Metabolomic Study of High-Fat Diet-Induced Obese (DIO) and DIO Plus CCl4-Induced NASH Mice and the Effect of Obeticholic Acid
Abstract
:1. Introduction
2. Results
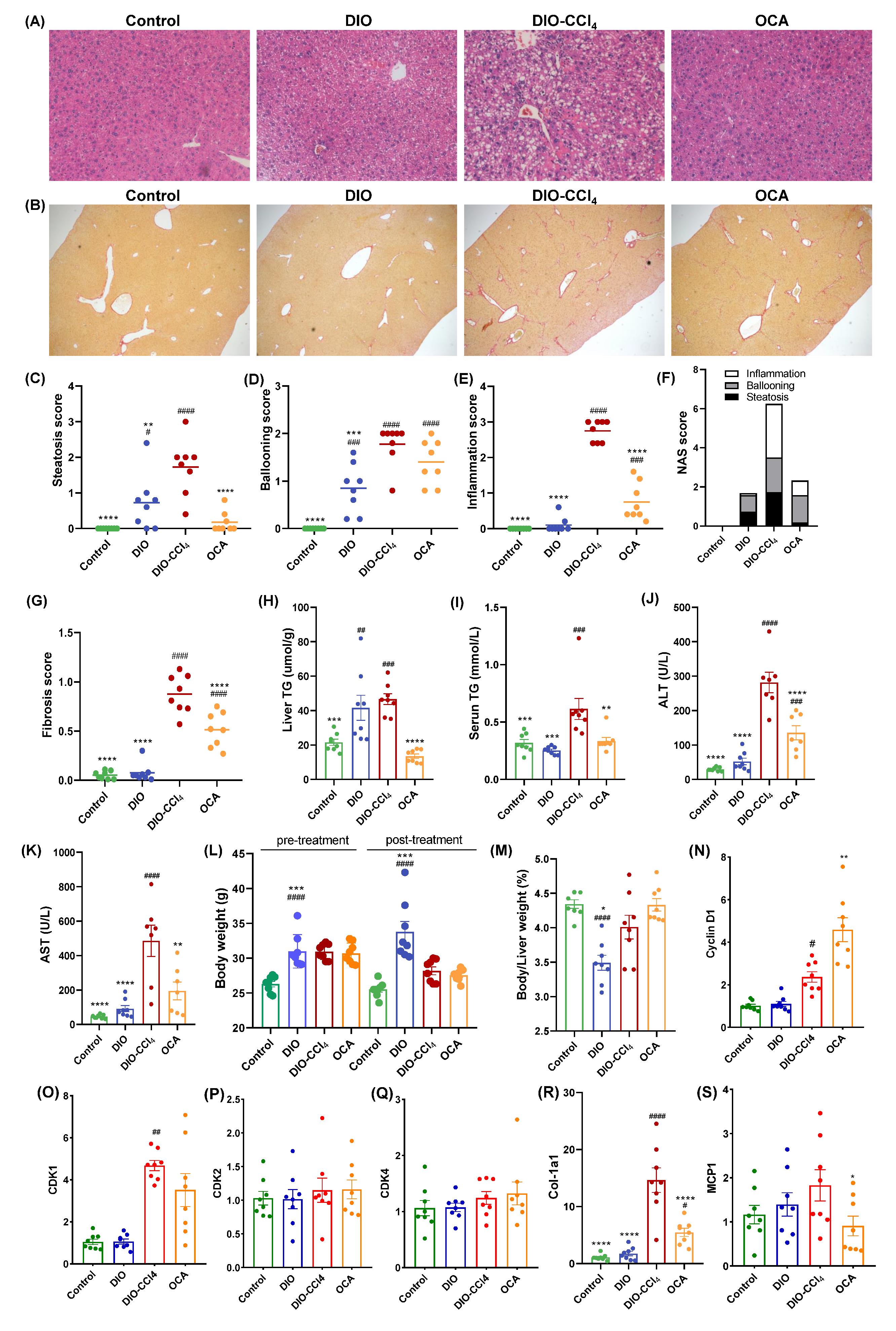
2.1. The Histological Characteristics of NAFLD Models and OCA Group
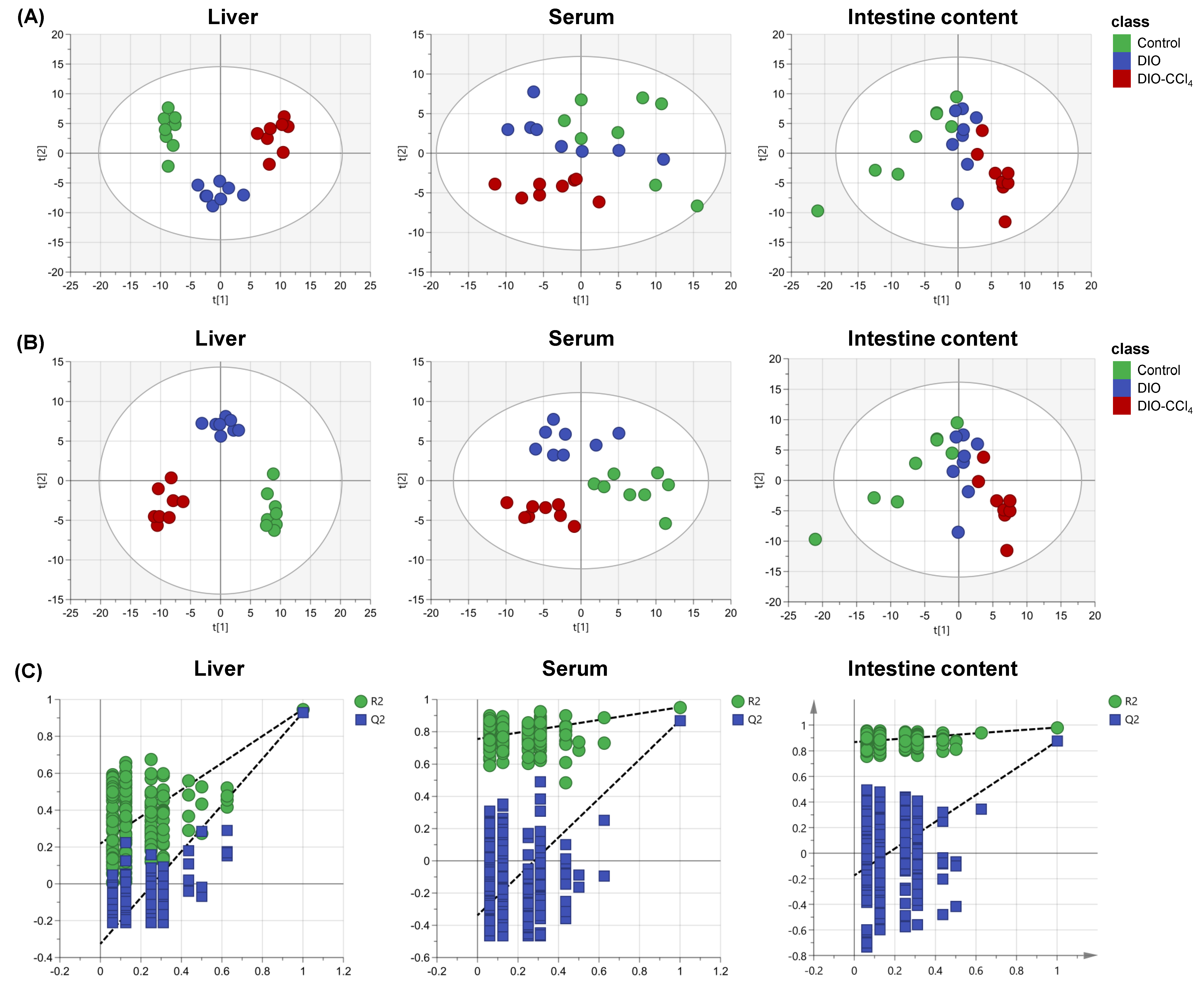
2.2. Metabolic Characteristics of NAFLD Models and Control Group
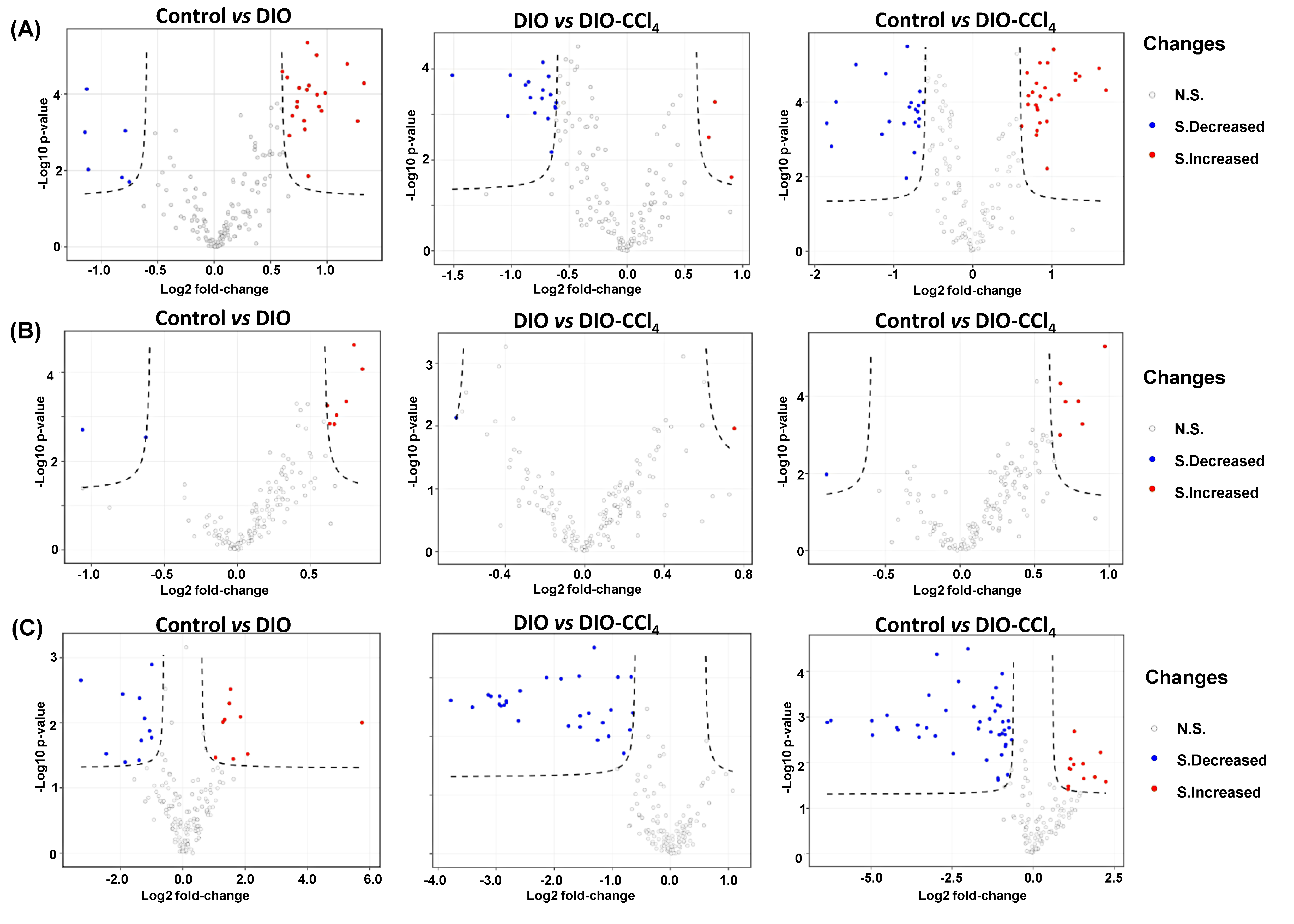
2.3. Metabolic Alteration Between NAFLD Models and Control Group
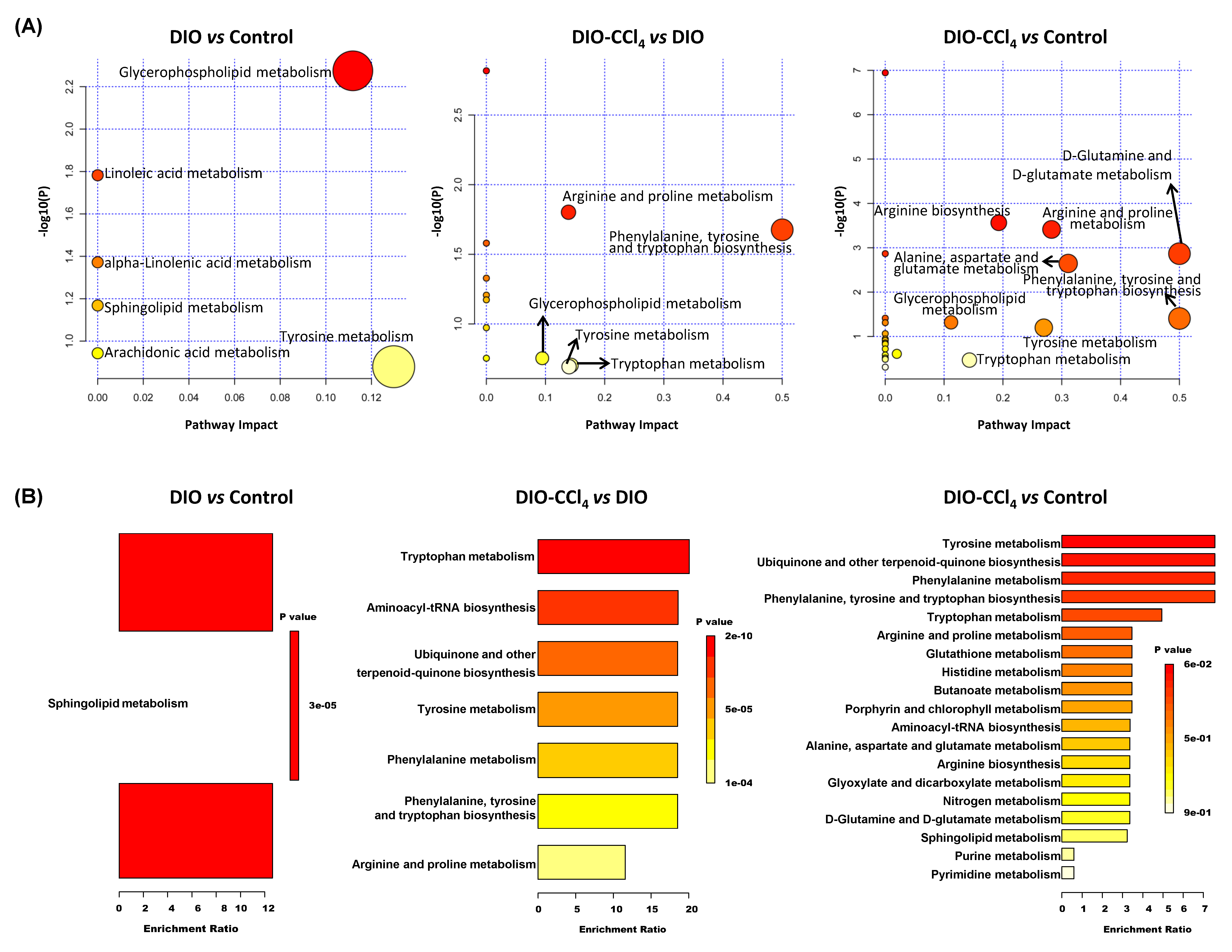
2.4. Metabolic Pathways and Metabolite Enrichment Analysis Distinguished NASH Models
2.5. Correlation Analysis between the Differential Metabolites and NAFLD-Associated Parameters
2.6. Amelioration of Obeticholic Acid (OCA) on NASH
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Ethics Statement and Animals
4.3. Biochemical Analysis, Histology and NAFLD Activity Score and Fibrosis Evaluation
4.4. RNA Isolation and Real-Time PCR
4.5. Metabolomics
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Anstee, Q.M.; Reeves, H.L.; Kotsiliti, E.; Govaere, O.; Heikenwalder, M. From NASH to HCC: Current concepts and future challenges. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 411–428. [Google Scholar] [CrossRef] [PubMed]
- Banales, J.M.; Inarrairaegui, M.; Arbelaiz, A.; Milkiewicz, P.; Muntane, J.; Munoz-Bellvis, L.; La Casta, A.; Gonzalez, L.M.; Arretxe, E.; Alonso, C.; et al. Serum Metabolites as Diagnostic Biomarkers for Cholangiocarcinoma, Hepatocellular Carcinoma, and Primary Sclerosing Cholangitis. Hepatology 2019, 70, 547–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blond, E.; Disse, E.; Cuerq, C.; Drai, J.; Valette, P.J.; Laville, M.; Thivolet, C.; Simon, C.; Caussy, C. EASL-EASD-EASO clinical practice guidelines for the management of non-alcoholic fatty liver disease in severely obese people: Do they lead to over-referral? Diabetologia 2017, 60, 1218–1222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, T.G.; Kim, S.M.; Caussy, C.; Fu, T.; Guo, J.; Bassirian, S.; Singh, S.; Madamba, E.V.; Bettencourt, R.; Richards, L.; et al. A Universal Gut-Microbiome-Derived Signature Predicts Cirrhosis. Cell Metab. 2020, 32, 1–11. [Google Scholar] [CrossRef]
