The Metabolic Response of Brachypodium Roots to the Interaction with Beneficial Bacteria Is Affected by the Plant Nutritional Status
Abstract
:1. Introduction
2. Results
2.1. Polar Metabolites
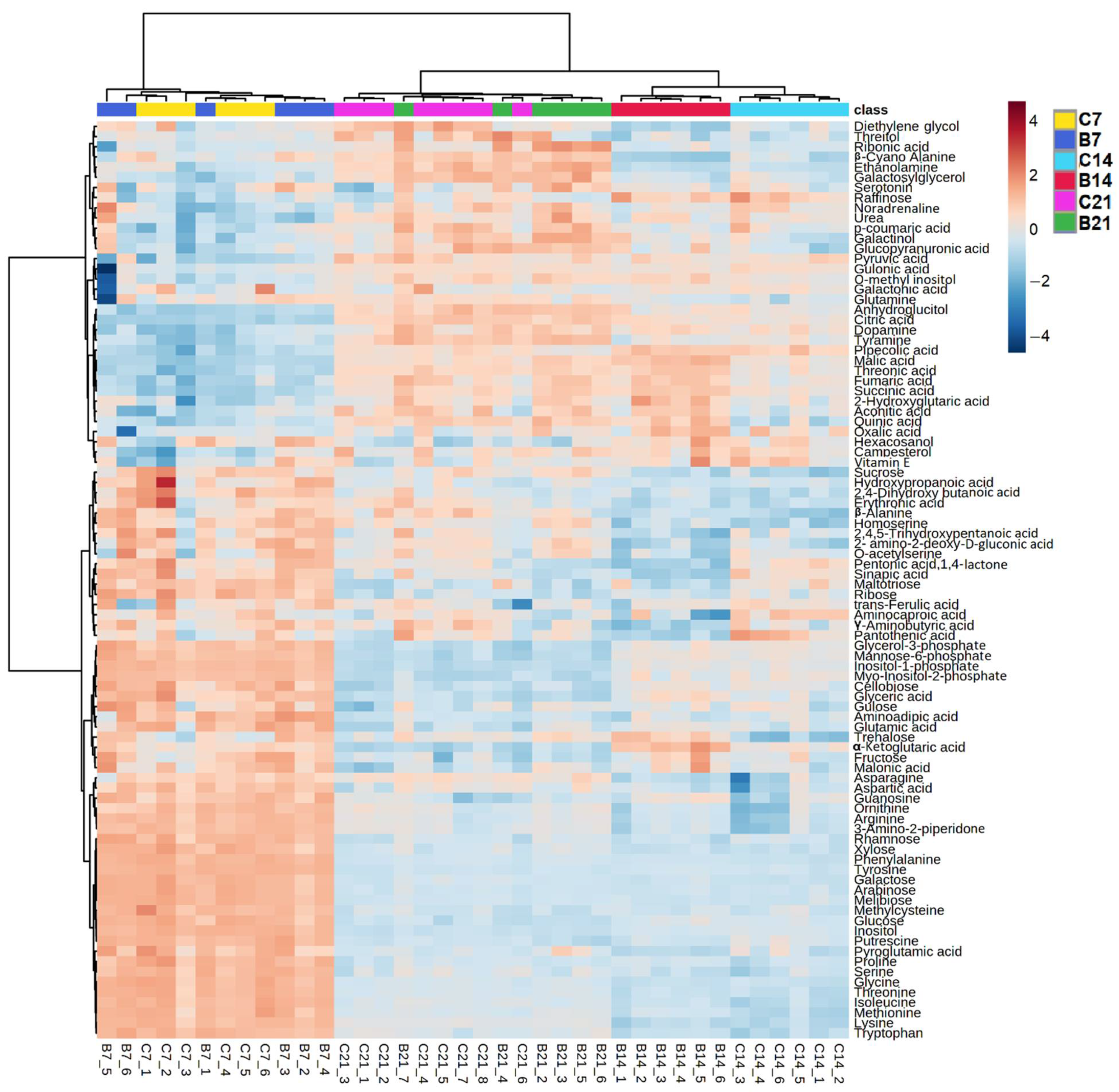
2.1.1. Root Polar Metabolites
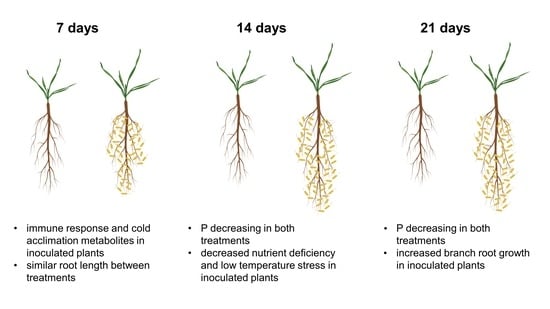
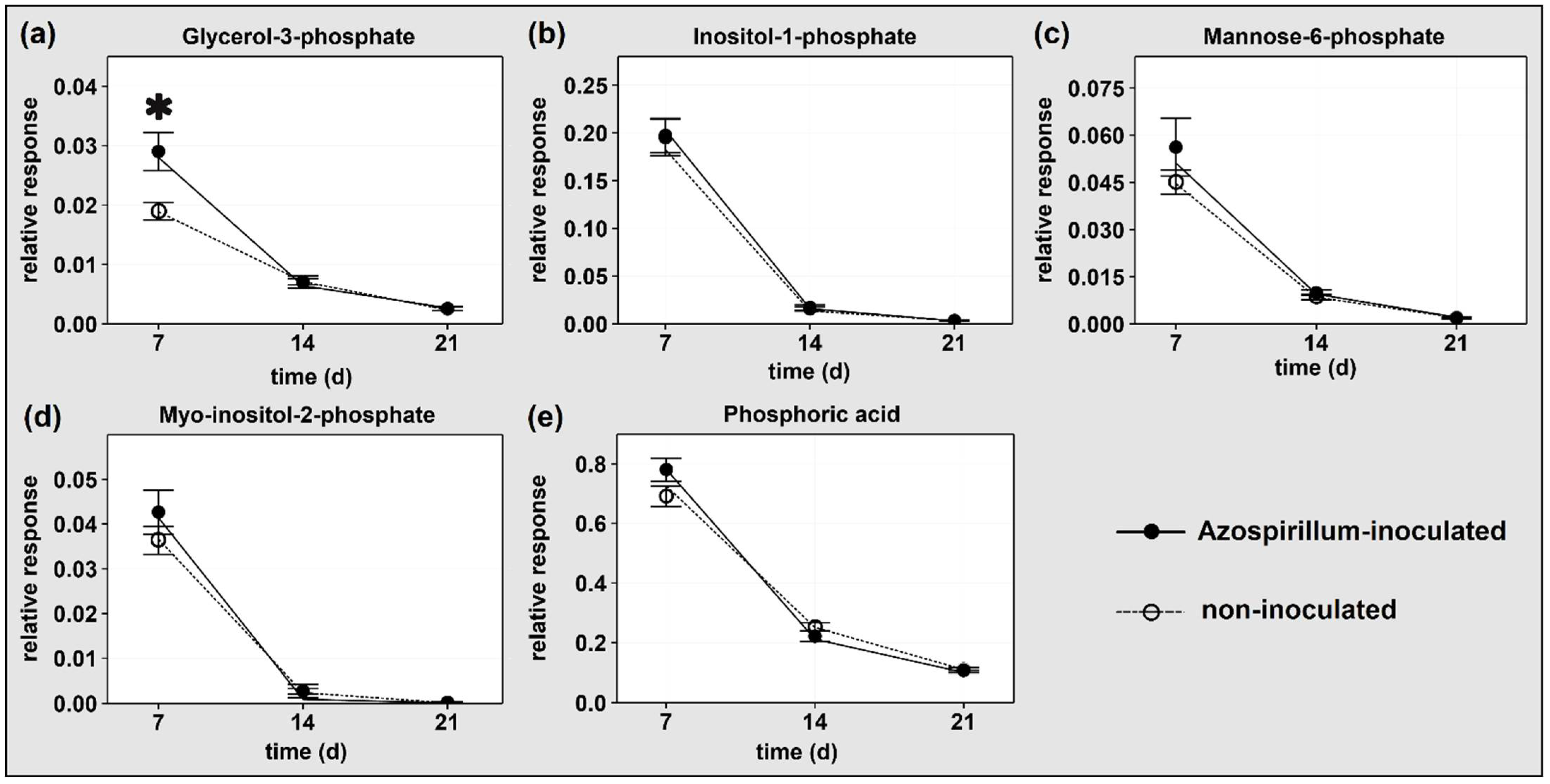
2.1.2. P-Containing Compounds
2.1.3. Comparison between Polar Metabolites of Inoculated and Non-Inoculated Plants
2.2. Lipids
2.2.1. Root Lipids
2.2.2. Root Glycerophospholipids and Galactolipids
2.2.3. Comparison between Lipids of Inoculated and Non-Inoculated Plants
3. Discussion
3.1. Polar Metabolites
3.1.1. Plants Consumed Phosphorylated Compounds to Face the Increasing P Deficiency
3.1.2. Azospirillum Elicited Different Responses in Plants at Different Stages
3.2. Lipids
3.2.1. P Deficiency Strongly Remodeled Root Lipid Profiles of Both Treatments
3.2.2. Azospirillum Inoculation Had a Limited Effect on Brachypodium’s Root Lipids
4. Materials and Methods
4.1. Plant Growth Conditions
4.2. Polar Metabolites Extraction from Roots
4.3. Lipid Extraction from Roots
4.4. GC-MS Analysis of Polar Metabolites
4.5. LC-MS Analysis of Lipids
4.6. Statistical Analyses and Data Visualization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Metabolite | Stress | Plant | PGP Bacteria Interaction | Role(s) |
|---|---|---|---|---|
| Aminocaproic acid | salinity | barley [85] | no | unknown |
| Campesterol | low temperature | rice [86,87], Maize [88] | no | brassinosteroids precursor |
| Campesterol | microbial attack | rice [27], barley [25] | no | brassinolide precursor |
| Diethylene glycol | Pb toxicity | Sedum alfredii [89] | no | unknown |
| Diethylene glycol | drought | Nicotiana benthamiana [90] | no | unknown |
| Glycerol-3-phosphate | pathogen attack | wheat [91] | no | systemic acquired resistance induction |
| Hexacosanol | pathogen attack | Asterids [92,93] | no | unknown |
| Hexacosanol | pathogen attack | A. thaliana [23] | no | wax barriers formation |
| Malic acid | P deficiency | wheat [40], barley [5], maize [94] | no | substrate P mobilization |
| Pantothenic acid | waterlogging | cucumber [95] | no | unknown |
| Pantothenic acid | drought | sorghum [96] | no | unknown |
| Pentonic acid, 1,4-lactone | low temperature | A. thaliana [97] | no | unknown |
| Pentonic acid, 1,4-lactone | P deficiency | Camelia sinensis [49] | no | unknown |
| Putrescine | P deficiency | rice [98] | no | growth inhibition |
| Pyroglutamic acid | no | Lolium multiflorum [99], tomato [100] | Pseudomonas putida [99], Pseudomonas fluorescens [100] | C source for PGP bacteria [99], bacterial chemotaxis [100] |
| Pyroglutamic acid | drought | lettuce [101] | no | photosynthetic rate improvement, ROS scavenging |
| Ribonic acid | no | sugarcane [102] | Herbaspirillum seropedicae, Gluconacetobacter diazotrophicus | unknown |
| Ribonic acid | P deficiency | Camelia sinensis [49], oat [103] | no | unknown |
| Serotonin | nutrient deficiency | rice [50] | no | ROS scavenging, nutrient recycling |
| Serotonin | no | barley [104] | no | auxin metabolism modification |
| Sinapic acid | pathogen attack | Aegilops variabilis [44] | no | unknown |
| Sinapic acid | no | wheat [43] | no | Antioxidant and antimicrobial activity |
| Trehalose | no | Maize [14] | Herbaspirillum seropedicae, Azospirillum brasilense | signaling during the interaction with PGP bacteria |
| Trehalose | drought | maize [105] | genetically modified A. brasilense | osmotic stress tolerance, root elongation |
| Trehalose | P deficiency | rice [106] | no | root elongation |
| Xylose | no | rice [107] | Corynebacterium sp., Rhizobium sp. | C source for PGP bacteria |
| Xylose | salinity | Wheat [108] | no | unknown |
| α-ketoglutaric acid | P deficiency | rice [109] | no | unknown |
References
