.D.; Smithers, B.M.; Gotley, D.C.; Drew, P.A.; Watson, D.I.; Tyagi, S.; Hayward, N.K.; Whiteman, D.C.; Australian Cancer, S.; et al. Whole genome expression array profiling highlights differences in mucosal defense genes in Barrett’s esophagus and esophageal adenocarcinoma. PLoS ONE 2011, 6, e22513. [Google Scholar] [CrossRef]
- Sumantran, V.N.; Mishra, P.; Bera, R.; Sudhakar, N. Microarray Analysis of Differentially-Expressed Genes Encoding CYP450 and Phase II Drug Metabolizing Enzymes in Psoriasis and Melanoma. Pharmaceutics 2016, 8, 4. [Google Scholar] [CrossRef]
- Gao, Y.; Yi, X.; Ding, Y. Combined Transcriptomic Analysis Revealed AKR1B10 Played an Important Role in Psoriasis through the Dysregulated Lipid Pathway and Overproliferation of Keratinocyte. Biomed. Res. Int. 2017, 2017, 8717369. [Google Scholar] [CrossRef]
- Jumper, N.; Hodgkinson, T.; Arscott, G.; Har-Shai, Y.; Paus, R.; Bayat, A. The Aldo-Keto Reductase AKR1B10 Is Up-Regulated in Keloid Epidermis, Implicating Retinoic Acid Pathway Dysregulation in the Pathogenesis of Keloid Disease. J. Investig. Dermatol. 2016, 136, 1500–1512. [Google Scholar] [CrossRef]
- Jumper, N.; Hodgkinson, T.; Paus, R.; Bayat, A. Site-specific gene expression profiling as a novel strategy for unravelling keloid disease pathobiology. PLoS ONE 2017, 12, e0172955. [Google Scholar] [CrossRef]
- Ghosh, D.; Ding, L.; Sivaprasad, U.; Geh, E.; Biagini Myers, J.; Bernstein, J.A.; Khurana Hershey, G.K.; Mersha, T.B. Multiple Transcriptome Data Analysis Reveals Biologically Relevant Atopic Dermatitis Signature Genes and Pathways. PLoS ONE 2015, 10, e0144316. [Google Scholar] [CrossRef]
- Soares, C.T.; Fachin, L.R.V.; Trombone, A.P.F.; Rosa, P.S.; Ghidella, C.C.; Belone, A.F.F. Potential of AKR1B10 as a Biomarker and Therapeutic Target in Type 2 Leprosy Reaction. Front. Med. (Lausanne) 2018, 5, 263. [Google Scholar] [CrossRef]
- Matsunaga, T.; El-Kabbani, O.; Hara, A. Aldo-keto reductases as new therapeutic targets for colon cancer chemoresistance. In Molecular Mechanisms of Tumor Cell Resistance to Chemotherapy: Targeted Therapies to Reverse Resistance; Bonavida, B., Ed.; Springer: New York, NY, USA, 2013; pp. 109–133. [Google Scholar]
- Penning, T.M. Aldo-Keto Reductase Regulation by the Nrf2 System: Implications for Stress Response, Chemotherapy Drug Resistance, and Carcinogenesis. Chem. Res. Toxicol. 2017, 30, 162–176. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Imanishi, K.; Tokui, N.; Okamoto, T.; Okamoto, A.; Hatakeyama, S.; Yoneyama, T.; Koie, T.; Kamimura, N.; Ohyama, C. Carboplatin-gemcitabine combination chemotherapy upregulates AKR1B10 expression in bladder cancer. Int. J. Clin. Oncol. 2013, 18, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, T.; Yamane, Y.; Iida, K.; Endo, S.; Banno, Y.; El-Kabbani, O.; Hara, A. Involvement of the aldo-keto reductase, AKR1B10, in mitomycin-c resistance through reactive oxygen species-dependent mechanisms. Anticancer Drugs 2011, 22, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, T.; Kawabata, S.; Yanagihara, Y.; Kezuka, C.; Kato, M.; Morikawa, Y.; Endo, S.; Chen, H.; Iguchi, K.; Ikari, A. Pathophysiological roles of autophagy and aldo-keto reductases in development of doxorubicin resistance in gastrointestinal cancer cells. Chem. Biol. Interact. 2019, 314, 108839. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, T.; Saito, H.; Endo, S.; Iguchi, K.; Soda, M.; El-Kabbani, O.; Hara, A.; Ikari, A. Roles of aldo-keto reductases 1B10 and 1C3 and ATP-binding cassette transporter in docetaxel tolerance. Free Radic. Res. 2016, 50, 1296–1308. [Google Scholar] [CrossRef]