- Xin, D.; Zong-Shun, L.; Bang-Mao, W.; Lu, Z. Expression of intestinal tight junction proteins in patients with non-alcoholic fatty liver disease. Hepatogastroenterology 2014, 61, 136–140. [Google Scholar]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [Green Version]
- Rinschen, M.M.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol. 2019, 20, 353–367. [Google Scholar] [CrossRef]
- Pezzatti, J.; Boccard, J.; Codesido, S.; Gagnebin, Y.; Joshi, A.; Picard, D.; Gonzalez-Ruiz, V.; Rudaz, S. Implementation of liquid chromatography-high resolution mass spectrometry methods for untargeted metabolomic analyses of biological samples: A tutorial. Anal. Chim. Acta 2020, 1105, 28–44. [Google Scholar] [CrossRef]
- Sreekumar, A.; Poisson, L.M.; Rajendiran, T.M.; Khan, A.P.; Cao, Q.; Yu, J.; Laxman, B.; Mehra, R.; Lonigro, R.J.; Li, Y.; et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 2009, 457, 910–914. [Google Scholar] [CrossRef] [Green Version]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C. Organisms often respond in complex and unpredictable ways to stimuli that cause disease or injury. By measuring and mathematically modelling changes in the levels of products of metabolism found in biological fluids and tissues, metabonomics offers fresh insight into the effects of diet, drugs and disease. Nature 2008, 455, 1054–1056. [Google Scholar]
- Magistretti, P.J.; Allaman, I. Lactate in the brain: From metabolic end-product to signalling molecule. Nat. Rev. Neurosci. 2018, 19, 235–249. [Google Scholar] [CrossRef]
- Liu, P.-S.; Wang, H.; Li, X.; Chao, T.; Teav, T.; Christen, S.; Di Conza, G.; Cheng, W.-C.; Chou, C.-H.; Vavakova, M.; et al. α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 2017, 18, 985–994. [Google Scholar] [CrossRef]
- Giera, M.; Branco Dos Santos, F.; Siuzdak, G. Metabolite-Induced Protein Expression Guided by Metabolomics and Systems Biology. Cell Metab. 2018, 27, 270–272. [Google Scholar] [CrossRef] [Green Version]
- Hameed, B.; Terrault, N.A.; Gill, R.M.; Loomba, R.; Chalasani, N.; Hoofnagle, J.H.; Van Natta, M.L. Clinical and metabolic effects associated with weight changes and obeticholic acid in non-alcoholic steatohepatitis. Aliment. Pharmacol. Ther. 2018, 47, 645–656. [Google Scholar] [CrossRef]