- Schillaci, M.; Arsova, B.; Walker, R.; Smith, P.M.C.; Nagel, K.A.; Roessner, U.; Watt, M. Time-resolution of the shoot and root growth of the model cereal Brachypodium in response to inoculation with Azospirillum bacteria at low phosphorus and temperature. Plant Growth Regul. 2021, 93, 149–162. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Strand, Å.; Hurry, V.; Henkes, S.; Huner, N.; Gustafsson, P.; Gardeström, P.; Stitt, M. Acclimation of Arabidopsis Leaves Developing at Low Temperatures. Increasing Cytoplasmic Volume Accompanies Increased Activities of Enzymes in the Calvin Cycle and in the Sucrose-Biosynthesis Pathway. Plant Physiol. 1999, 119, 1387–1398. [Google Scholar] [CrossRef] [Green Version]
- Plaxton, W.C.; Tran, H.T. Metabolic Adaptations of Phosphate-Starved Plants. Plant Physiol. 2011, 156, 1006–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.Y.; Roessner, U.; Eickmeier, I.; Genc, Y.; Callahan, D.L.; Shirley, N.; Langridge, P.; Bacic, A. Metabolite Profiling Reveals Distinct Changes in Carbon and Nitrogen Metabolism in Phosphate-Deficient Barley Plants (Hordeum vulgare L.). Plant Cell Physiol. 2008, 49, 691–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grant, C.A.; Flaten, D.N.; Tomasiewicz, D.J.; Sheppard, S.C. The importance of early season phosphorus nutrition. Can. J. Plant Sci. 2001, 81, 211–224. [Google Scholar] [CrossRef]
- Sharkey, T.D.; Stitt, M.; Heineke, D.; Gerhardt, R.; Raschke, K.; Heldt, H.W. Limitation of photosynthesis by carbon metabolism: II. O2-insensitive CO2 uptake results from limitation of triose phosphate utilization. Plant Physiol. 1986, 81, 1123–1129. [Google Scholar] [CrossRef]
- Hurry, V.; Strand, Å.; Furbank, R.; Stitt, M. The role of inorganic phosphate in the development of freezing tolerance and the acclimatization of photosynthesis to low temperature is revealed by the pho mutants of Arabidopsis thaliana. Plant J. 2000, 24, 383–396. [Google Scholar] [CrossRef]
- Schlüter, U.; Colmsee, C.; Scholz, U.; Bräutigam, A.; Weber, A.P.M.; Zellerhoff, N.; Bucher, M.; Fahnenstich, H.; Sonnewald, U. Adaptation of maize source leaf metabolism to stress related disturbances in carbon, nitrogen and phosphorus balance. BMC Genom. 2013, 14, 442. [Google Scholar] [CrossRef] [Green Version]
- Aeron, A.; Kumar, S.; Pandey, P.; Maheshwari, D. Emerging role of plant growth promoting rhizobacteria in agrobiology. In Bacteria in Agrobiology: Crop Ecosystems; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1–36. [Google Scholar]
- Glick, B.R. Promotion of plant growth by soil bacteria that regulate plant ethylene levels. In Proceedings of the 33rd Annual Meeting of the Plant Growth Regulation Society of America, Quebec City, QC, Canada, 9–13 July 2006. [Google Scholar]
- Goswami, D.; Thakker, J.; Dhandhukia, P.C. Portraying mechanics of plant growth promoting rhizobacteria (PGPR): A review. Cogent Food Agric. 2016, 2, 1127500. [Google Scholar] [CrossRef]
- Agtuca, B.J.; Stopka, S.A.; Tuleski, T.R.; Amaral, F.P.D.; Evans, S.; Liu, Y.; Xu, D.; Monteiro, R.A.; Koppenaal, D.W.; Paša-Tolić, L.; et al. In-Situ Metabolomic Analysis of Setaria viridis Roots Colonized by Beneficial Endophytic Bacteria. Mol. Plant Microbe Interact. 2020, 33, 272–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brusamarello-Santos, L.C.; Gilard, F.; Brulé, L.; Quilleré, I.; Gourion, B.; Ratet, P.; de Souza, E.M.; Lea, P.J.; Hirel, B. Metabolic profiling of two maize (Zea mays L.) inbred lines inoculated with the nitrogen fixing plant-interacting bacteria Herbaspirillum seropedicae and Azospirillum brasilense. PLoS ONE 2017, 12, e0174576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamam, A.; Sanguin, H.; Bellvert, F.; Meiffren, G.; Comte, G.; Wisniewski-Dyé, F.; Bertrand, C.; Prigent-Combaret, C. Plant secondary metabolite profiling evidences strain-dependent effect in the Azospirillum–Oryza sativa association. Phytochemistry 2013, 87, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Curzi, M.J.; Ribaudo, C.M.; Trinchero, G.D.; Curá, J.A.; Pagano, E.A. Changes in the content of organic and amino acids and ethylene production of rice plants in response to the inoculation with Herbaspirillum seropedicae. J. Plant Interact. 2008, 3, 163–173. [Google Scholar] [CrossRef]