- Heibein, A.D.; Guo, B.; Sprowl, J.A.; Maclean, D.A.; Parissenti, A.M. Role of aldo-keto reductases and other doxorubicin pharmacokinetic genes in doxorubicin resistance, DNA binding, and subcellular localization. BMC Cancer 2012, 12, 381. [Google Scholar] [CrossRef]
- Bacolod, M.D.; Lin, S.M.; Johnson, S.P.; Bullock, N.S.; Colvin, M.; Bigner, D.D.; Friedman, H.S. The gene expression profiles of medulloblastoma cell lines resistant to preactivated cyclophosphamide. Curr. Cancer Drug Targets 2008, 8, 172–179. [Google Scholar] [CrossRef][Green Version]
- Singh, A.; Misra, V.; Thimmulappa, R.K.; Lee, H.; Ames, S.; Hoque, M.O.; Herman, J.G.; Baylin, S.B.; Sidransky, D.; Gabrielson, E.; et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006, 3, e420. [Google Scholar] [CrossRef]
- Shibata, T.; Kokubu, A.; Gotoh, M.; Ojima, H.; Ohta, T.; Yamamoto, M.; Hirohashi, S. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology 2008, 135, 1358–1368. [Google Scholar] [CrossRef]
- Kang, J.S.; Nam, L.B.; Yoo, O.K.; Keum, Y.S. Molecular mechanisms and systemic targeting of NRF2 dysregulation in cancer. Biochem. Pharmacol. 2020, 177, 114002. [Google Scholar] [CrossRef]
- Zhong, L.; Shen, H.; Huang, C.; Jing, H.; Cao, D. AKR1B10 induces cell resistance to daunorubicin and idarubicin by reducing C13 ketonic group. Toxicol. Appl. Pharmacol. 2011, 255, 40–47. [Google Scholar] [CrossRef]
- Yan, R.; Zu, X.; Ma, J.; Liu, Z.; Adeyanju, M.; Cao, D. Aldo-keto reductase family 1 B10 gene silencing results in growth inhibition of colorectal cancer cells: Implication for cancer intervention. Int. J. Cancer 2007, 121, 2301–2306. [Google Scholar] [CrossRef]
- Huang, L.; He, R.; Luo, W.; Zhu, Y.S.; Li, J.; Tan, T.; Zhang, X.; Hu, Z.; Luo, D. Aldo-Keto Reductase Family 1 Member B10 Inhibitors: Potential Drugs for Cancer Treatment. Recent Pat. Anticancer Drug Discov. 2016, 11, 184–196. [Google Scholar] [CrossRef]
- Soda, M.; Hu, D.; Endo, S.; Takemura, M.; Li, J.; Wada, R.; Ifuku, S.; Zhao, H.T.; El-Kabbani, O.; Ohta, S.; et al. Design, synthesis and evaluation of caffeic acid phenethyl ester-based inhibitors targeting a selectivity pocket in the active site of human aldo-keto reductase 1B10. Eur. J. Med. Chem. 2012, 48, 321–329. [Google Scholar] [CrossRef]
- Takemura, M.; Endo, S.; Matsunaga, T.; Soda, M.; Zhao, H.T.; El-Kabbani, O.; Tajima, K.; Iinuma, M.; Hara, A. Selective inhibition of the tumor marker aldo-keto reductase family member 1B10 by oleanolic acid. J. Nat. Prod. 2011, 74, 1201–1206. [Google Scholar] [CrossRef]
- Endo, S.; Matsunaga, T.; Soda, M.; Tajima, K.; Zhao, H.T.; El-Kabbani, O.; Hara, A. Selective inhibition of the tumor marker AKR1B10 by antiinflammatory N-phenylanthranilic acids and glycyrrhetic acid. Biol. Pharm. Bull. 2010, 33, 886–890. [Google Scholar] [CrossRef]
- Hintzpeter, J.; Seliger, J.M.; Hofman, J.; Martin, H.J.; Wsol, V.; Maser, E. Inhibition of human anthracycline reductases by emodin—A possible remedy for anthracycline resistance. Toxicol. Appl. Pharmacol. 2016, 293, 21–29. [Google Scholar] [CrossRef]