- Kubota, N.; Kado, S.; Kano, M.; Masuoka, N.; Nagata, Y.; Kobayashi, T.; Miyazaki, K.; Ishikawa, F. A high-fat diet and multiple administration of carbon tetrachloride induces liver injury and pathological features associated with non-alcoholic steatohepatitis in mice. Clin. Exp. Pharmcol. Physiol. 2013, 40, 422–430. [Google Scholar] [CrossRef]
- Wigger, L.; Casals-Casas, C.; Baruchet, M.; Trang, K.B.; Pradervand, S.; Naldi, A.; Desvergne, B. System analysis of cross-talk between nuclear receptors reveals an opposite regulation of the cell cycle by LXR and FXR in human HepaRG liver cells. PLoS ONE 2019, 14, e0220894. [Google Scholar] [CrossRef] [Green Version]
- Jouihan, H.; Will, S.; Guionaud, S.; Boland, M.L.; Oldham, S.; Ravn, P.; Celeste, A.; Trevaskis, J.L. Superior reductions in hepatic steatosis and fibrosis with co-administration of a glucagon-like peptide-1 receptor agonist and obeticholic acid in mice. Mol. Metab. 2017, 6, 1360–1370. [Google Scholar] [CrossRef]
- Younossi, Z.; Anstee, Q.M.; Marietti, M.; Hardy, T.; Henry, L.; Eslam, M.; George, J.; Bugianesi, E. Global burden of NAFLD and NASH: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 11–20. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [Green Version]
- Siddiqui, M.S.; Van Natta, M.L.; Connelly, M.A.; Vuppalanchi, R.; Neuschwander-Tetri, B.A.; Tonascia, J.; Guy, C.; Loomba, R.; Dasarathy, S.; Wattacheril, J.; et al. Impact of obeticholic acid on the lipoprotein profile in patients with non-alcoholic steatohepatitis. J. Hepatol. 2020, 72, 25–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Younossi, Z.M.; Ratziu, V.; Loomba, R.; Rinella, M.; Anstee, Q.M.; Goodman, Z.; Bedossa, P.; Geier, A.; Beckebaum, S.; Newsome, P.N.; et al. Obeticholic acid for the treatment of non-alcoholic steatohepatitis: Interim analysis from a multicentre, randomised, placebo-controlled phase 3 trial. Lancet 2019, 394, 2184–2196. [Google Scholar] [CrossRef] [Green Version]
- Mudaliar, S.; Henry, R.R.; Sanyal, A.J.; Morrow, L.; Marschall, H.U.; Kipnes, M.; Adorini, L.; Sciacca, C.I.; Clopton, P.; Castelloe, E.; et al. Efficacy and Safety of the Farnesoid X Receptor Agonist Obeticholic Acid in Patients With Type 2 Diabetes and Nonalcoholic Fatty Liver Disease. Gastroenterology 2013, 145, 574–582.e571. [Google Scholar] [CrossRef] [PubMed]