- Valette, M.; Rey, M.; Gerin, F.; Comte, G.; Wisniewski-Dyé, F. A common metabolomic signature is observed upon inoculation of rice roots with various rhizobacteria. J. Integr. Plant Biol. 2020, 62, 228–246. [Google Scholar] [CrossRef]
- Gagné-Bourque, F.; Bertrand, A.; Claessens, A.; Aliferis, K.A.; Jabaji, S. Alleviation of Drought Stress and Metabolic Changes in Timothy (Phleum pratense L.) Colonized with Bacillus subtilis B26. Front. Plant Sci. 2016, 7, 584. [Google Scholar] [CrossRef] [Green Version]
- Planchamp, C.; Glauser, G.; Mauch-Mani, B. Root inoculation with Pseudomonas putida KT2440 induces transcriptional and metabolic changes and systemic resistance in maize plants. Front. Plant Sci. 2015, 5, 719. [Google Scholar] [CrossRef] [Green Version]
- Catalan, P.; Chalhoub, B.; Chochois, V.; Garvin, D.F.; Hasterok, R.; Manzaneda, A.J.; Mur, L.A.; Pecchioni, N.; Rasmussen, S.K.; Vogel, J.P. Update on the genomics and basic biology of Brachypodium: International Brachypodium Initiative (IBI). Trends Plant Sci. 2014, 19, 414–418. [Google Scholar] [CrossRef] [Green Version]
- Rothballer, M.; Schmid, M.; Fekete, A.; Hartmann, A. Comparative in situ analysis of ipdC-gfpmut3 promoter fusions of Azospirillum brasilense strains Sp7 and Sp245. Environ. Microbiol. 2005, 7, 1839–1846. [Google Scholar] [CrossRef]
- Plaxton, W.; Carswell, M.C. Metabolic aspects of the phosphate starvation response in plants. In Plant Responses to Environmental Stresses; Routledge: London, UK, 1999. [Google Scholar]
- Shanmugarajah, K.; Linka, N.; Gräfe, K.; Smits, S.H.J.; Weber, A.P.M.; Zeier, J.; Schmitt, L. ABCG1 contributes to suberin formation in Arabidopsis thaliana roots. Sci. Rep. 2019, 9, 11381. [Google Scholar] [CrossRef]
- Zimmermann, W.; Eeemüller, E. Degradation of Raspberry Suberin by Fusarium solani f. sp. Pisi and Armillaria mellea. J. Phytopathol. 1984, 110, 192–199. [Google Scholar] [CrossRef]
- Ali, S.S.; Kumar, G.B.S.; Khan, M.; Doohan, F. Brassinosteroid Enhances Resistance to Fusarium Diseases of Barley. Phytopathology 2013, 103, 1260–1267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bajguz, A.; Hayat, S. Effects of brassinosteroids on the plant responses to environmental stresses. Plant Physiol. Biochem. 2009, 47, 1–8. [Google Scholar] [CrossRef]
- Nakashita, H.; Yasuda, M.; Nitta, T.; Asami, T.; Fujioka, S.; Arai, Y.; Sekimata, K.; Takatsuto, S.; Yamaguchi, I.; Yoshida, S. Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 2003, 33, 887–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chanda, B.; Xia, Y.; Mandal, M.K.; Yu, K.; Sekine, K.-T.; Gao, Q.-M.; Selote, D.; Hu, Y.; Stromberg, A.; Navarre, D.; et al. Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nat. Genet. 2011, 43, 421–427. [Google Scholar] [CrossRef]
- Shah, J.; Zeier, J. Long-distance communication and signal amplification in systemic acquired resistance. Front. Plant Sci. 2013, 4, 30. [Google Scholar] [CrossRef] [Green Version]
- Yokota, T. The structure, biosynthesis and function of brassinosteroids. Trends Plant Sci. 1997, 2, 137–143. [Google Scholar] [CrossRef]
- Yang, C.-J.; Zhang, C.; Lu, Y.-N.; Jin, J.-Q.; Wang, X.-L. The Mechanisms of Brassinosteroids’ Action: From Signal Transduction to Plant Development. Mol. Plant 2011, 4, 588–600. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Lynch, J.P. The contribution of lateral rooting to phosphorus acquisition efficiency in maize (Zea mays) seedlings. Funct. Plant Biol. 2004, 31, 949–958. [Google Scholar] [CrossRef] [PubMed]