- Hara, A.; Endo, S.; Matsunaga, T.; Soda, M.; El-Kabbani, O.; Yashiro, K. Inhibition of aldo-keto reductase family 1 member B10 by unsaturated fatty acids. Arch. Biochem. Biophys. 2016, 609, 69–76. [Google Scholar] [CrossRef]
- Seliger, J.M.; Cicek, S.S.; Witt, L.T.; Martin, H.J.; Maser, E.; Hintzpeter, J. Selective Inhibition of Human AKR1B10 by n-Humulone, Adhumulone and Cohumulone Isolated from Humulus lupulus Extract. Molecules 2018, 23, 3041. [Google Scholar] [CrossRef]
- Seliger, J.M.; Misuri, L.; Maser, E.; Hintzpeter, J. The hop-derived compounds xanthohumol, isoxanthohumol and 8-prenylnaringenin are tight-binding inhibitors of human aldo-keto reductases 1B1 and 1B10. J. Enzyme Inhib. Med. Chem. 2018, 33, 607–614. [Google Scholar] [CrossRef]
- Bateman, R.L.; Rauh, D.; Tavshanjian, B.; Shokat, K.M. Human carbonyl reductase 1 is an S-nitrosoglutathione reductase. J. Biol. Chem. 2008, 283, 35756–35762. [Google Scholar] [CrossRef]
- Hara, A.; Endo, S.; Matsunaga, T.; El-Kabbani, O.; Miura, T.; Nishinaka, T.; Terada, T. Human carbonyl reductase 1 participating in intestinal first-pass drug metabolism is inhibited by fatty acids and acyl-CoAs. Biochem. Pharmacol. 2017, 138, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Hara, A.; Endo, S.; Matsunaga, T.; Soda, M.; Yashiro, K.; El-Kabbani, O. Long-chain fatty acids inhibit human members of the aldo-keto reductase 1C subfamily. J. Biochem. 2017, 162, 371–379. [Google Scholar] [CrossRef]
- Endo, S.; Xia, S.; Suyama, M.; Morikawa, Y.; Oguri, H.; Hu, D.; Ao, Y.; Takahara, S.; Horino, Y.; Hayakawa, Y.; et al. Synthesis of Potent and Selective Inhibitors of Aldo-Keto Reductase 1B10 and Their Efficacy against Proliferation, Metastasis, and Cisplatin Resistance of Lung Cancer Cells. J. Med. Chem. 2017, 60, 8441–8455. [Google Scholar] [CrossRef]
- Cousido-Siah, A.; Ruiz, F.X.; Fanfrlik, J.; Gimenez-Dejoz, J.; Mitschler, A.; Kamlar, M.; Vesely, J.; Ajani, H.; Pares, X.; Farres, J.; et al. IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors. ACS Chem. Biol. 2016, 11, 2693–2705. [Google Scholar] [CrossRef]
- Chen, W.; Chen, X.; Zhou, S.; Zhang, H.; Wang, L.; Xu, J.; Hu, X.; Yin, W.; Yan, G.; Zhang, J. Design and synthesis of polyhydroxy steroids as selective inhibitors against AKR1B10 and molecular docking. Steroids 2016, 110, 1–8. [Google Scholar] [CrossRef]
- Geng, N.; Jin, Y.Y.; Zhu, S.X.; Li, Y.R.; Zheng, L.Y.; Zhu, W.J.; Li, Y.W.; Han, C.; Dou, X.G.; Bai, H. [Aldo-keto reductase family 1 B10 participates in the regulation of hepatoma cell cycle through p27/p-Rb signaling pathway]. Zhonghua Gan Zang Bing Za Zhi 2020, 28, 861–867. [Google Scholar] [CrossRef]
- Shi, L.; Guo, S.; Zhang, S.; Gao, X.; Liu, A.; Wang, Q.; Zhang, T.; Zhang, Y.; Wen, A. Glycyrrhetinic acid attenuates disturbed vitamin a metabolism in non-alcoholic fatty liver disease through AKR1B10. Eur. J. Pharmacol. 2020, 883, 173167. [Google Scholar] [CrossRef]
- Gimenez-Dejoz, J.; Weber, S.; Barski, O.A.; Moller, G.; Adamski, J.; Pares, X.; Porte, S.; Farres, J. Characterization of AKR1B16, a novel mouse aldo-keto reductase. Chem. Biol. Interact. 2017, 276, 182–193. [Google Scholar] [CrossRef]
- Pastel, E.; Pointud, J.C.; Volat, F.; Martinez, A.; Lefrancois-Martinez, A.M. Aldo-Keto Reductases 1B in Endocrinology and Metabolism. Front. Pharmacol. 2012, 3, 148. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).