- Forman, B.M.; Goode, E.; Chen, J.; Oro, A.E.; Bradley, D.J.; Perlmann, T.; Noonan, D.J.; Burka, L.T.; Mcmorris, T.; Lamph, W.W.; et al. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 1995, 81, 687–693. [Google Scholar] [CrossRef] [Green Version]
- Neuschwander-Tetri, B.A.; Loomba, R.; Sanyal, A.J.; Lavine, J.E.; Van Natta, M.L.; Abdelmalek, M.F.; Chalasani, N.; Dasarathy, S.; Diehl, A.M.; Hameed, B.; et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): A multicentre, randomised, placebo-controlled trial. Lancet 2015, 385, 956–965. [Google Scholar] [CrossRef] [Green Version]
- Lian, S.; Li, W.; Wang, D.; Xu, B.; Guo, X.; Yang, H.; Wang, J. Effects of prenatal cold stress on maternal serum metabolomics in rats. Life Sci. 2020, 246, 117432. [Google Scholar] [CrossRef]
- Canfora, E.E.; Meex, R.C.R.; Venema, K.; Blaak, E.E. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat. Rev. Endocrinol. 2019, 15, 261–273. [Google Scholar] [CrossRef]
- Brandl, K.; Schnabl, B. Intestinal microbiota and nonalcoholic steatohepatitis. Curr. Opin. Gastroenterol. 2017, 33, 128–133. [Google Scholar] [CrossRef] [Green Version]
- Gorden, D.L.; Myers, D.S.; Ivanova, P.T.; Fahy, E.; Maurya, M.R.; Gupta, S.; Min, J.; Spann, N.J.; Mcdonald, J.G.; Kelly, S.L.; et al. Biomarkers of NAFLD progression: A lipidomics approach to an epidemic. J. Lipid Res. 2015, 56, 722–736. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Agellon, L.B.; Allen, T.M.; Umeda, M.; Jewell, L.; Mason, A.; Vance, D.E. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab. 2006, 3, 321–331. [Google Scholar] [CrossRef] [Green Version]
- Van Der Veen, J.N.; Kennelly, J.P.; Wan, S.; Vance, J.E.; Vance, D.E.; Jacobs, R.L. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim. Biophys. Acta (BBA) Biomembr. 2017, 1859, 1558–1572. [Google Scholar] [CrossRef]
- Puri, P.; Baillie, R.A.; Wiest, M.M.; Mirshahi, F.; Choudhury, J.; Cheung, O.; Sargeant, C.; Contos, M.J.; Sanyal, A.J. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology 2007, 46, 1081–1090. [Google Scholar] [CrossRef]
- Chocian, G.; Chabowski, A.; Żendzian-Piotrowska, M.; Harasim, E.; Łukaszuk, B.; Górski, J. High fat diet induces ceramide and sphingomyelin formation in rat’s liver nuclei. Mol. Cell. Biochem. 2010, 340, 125–131. [Google Scholar] [CrossRef]