- Lunn, J.E.; Delorge, I.; Figueroa, C.M.; Van Dijck, P.; Stitt, M. Trehalose metabolism in plants. Plant J. 2014, 79, 544–567. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, C.A.; Paul, M.J.; Foyer, C.H. Metabolite transport and associated sugar signalling systems underpinning source/sink interactions. Biochim. Biophys. Acta Bioenerg. 2016, 1857, 1715–1725. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Kumar, P.; Gautam, V.; Rengasamy, B.; Adhikari, B.; Udayakumar, M.; Sarkar, A.K. Root transcriptome of two contrasting indica rice cultivars uncovers regulators of root development and physiological responses. Sci. Rep. 2016, 6, 39266. [Google Scholar] [CrossRef] [Green Version]
- Hammond, J.P.; White, P.J. Sugar Signaling in Root Responses to Low Phosphorus Availability. Plant Physiol. 2011, 156, 1033–1040. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, S.; Parsons, A.J.; Jones, C.S. Metabolomics of forage plants: A review. Ann. Bot. 2012, 110, 1281–1290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrold, S.; Tabatabai, M. Release of inorganic phosphorus from soils by low-molecular-weight organic acids. Commun. Soil Sci. Plant Anal. 2006, 37, 1233–1245. [Google Scholar] [CrossRef]
- Hocking, P.J. Organic acids exuded from roots in phosphorus uptake and aluminum tolerance of plants in acid soils. Adv. Agron. 2001, 74, 63–97. [Google Scholar]
- Neumann, G.; Römheld, V. Root excretion of carboxylic acids and protons in phosphorus-deficient plants. Plant Soil 1999, 211, 121–130. [Google Scholar] [CrossRef]
- Jacoby, R.P.; Millar, A.H.; Taylor, N.L. Investigating the Role of Respiration in Plant Salinity Tolerance by Analyzing Mitochondrial Proteomes from Wheat and a Salinity-Tolerant Amphiploid (Wheat × Lophopyrum elongatum). J. Proteome Res. 2013, 12, 4807–4829. [Google Scholar] [CrossRef] [Green Version]
- Morgan, P.W.; Drew, M.C. Ethylene and plant responses to stress. Physiol. Plant. 1997, 100, 620–630. [Google Scholar] [CrossRef]
- Jeong, E.-Y.; Sung, B.-K.; Song, H.-Y.; Yang, J.-Y.; Kim, D.-K.; Lee, H.-S. Antioxidative and Antimicrobial Activities of Active Materials Derived from Triticum aestivum Sprouts. J. Korean Soc. Appl. Biol. Chem. 2010, 53, 519–524. [Google Scholar] [CrossRef]
- Huang, Q.; Li, L.; Zheng, M.; Chen, F.; Long, H.; Deng, G.; Pan, Z.; Liang, J.; Li, Q.; Yu, M.; et al. The Tryptophan decarboxylase 1 Gene from Aegilops variabilis No.1 Regulate the Resistance against Cereal Cyst Nematode by Altering the Downstream Secondary Metabolite Contents rather than Auxin Synthesis. Front. Plant Sci. 2018, 9, 1297. [Google Scholar] [CrossRef] [PubMed]
- Selvi, K.; Paul, J.; Vijaya, V.; Saraswathi, K. Analyzing the efficacy of phosphate solubilizing microorganisms by enrichment culture techniques. Biochem. Mol. Biol. J. 2017, 3. [Google Scholar] [CrossRef] [Green Version]
- Echevarría, C.; Maurĩno, S.G.; Maldonado, J.M. Reversible inactivation of maize leaf nitrate reductase. Phytochemistry 1984, 23, 2155–2158. [Google Scholar] [CrossRef]
- Machingura, M.; Salomon, E.; Jez, J.M.; Ebbs, S.D. The β-cyanoalanine synthase pathway: Beyond cyanide detoxification. Plant Cell Environ. 2016, 39, 2329–2341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dias, D.; Hill, C.B.; Jayasinghe, N.S.; Atieno, J.; Sutton, T.; Roessner, U. Quantitative profiling of polar primary metabolites of two chickpea cultivars with contrasting responses to salinity. J. Chromatogr. B 2015, 1000, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Ding, Z.; Jia, S.; Wang, Y.; Xiao, J.; Zhang, Y. Phosphate stresses affect ionome and metabolome in tea plants. Plant Physiol. Biochem. 2017, 120, 30–39. [Google Scholar] [CrossRef]
- Kang, K.; Kim, Y.-S.; Park, S.; Back, K. Senescence-Induced Serotonin Biosynthesis and Its Role in Delaying Senescence in Rice Leaves. Plant Physiol. 2009, 150, 1380–1393. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Yu, J.; Peng, Y.; Huang, B. Metabolic pathways regulated by abscisic acid, salicylic acid and γ-aminobutyric acid in association with improved drought tolerance in creeping bentgrass (Agrostis stolonifera). Physiol. Plant. 2017, 159, 42–58. [Google Scholar] [CrossRef]
- Pacovsky, R.S. Influence of inoculation with Azospirillum brasilense and Glomus fasciculatum on sorghum nutrition. Plant Soil 1988, 110, 283–287. [Google Scholar] [CrossRef]
- Ramakrishna, A.; Giridhar, P.; Ravishankar, G.A. Phytoserotonin: A review. Plant Signal. Behav. 2011, 6, 800. [Google Scholar]
- Hayashi, K.; Fujita, Y.; Ashizawa, T.; Suzuki, F.; Nagamura, Y.; Hayano-Saito, Y. Serotonin attenuates biotic stress and leads to lesion browning caused by a hypersensitive response to Magnaporthe oryzae penetration in rice. Plant J. 2016, 85, 46–56. [Google Scholar] [CrossRef] [Green Version]
- Pelagio-Flores, R.; Ortíz-Castro, R.; Méndez-Bravo, A.; Macías-Rodríguez, L.; López-Bucio, J. Serotonin, a Tryptophan-Derived Signal Conserved in Plants and Animals, Regulates Root System Architecture Probably Acting as a Natural Auxin Inhibitor in Arabidopsis thaliana. Plant Cell Physiol. 2011, 52, 490–508. [Google Scholar] [CrossRef] [Green Version]
- Wan, J.; Zhang, P.; Sun, L.; Li, S.; Wang, R.; Zhou, H.; Wang, W.; Xu, J. Involvement of reactive oxygen species and auxin in serotonin-induced inhibition of primary root elongation. J. Plant Physiol. 2018, 229, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Tjellström, H.; Andersson, M.X.; Larsson, K.E.; Sandelius, A.S. Membrane phospholipids as a phosphate reserve: The dynamic nature of phospholipid-to-digalactosyl diacylglycerol exchange in higher plants. Plant Cell Environ. 2008, 31, 1388–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, B.; Xu, C.; Benning, C. Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc. Natl. Acad. Sci. USA 2002, 99, 5732–5737. [Google Scholar] [CrossRef] [Green Version]
- Kelly, A.A.; Dörmann, P. DGD2, an Arabidopsis Gene Encoding a UDP-Galactose-dependent Digalactosyldiacylglycerol Synthase Is Expressed during Growth under Phosphate-limiting Conditions. J. Biol. Chem. 2002, 277, 1166–1173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.; Ahmad, M.Z.; Chen, B.; Manan, S.; Zhang, Y.; Jin, H.; Wang, X.; Zhao, J. Lipidomic and transcriptomic profiling of developing nodules reveals the essential roles of active glycolysis and fatty acid and membrane lipid biosynthesis in soybean nodulation. Plant J. 2020, 103, 1351–1371. [Google Scholar] [CrossRef]
- Abeer, H.; Abdallah, E.; Alqarawi, A.; Al-Huqail, A.A.; Alshalawi, S.; Wirth, S.; Dilfuza, E. Impact of plant growth promoting Bacillus subtilis on growth and physiological parameters of Bassia indica (Indian bassia) grown udder salt stress. Pak. J. Bot. 2015, 47, 1735–1741. [Google Scholar]
- Calderon-Vazquez, C.; Ibarra-Laclette, E.; Caballero-Perez, J.; Herrera-Estrella, L.R. Transcript profiling of Zea mays roots reveals gene responses to phosphate deficiency at the plant- and species-specific levels. J. Exp. Bot. 2008, 59, 2479–2497. [Google Scholar] [CrossRef] [Green Version]
- Siebers, M.; Brands, M.; Wewer, V.; Duan, Y.; Hölzl, G.; Dörmann, P. Lipids in plant-microbe interactions. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2016, 1861, 1379–1395. [Google Scholar] [CrossRef]
- Cao, D.; Lutz, A.; Hill, C.B.; Callahan, D.L.; Roessner, U. A Quantitative Profiling Method of Phytohormones and Other Metabolites Applied to Barley Roots Subjected to Salinity Stress. Front. Plant Sci. 2017, 7, 2070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereyra, M.; Zalazar, C.; Barassi, C. Root phospholipids in Azospirillum-inoculated wheat seedlings exposed to water stress. Plant Physiol. Biochem. 2006, 44, 873–879. [Google Scholar] [CrossRef] [PubMed]
- Gika, H.G.; Theodoridis, G.A.; Wilson, I.D. Metabolic Profiling: Status, Challenges, and Perspective. In Metabolic Profiling. Methods in Molecular Biology; Theodoridis, G., Gika, H., Wilson, I., Eds.; Humana Press: New York, NY, USA, 2018; Volume 1738. [Google Scholar] [CrossRef]