- Chun, L.; Junlin, Z.; Aimin, W.; Niansheng, L.; Benmei, C.; Minxiang, L. Inhibition of ceramide synthesis reverses endothelial dysfunction and atherosclerosis in streptozotocin-induced diabetic rats. Diabetes Res. Clin. Pract. 2011, 93, 77–85. [Google Scholar] [CrossRef]
- Watson, L.M.; Coghlan, M.; Hundal, S.H. Modulating serine palmitoyl transferase (SPT) expression and activity unveils a crucial role in lipid-induced insulin resistance in rat skeletal muscle cells. Biochem. J. 2009, 417, 791–801. [Google Scholar] [CrossRef] [Green Version]
- Alonso, C.; Fernández-Ramos, D.; Varela-Rey, M.; Martínez-Arranz, I.; Navasa, N.; Van Liempd, S.M.; Lavín Trueba, J.L.; Mayo, R.; Ilisso, C.P.; De Juan, V.G.; et al. Metabolomic Identification of Subtypes of Nonalcoholic Steatohepatitis. Gastroenterology 2017, 152, 1449–1461.e1447. [Google Scholar] [CrossRef] [Green Version]
- Rom, O.; Liu, Y.; Liu, Z.; Zhao, Y.; Wu, J.; Ghrayeb, A.; Villacorta, L.; Fan, Y.; Chang, L.; Wang, L.; et al. Glycine-based treatment ameliorates NAFLD by modulating fatty acid oxidation, glutathione synthesis, and the gut microbiome. Sci. Transl. Med. 2020, 12, eaaz2841. [Google Scholar] [CrossRef]
- Gaggini, M.; Carli, F.; Rosso, C.; Buzzigoli, E.; Marietti, M.; Della Latta, V.; Ciociaro, D.; Abate, M.L.; Gambino, R.; Cassader, M.; et al. Altered amino acid concentrations in NAFLD: Impact of obesity and insulin resistance. Hepatology 2018, 67, 145–158. [Google Scholar] [CrossRef] [Green Version]
- Schooneman, M.G.; Vaz, F.M.; Houten, S.M.; Soeters, M.R. Acylcarnitines: Reflecting or Inflicting Insulin Resistance? Diabetes 2013, 62, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Honda, T.; Ishigami, M.; Luo, F.; Lingyun, M.; Ishizu, Y.; Kuzuya, T.; Hayashi, K.; Nakano, I.; Ishikawa, T.; Feng, G.G.; et al. Branched-chain amino acids alleviate hepatic steatosis and liver injury in choline-deficient high-fat diet induced NASH mice. Metabolism 2017, 69, 177–187. [Google Scholar] [CrossRef]
- Cole, L.K.; Vance, J.E.; Vance, D.E. Phosphatidylcholine biosynthesis and lipoprotein metabolism. Biochim. Biophys. Acta 2012, 1821, 754–761. [Google Scholar] [CrossRef] [PubMed]
- Maricic, I.; Girardi, E.; Zajonc, D.M.; Kumar, V. Recognition of Lysophosphatidylcholine by Type II NKT Cells and Protection from an Inflammatory Liver Disease. J. Immunol. 2014, 193, 4580–4589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, W.; Menke, A.L.; Driessen, A.; Koek, G.H.; Lindeman, J.H.; Stoop, R.; Havekes, L.M.; Kleemann, R.; van den Hoek, A.M. Establishment of a general NAFLD scoring system for rodent models and comparison to human liver pathology. PLoS ONE 2014, 9, e115922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunt, E.M.; Kleiner, D.E.; Wilson, L.A.; Belt, P.; Neuschwander-Tetri, B.A.; Network, N.C.R. Nonalcoholic Fatty Liver Disease (NAFLD) Activity Score and the Histopathologic Diagnosis in NAFLD: Distinct Clinicopathologic Meanings. Hepatology 2011, 53, 810–820. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [Green Version]
- Pang, Z.; Chong, J.; Li, S.; Xia, J. MetaboAnalystR 3.0: Toward an Optimized Workflow for Global Metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef]
- Li, L.; Chang, L.; Zhang, X.; Ning, Z.; Mayne, J.; Ye, Y.; Stintzi, A.; Liu, J.; Figeys, D. Berberine and its structural analogs have differing effects on functional profiles of individual gut microbiomes. Gut Microbes 2020, 1–14. [Google Scholar] [CrossRef]

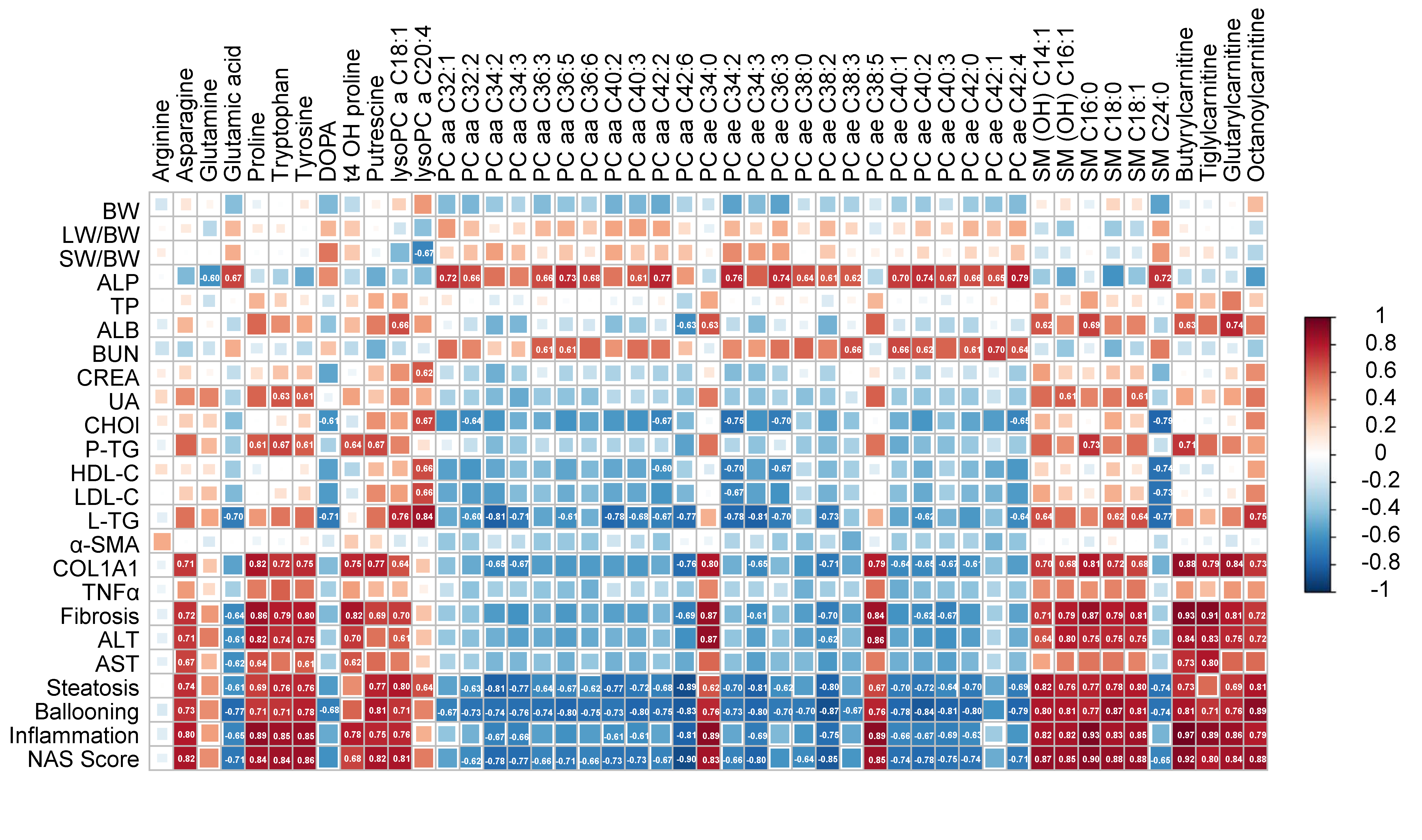

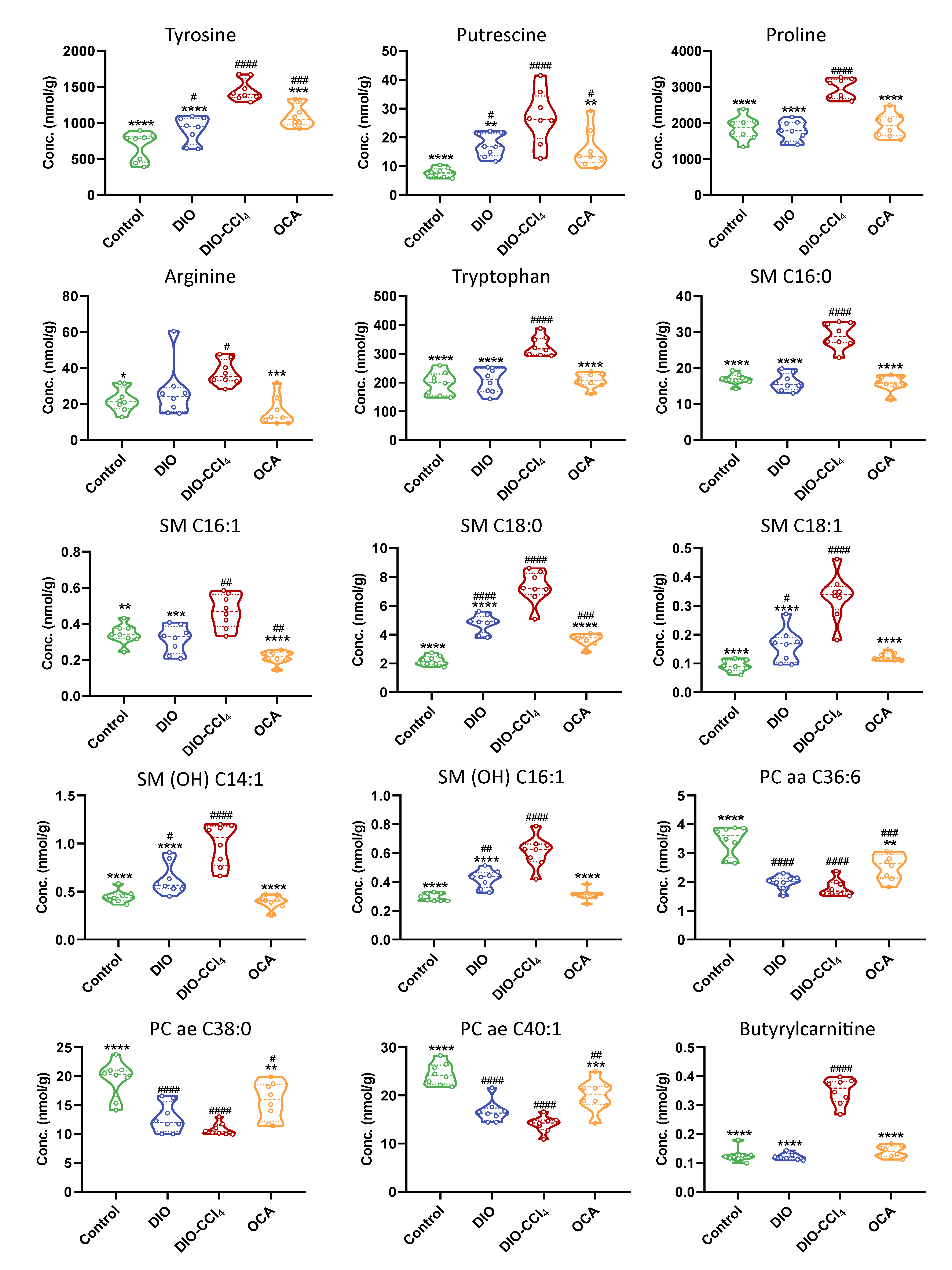
| DIO vs. Control | DIO−CCl4 vs. DIO | DIO−CCl4 vs. Control | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Metabolite | Log2 FC | p Value | Change | Log2 FC | p Value | Change | Log2 FC | p Value | Change |
| arginine | 0.34 | 0.32 | - | 0.40 | 0.21 | - | 0.74 | 0.002 | ↑ |
| asparagine | 0.20 | 0.20 | - | 0.53 | 0.00 | - | 0.73 | 0.0003 | ↑ |
| glutamine | 0.45 | 0.14 | - | 0.39 | 0.28 | - | 0.84 | 0.011 | ↑ |
| glutamic acid | −0.44 | 0.02 | - | −0.47 | 0.06 | - | −0.92 | 0.00004 | ↓ |
| proline | −0.06 | 0.76 | - | 0.73 | 0.0001 | ↑ | 0.67 | 0.0001 | ↑ |
| tryptophan | 0.06 | 0.68 | - | 0.66 | 0.0004 | ↑ | 0.73 | 0.0002 | ↑ |
| tyrosine | 0.42 | 0.03 | - | 0.68 | 0.0001 | ↑ | 1.10 | 0.00002 | ↑ |
| DOPA | −0.66 | 0.05 | ↓ | −0.28 | 0.28 | - | −0.94 | 0.006 | ↓ |
| t4 OH proline | 0.00 | 0.98 | - | 0.68 | 0.001 | ↑ | 0.68 | 0.0001 | ↑ |