- Dickinson, E.; Rusilowicz, M.J.; Dickinson, M.; Charlton, A.J.; Bechtold, U.; Mullineaux, P.M.; Wilson, J. Integrating transcriptomic techniques and k-means clustering in metabolomics to identify markers of abiotic and biotic stress in Medicago truncatula. Metabolomics 2018, 14, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gioia, T.; Galinski, A.; Lenz, H.; Müller, C.; Lentz, J.; Heinz, K.; Briese, C.; Putz, A.; Fiorani, F.; Watt, M.; et al. GrowScreen-PaGe, a non-invasive, high-throughput phenotyping system based on germination paper to quantify crop phenotypic diversity and plasticity of root traits under varying nutrient supply. Funct. Plant Biol. 2017, 44, 76–93. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.B.; Roessner, U. Metabolic profiling of plants by GC-MS. In The Handbook of Plant Metabolomics: Metabolite Profiling and Networking; Wiley-VCH: Hoboken, NJ, USA, 2013; pp. 3–23. [Google Scholar]
- Cheong, B.E.; Onyemaobi, O.; Ho, W.W.H.; Ben Biddulph, T.; Rupasinghe, T.W.T.; Roessner, U.; Dolferus, R. Phenotyping the Chilling and Freezing Responses of Young Microspore Stage Wheat Spikes Using Targeted Metabolome and Lipidome Profiling. Cells 2020, 9, 1309. [Google Scholar] [CrossRef]
- Shiva, S.; Enninful, R.; Roth, M.R.; Tamura, P.; Jagadish, K.; Welti, R. An efficient modified method for plant leaf lipid extraction results in improved recovery of phosphatidic acid. Plant Methods 2018, 14, 14. [Google Scholar] [CrossRef] [Green Version]
- Kehelpannala, C.; Rupasinghe, T.W.T.; Hennessy, T.; Bradley, D.; Ebert, B.; Roessner, U. A comprehensive comparison of four methods for extracting lipids from Arabidopsis tissues. Plant Methods 2020, 16, 155. [Google Scholar] [CrossRef]
- Hillyer, K.E.; Dias, D.; Lutz, A.; Wilkinson, S.P.; Roessner, U.; Davy, S.K. Metabolite profiling of symbiont and host during thermal stress and bleaching in the coral Acropora aspera. Coral Reefs 2017, 36, 105–118. [Google Scholar] [CrossRef]
- Kehelpannala, C.; Rupasinghe, T.; Pasha, A.; Esteban, E.; Hennessy, T.; Bradley, D.; Ebert, B.; Provart, N.J.; Roessner, U. An Arabidopsis lipid map reveals differences between tissues and dynamic changes throughout development. Plant J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.T.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.S.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Yu, D.; Rupasinghe, T.W.; Boughton, B.A.; Natera, S.H.; Hill, C.B.; Tarazona, P.; Feussner, I.; Roessner, U. A high-resolution HPLC-QqTOF platform using parallel reaction monitoring for in-depth lipid discovery and rapid profiling. Anal. Chim. Acta 2018, 1026, 87–100. [Google Scholar] [CrossRef]
- Ihaka, R.; Gentleman, R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [Green Version]
- Valero-Mora, P.M. ggplot2: Elegant graphics for data analysis. J. Stat. Softw. 2010, 35. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, A.; Donn, S.; Ryan, P.R.; Mathesius, U.; Devilla, R.; Jones, A.; Watt, M. Microbiome and Exudates of the Root and Rhizosphere of Brachypodium distachyon, a Model for Wheat. PLoS ONE 2016, 11, e0164533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olanrewaju, O.S.; Ayangbenro, A.S.; Glick, B.R.; Babalola, O.O.; Ayangbenro, A. Plant health: Feedback effect of root exudates-rhizobiome interactions. Appl. Microbiol. Biotechnol. 2019, 103, 1155–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casanovas, E.M.; Barassi, C.A.; Sueldo, R.J. Azospirillum inoculation mitigates water stress effects in maize seedlings. Cereal Res. Commun. 2002, 30, 343–350. [Google Scholar] [CrossRef]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [Green Version]