| putrescine | 1.14 | 0.001 | ↑ | 0.65 | 0.03 | - | 1.79 | 0.002 | ↑ |
| DIO vs. Control | DIO−CCl4 vs. DIO | DIO−CCl4 vs. Control | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Metabolite | Log2 FC | p Value | Change | Log2 FC | p Value | Change | Log2 FC | p Value | Change |
| lysoPC a C18:1 | 0.27 | 0.18 | - | 0.43 | 0.02 | - | 0.70 | 0.0002 | ↑ |
| lysoPC a C20:4 | 0.62 | 0.09 | - | 0.05 | 0.87 | - | 0.68 | 0.0003 | ↑ |
| PC aa C32:1 | −0.95 | 0.0003 | ↓ | 0.14 | 0.16 | - | −0.81 | 0.001 | ↓ |
| PC aa C32:2 | −0.80 | 0.001 | ↓ | 0.00 | 1.00 | - | −0.81 | 0.001 | ↓ |
| PC aa C34:2 | −0.38 | 0.02 | - | −0.33 | 0.09 | - | −0.70 | 0.0001 | ↓ |
| PC aa C34:3 | −0.45 | 0.02 | - | −0.37 | 0.06 | - | −0.82 | 0.0002 | ↓ |
| PC aa C36:3 | −0.69 | 0.0004 | ↓ | −0.12 | 0.20 | - | −0.82 | 0.0001 | ↓ |
| PC aa C36:5 | −1.33 | 0.0001 | ↓ | −0.36 | 0.00 | - | −1.69 | 0.00005 | ↓ |
| PC aa C36:6 | −0.80 | 0.0005 | ↓ | −0.14 | 0.21 | - | −0.94 | 0.0003 | ↓ |
| PC aa C40:2 | −0.50 | 0.003 | - | −0.20 | 0.09 | - | −0.70 | 0.0001 | ↓ |
| PC aa C40:3 | −0.98 | 0.0001 | ↓ | −0.32 | 0.03 | - | −1.30 | 0.00003 | ↓ |
| PC aa C42:2 | −0.83 | 0.000005 | ↓ | −0.03 | 0.63 | - | −0.85 | 0.00001 | ↓ |
| PC aa C42:6 | −0.38 | 0.02 | - | −0.71 | 0.003 | ↓ | −1.09 | 0.0001 | ↓ |
| PC ae C34:0 | 0.03 | 0.64 | - | 0.60 | 0.001 | - | 0.62 | 0.0001 | ↑ |
| PC ae C34:2 | −0.91 | 0.0001 | ↓ | −0.04 | 0.75 | - | −0.95 | 0.00001 | ↓ |
| PC ae C34:3 | −0.51 | 0.004 | - | −0.34 | 0.05 | - | −0.85 | 0.0001 | ↓ |
| PC ae C36:3 | −0.82 | 0.0001 | ↓ | 0.06 | 0.67 | - | −0.76 | 0.0001 | ↓ |
| PC ae C38:0 | −0.61 | 0.003 | - | −0.24 | 0.09 | - | −0.85 | 0.0004 | ↓ |
| PC ae C38:2 | −0.84 | 0.0001 | ↓ | −0.76 | 0.001 | ↓ | −1.60 | 0.00001 | ↓ |
| PC ae C38:3 | −0.67 | 0.001 | ↓ | −0.13 | 0.32 | - | −0.80 | 0.0001 | ↓ |
| PC ae C38:5 | 0.07 | 0.36 | - | 0.80 | 0.001 | ↑ | 0.87 | 0.0004 | ↑ |
| PC ae C40:1 | −0.56 | 0.00 | - | −0.25 | 0.02 | - | −0.81 | 0.00003 | ↓ |
| PC ae C40:2 | −0.91 | 0.00001 | ↓ | −0.40 | 0.001 | - | −1.30 | 0.00002 | ↓ |
| PC ae C40:3 | −0.65 | 0.00004 | ↓ | −0.38 | 0.01 | - | −1.02 | 0.000004 | ↓ |
| PC ae C42:0 | −0.73 | 0.0002 | ↓ | −0.26 | 0.003 | - | −0.99 | 0.0001 | ↓ |
| PC ae C42:1 | −0.73 | 0.0002 | ↓ | 0.12 | 0.41 | - | −0.62 | 0.0004 | ↓ |
| PC ae C42:4 | −1.18 | 0.00002 | ↓ | −0.18 | 0.04 | - | −1.35 | 0.00002 | ↓ |
| DIO vs. Control | DIO−CCl4 vs. DIO | DIO−CCl4 vs. Control | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Metabolite | Log2 FC | p Value | Change | Log2 FC | p Value | Change | Log2 FC | p Value | Change |
| SM OH C14:1 | 0.49 | 0.02 | - | 0.65 | 0.01 | ↑ | 1.15 | 0.001 | ↑ |
| SM OH C16:1 | 0.52 | 0.001 | - | 0.53 | 0.002 | - | 1.06 | 0.0003 | ↑ |
| SM C16:0 | −0.07 | 0.52 | - | 0.85 | 0.0002 | ↑ | 0.78 | 0.0001 | ↑ |
| SM C18:0 | 1.13 | 0.0001 | ↑ | 0.60 | 0.001 | - | 1.73 | 0.0001 | ↑ |
| SM C18:1 | 0.82 | 0.02 | ↑ | 1.03 | 0.001 | ↑ | 1.85 | 0.0004 | ↑ |
| SM C24:0 | −0.75 | 0.0001 | ↓ | 0.06 | 0.74 | - | −0.69 | 0.00002 | ↓ |
| Butyrylcarnitine | −0.03 | 0.76 | - | 1.51 | 0.0001 | ↑ | 1.48 | 0.00001 | ↑ |
| Tiglylcarnitine | −0.05 | 0.91 | - | 0.88 | 0.0002 | ↑ | 0.83 | 0.000003 | ↓ |
| Glutarylcarnitine | 0.07 | 0.63 | - | 0.61 | 0.001 | - | 0.68 | 0.0004 | ↓ |
| Octanoylcarnitine | 0.46 | 0.01 | - | 0.34 | 0.02 | - | 0.80 | 0.0001 | ↓ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, N.; Huang, S.; Zhang, Q.; Zhao, Z.; Qu, H.; Ning, M.; Leng, Y.; Liu, J. Metabolomic Study of High-Fat Diet-Induced Obese (DIO) and DIO Plus CCl4-Induced NASH Mice and the Effect of Obeticholic Acid. Metabolites 2021, 11, 374. https://doi.org/10.3390/metabo11060374
Zhu N, Huang S, Zhang Q, Zhao Z, Qu H, Ning M, Leng Y, Liu J. Metabolomic Study of High-Fat Diet-Induced Obese (DIO) and DIO Plus CCl4-Induced NASH Mice and the Effect of Obeticholic Acid. Metabolites. 2021; 11(6):374. https://doi.org/10.3390/metabo11060374
Chicago/Turabian StyleZhu, Nanlin, Suling Huang, Qingli Zhang, Zhuohui Zhao, Hui Qu, Mengmeng Ning, Ying Leng, and Jia Liu. 2021. "Metabolomic Study of High-Fat Diet-Induced Obese (DIO) and DIO Plus CCl4-Induced NASH Mice and the Effect of Obeticholic Acid" Metabolites 11, no. 6: 374. https://doi.org/10.3390/metabo11060374
APA StyleZhu, N., Huang, S., Zhang, Q., Zhao, Z., Qu, H., Ning, M., Leng, Y., & Liu, J. (2021). Metabolomic Study of High-Fat Diet-Induced Obese (DIO) and DIO Plus CCl4-Induced NASH Mice and the Effect of Obeticholic Acid. Metabolites, 11(6), 374. https://doi.org/10.3390/metabo11060374