- Yuwono, T.; Handayani, D.; Soedarsono, J. The role of osmotolerant rhizobacteria in rice growth under different drought conditions. Aust. J. Agric. Res. 2005, 56, 715–721. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, X.; Xu, Q.; Mei, X.; Yuan, H.; Jiabu, D.; Sang, Z.; Nyima, T. Metabolite profiling in two contrasting Tibetan hulless barley cultivars revealed the core salt-responsive metabolome and key salt-tolerance biomarkers. AoB Plants 2019, 11, plz021. [Google Scholar] [CrossRef] [Green Version]
- Fujii, S.; Saka, H. The Promotive Effect of Brassinolide on Lamina Joint-Cell Elongation, Germination and Seedling Growth under Low-Temperature Stress in Rice (Oryza sativa L.). Plant Prod. Sci. 2001, 4, 210–214. [Google Scholar] [CrossRef] [Green Version]
- Hotta, Y.; Tanaka, T.; Luo, B.; Takeuchi, Y.; Konnai, M. Improvement of Cold Resistance in Rice Seedlings by 5-Aminolevulinic Acid. J. Pestic. Sci. 1998, 23, 29–33. [Google Scholar] [CrossRef] [Green Version]
- He, R.-Y.; Wang, G.-J.; Wang, X.-S. Effects of brassinolide on growth and chilling resistance of maize seedlings. In Brassinosteroids; ACS Publications: Washington, DC, USA, 1991. [Google Scholar]
- Luo, Q.; Wang, S.; Sun, L.-N.; Wang, H.; Bao, T.; Adeel, M. Identification of root exudates from the Pb-accumulator Sedum alfredii under Pb stresses and assessment of their roles. J. Plant Interact. 2017, 12, 272–278. [Google Scholar] [CrossRef]
- Dastogeer, K.M.G.; Li, H.; Sivasithamparam, K.; Jones, M.; Du, X.; Ren, Y.; Wylie, S.J. Metabolic responses of endophytic Nicotiana benthamiana plants experiencing water stress. Environ. Exp. Bot. 2017, 143, 59–71. [Google Scholar] [CrossRef]
- Yang, Y.; Zhao, J.; Liu, P.; Xing, H.; Li, C.; Wei, G.; Kang, Z. Glycerol-3-Phosphate Metabolism in Wheat Contributes to Systemic Acquired Resistance against Puccinia striiformis f. sp. tritici. PLoS ONE 2013, 8, e81756. [Google Scholar] [CrossRef]
- Castilho, P.; Savluchinske-Feio, S.; Weinhold, T.S.; Gouveia, S.C. Evaluation of the antimicrobial and antioxidant activities of essential oils, extracts and their main components from oregano from Madeira Island, Portugal. Food Control 2012, 23, 552–558. [Google Scholar] [CrossRef]
- Mbosso, E.J.T.; Ngouela, S.; Nguedia, J.C.A.; Beng, V.P.; Rohmer, M.; Tsamo, E. In vitro antimicrobial activity of extracts and compounds of some selected medicinal plants from Cameroon. J. Ethnopharmacol. 2010, 128, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Gaume, A.; Machler, F.; De Leon, C.; Narro, L.; Frossard, E. Low-P tolerance by maize (Zea mays L.) genotypes: Significance of root growth, and organic acids and acid phosphatase root exudation. Plant Soil 2001, 228, 253–264. [Google Scholar] [CrossRef]
- Qi, X.-H.; Xu, X.-W.; Lin, X.-J.; Zhang, W.-J.; Chen, X.-H. Identification of differentially expressed genes in cucumber (Cucumis sativus L.) root under waterlogging stress by digital gene expression profile. Genomics 2012, 99, 160–168. [Google Scholar] [CrossRef] [Green Version]
- Pavli, O.I.; Vlachos, C.E.; Kalloniati, C.; Flemetakis, E.; Skaracis, G.N. Metabolite profiling reveals the effect of drought on sorghum (‘Sorghum bicolor’ L. Moench) metabolism. Plant Omics 2013, 6, 371–376. [Google Scholar]
- Chen, M.; Thelen, J.J. Acyl-lipid desaturase 1 primes cold acclimation response in Arabidopsis. Physiol. Plant. 2016, 158, 11–22. [Google Scholar] [CrossRef]
- Shih, C.Y.; Kao, C.H. Growth Inhibition in Suspension-Cultured Rice Cells under Phosphate Deprivation Is Mediated through Putrescine Accumulation. Plant Physiol. 1996, 111, 721–724. [Google Scholar] [CrossRef] [Green Version]
- Kuiper, I.; Kravchenko, L.V.; Bloemberg, G.V.; Lugtenberg, B.J.J. Pseudomonas putida Strain PCL1444, Selected for Efficient Root Colonization and Naphthalene Degradation, Effectively Utilizes Root Exudate Components. Mol. Plant Microbe Interact. 2002, 15, 734–741. [Google Scholar] [CrossRef] [Green Version]
- De Weert, S.; Vermeiren, H.; Mulders, I.H.M.; Kuiper, I.; Hendrickx, N.; Bloemberg, G.V.; Vanderleyden, J.; De Mot, R.; Lugtenberg, B.J.J. Flagella-Driven Chemotaxis towards Exudate Components Is an Important Trait for Tomato Root Colonization by Pseudomonas fluorescens. Mol. Plant Microbe Interact. 2002, 15, 1173–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiménez-Arias, D.; García-Machado, F.J.; Morales-Sierra, S.; Luis, J.C.; Suarez, E.; Hernández, M.; Valdés, F.; Borges, A.A. Lettuce plants treated with L-pyroglutamic acid increase yield under water deficit stress. Environ. Exp. Bot. 2019, 158, 215–222. [Google Scholar] [CrossRef] [Green Version]
- Aguiar, N.O.; Olivares, F.L.; Novotny, E.H.; Canellas, L.P. Changes in metabolic profiling of sugarcane leaves induced by endophytic diazotrophic bacteria and humic acids. PeerJ 2018, 6, e5445. [Google Scholar] [CrossRef]
- Wang, Y.; Lysøe, E.; Armarego-Marriott, T.; Erban, A.; Paruch, L.; Van Eerde, A.; Bock, R.; Liu-Clarke, J. Transcriptome and metabolome analyses provide insights into root and root-released organic anion responses to phosphorus deficiency in oat. J. Exp. Bot. 2018, 69, 3759–3771. [Google Scholar] [CrossRef] [Green Version]
- Csaba, G.; Pál, K. Effects of insulin, triiodothyronine, and serotonin on plant seed development. Protoplasma 1982, 110, 20–22. [Google Scholar] [CrossRef]
- Rodríguez-Salazar, J.; Suárez, R.; Caballero-Mellado, J.; Iturriaga, G. Trehalose accumulation in Azospirillum brasilense improves drought tolerance and biomass in maize plants. FEMS Microbiol. Lett. 2009, 296, 52–59. [Google Scholar] [CrossRef] [Green Version]
- Tyagi, W.; Rai, M. Root transcriptomes of two acidic soil adapted Indica rice genotypes suggest diverse and complex mechanism of low phosphorus tolerance. Protoplasma 2017, 254, 725–736. [Google Scholar] [CrossRef] [PubMed]
- Naher, U.; Radziah, O.; Halimi, M.; Shamsuddin, Z.; Razi, I.M. Effect of Inoculation on Root Exudates Carbon Sugar and Amino Acids Production of Different Rice Varieties. Res. J. Microbiol. 2008, 3, 580–587. [Google Scholar] [CrossRef] [Green Version]
- Guo, R.; Yang, Z.; Li, F.; Yan, C.; Zhong, X.; Liu, Q.; Xia, X.; Li, H.; Zhao, L. Comparative metabolic responses and adaptive strategies of wheat (Triticum aestivum) to salt and alkali stress. BMC Plant Biol. 2015, 15, 170. [Google Scholar] [CrossRef] [Green Version]
- Tawaraya, K.; Horie, R.; Saito, A.; Shinano, T.; Wagatsuma, T.; Saito, K.; Oikawa, A. Metabolite profiling of shoot extracts, root extracts, and root exudates of rice plant under phosphorus deficiency. J. Plant Nutr. 2013, 36, 1138–1159. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schillaci, M.; Kehelpannala, C.; Martinez-Seidel, F.; Smith, P.M.C.; Arsova, B.; Watt, M.; Roessner, U. The Metabolic Response of Brachypodium Roots to the Interaction with Beneficial Bacteria Is Affected by the Plant Nutritional Status. Metabolites 2021, 11, 358. https://doi.org/10.3390/metabo11060358
Schillaci M, Kehelpannala C, Martinez-Seidel F, Smith PMC, Arsova B, Watt M, Roessner U. The Metabolic Response of Brachypodium Roots to the Interaction with Beneficial Bacteria Is Affected by the Plant Nutritional Status. Metabolites. 2021; 11(6):358. https://doi.org/10.3390/metabo11060358
Chicago/Turabian StyleSchillaci, Martino, Cheka Kehelpannala, Federico Martinez-Seidel, Penelope M. C. Smith, Borjana Arsova, Michelle Watt, and Ute Roessner. 2021. "The Metabolic Response of Brachypodium Roots to the Interaction with Beneficial Bacteria Is Affected by the Plant Nutritional Status" Metabolites 11, no. 6: 358. https://doi.org/10.3390/metabo11060358
APA StyleSchillaci, M., Kehelpannala, C., Martinez-Seidel, F., Smith, P. M. C., Arsova, B., Watt, M., & Roessner, U. (2021). The Metabolic Response of Brachypodium Roots to the Interaction with Beneficial Bacteria Is Affected by the Plant Nutritional Status. Metabolites, 11(6), 358. https://doi.org/10.3390/metabo11060358